Abstract
Chromosomal translocations in lymphoid tumours can involve antigen-receptor loci undergoing V(D)J recombination. Here, we show that translocations are recovered from the joining of RAG-generated double-strand breaks (DSBs) on one chromosome to an endonuclease-generated DSB on a second chromosome, providing evidence for the participation of non-RAG DSBs in some lymphoid translocations. Surprisingly, translocations are increased in cells deficient for the nonhomologous end-joining (NHEJ) protein Ku70, implicating non-canonical joining pathways in their etiology.
Lymphocytes are unique among somatic cells in that they undergo programmed DSB formation to induce rearrangements of antigen-receptor genes1. The initial assembly of antigen receptors involves V(D)J recombination, a process in which the RAG recombinase cleaves recombination signal sequences (RSSs) to produce hairpin coding ends and blunt signal ends. The Ku70–Ku80 heterodimer and other NHEJ factors are critical for joining RAG-generated DSBs, and they also have a critical role in the repair of DSBs that arise from DNA-damaging agents2.
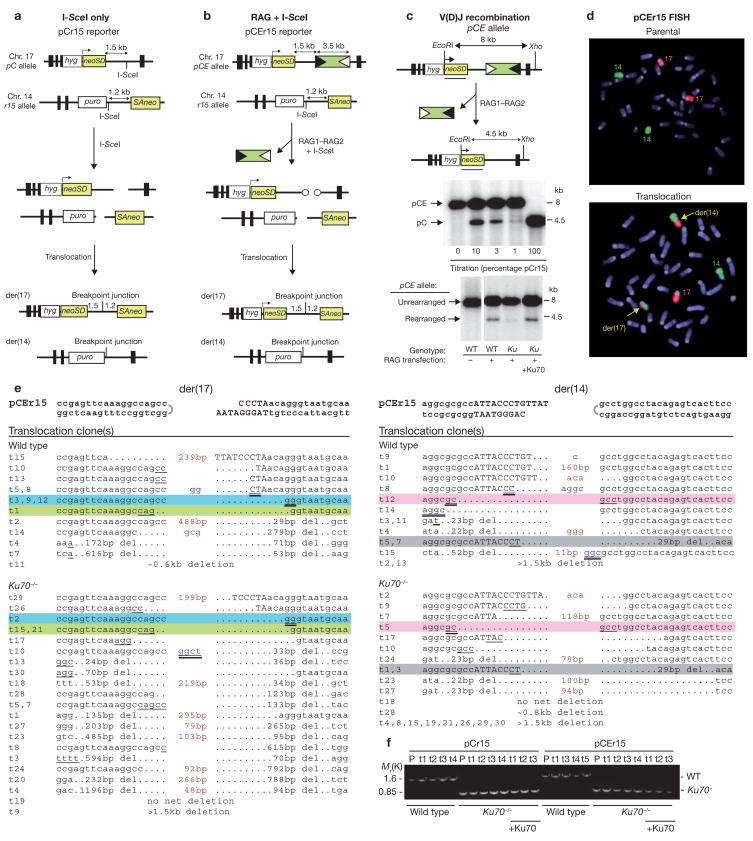
Errors in V(D)J recombination have been implicated in the reciprocal chromosomal translocations found in several lymphoid malignancies, including follicular and mantle cell lymphoma, and T cell leukaemias3,4. In some cases, the RAG recombinase may orchestrate interchromosomal recombination between an RSS at an antigen-receptor gene and an RSS-like sequence on the other participating chromosome. In other translocations, DSB formation in the second chromosome may occur by alternate means. To determine whether DSBs generated by the RAG recombinase can participate in translocations with a non-RAG generated DSB, we modified the previously developed reporter pCr15 in which the joining of two DSBs generated by the I-SceI endonuclease results in a translocation5. This reporter consists of a neomycin phosphotransferase gene (neo) split at its intron and targeted to chromosomes 17 and 14 in mouse embryonic stem (ES) cells (Fig. 1a). The intron segment on each chromosome is demarcated by an I-SceI site, such that cleavage by I-SceI followed by joining of chromosomes 17 and 14 generates a derivative chromosome, der(17), that contains the centromeric portion of chromosome 17 and the telomeric portion of chromosone 14 while forming a functional neo gene. In the new reporter, termed pCEr15 (CE, coding end), the chromosome 17 I-SceI site is replaced with RSS elements (Fig. 1b). Transient RAG1–RAG2 expression in the pCEr15 cells induces ≥5% rearrangement at these RSS elements (Fig. 1c and Table 1).
Figure 1.
Chromosomal translocations induced after DSB formation and NHEJ repair. (a) Translocation reporter pCr15. DSB formation at the I-SceI sites on chromosomes 17 and 14, followed by interchromosomal NHEJ, results in a chromosomal translocation and a neo+ gene on der(17) if joining occurs within the 2.7 kb neo intron. The non-palindromic I-SceI sites are present in opposite orientation, such that cleavage results in non-overlapping four-base 3# overhangs. (b) Translocation reporter pCEr15. RAG-recombinase cleavage to generate hairpin ends on chromosome 17 and I-SceI cleavage at the I-SceI site on chromosome 14, followed by interchromosomal NHEJ, results in a chromosomal translocation, similar to pCr15. Circles, hairpin ends; white triangles, 12-RSS; black triangles, 23-RSS. (c) V(D)J recombination at the pCE allele after RAG-recombinase expression. Compared to wild-type cells, V(D)J recombination is significantly reduced in Ku70−/− cells, but is restored by Ku70 expression. V(D)J recombination frequency was quantified by comparison with a standard curve established by diluting known quantities of DNA from cells containing the pCEr15 (8 kb) and pCr15 (4.5 kb) reporters. (d) Fluorescence in situ hybridization. Parental pCEr15 cells have normal chromosomes 14 (green) and 17 (red), whereas neo+ clones, as Ku70−/− clone t1 shown here, also have derivative chromosomes (yellow arrows). (e) Translocation junctions derived from the pCEr15 reporter. Junctions that occurred in both wild-type and Ku70−/− cells are highlighted with the same colour. Deletions that were not confirmed by sequencing are estimates based on Southern blotting. Hairpin ends (half circle), I-SceI site (capital letters), microhomology (underlined), insertions (red), and P nucleotides (blue) are indicated. Possible microhomologies from hairpin opening are double underlined. Long insertions include chromosome 17 RSS–spacer sequences (wild-type der(14)t1; Ku70−/− der(14)t7, der(17)t29; see Supplementary Information, Fig. 1b) and chromosome 14 sequences located within a few kilobases of the DSB which may have been templated (wild-type der(17)t15; Ku70−/− der(17)t1, t18, t20). Smaller templated insertions (<5 bp) are also possible (for example, wild-type der(14)t10). (f) Confirmation of the Ku70 genotype in parental (P) and multiple translocation clones (t). +Ku, Ku70 transient complementation.
Table 1.
Translocation induced by the I-SceI endonuclease with or without RAG recombinase.
| Construct | Genotype | Expression vector | Translocations (× 10−7) | P value | V(D)J recombination (× 10−2) | P value | Ratio of translocation versus V(D)J recombination (×10−7)e |
|---|---|---|---|---|---|---|---|
| PCr15 | WT | Vector | <0.1 | ||||
| I-SceI | 287.1 ± 90.5 | ||||||
| I-SceI + Ku70 | 363. 3 ±93. 3 | 0.24a | |||||
| PCr15 | Ku70−/− | Vector | <0.1 | ||||
| I-SceI | 789.0 ± 221.6 | <0.001a | |||||
| I-SceI + Ku70 | 323.3 ± 119.7 | 0.32a | |||||
| pCEr15 | WT | RAG | <0.03 | <0.001b | 5.43± 1.02 | ||
| I-SceI | <0.03 | <0.001b | 0 | <0.001d | |||
| RAG + I-SceI | 0.23 ± 0.16 | n.d. | 4.24 | ||||
| pCEr15 | Ku70−/− | RAG | <0.03 | <0.001c | 0.66 ± 0.73 | <0.001d | |
| I-SceI | <0.03 | <0.001c | 0 | ||||
| RAG + Ku70 | n.d. | 5.86 ± 1.51 | 0.58d | ||||
| RAG + I-SceI | 0.58 ± 0.34 | <0.001b | n.d. | 87.9 | |||
| RAG + I-SceI + Ku70 | 0.22 ±0.16 | 0.93b | n.d. | 3.75 |
Versus pCr15 wild-type I-SceI.
Versus pCEr15 wild-type I-SceI + RAG.
Versus pCEr15 Ku70−/− RAG + I-SceI.
Versus pCEr15 wild-type RAG.
Within each genotype, RAG + I-SceI versus RAG or RAG + I-SceI + Ku70 versus RAG + Ku70. ±, 1 s.d. from the mean, n.d., not determined.
To determine whether coding ends on chromosome 17 can join to I-SceI ends on chromosome 14 to result in a translocation, RAG1–RAG2 and I-SceI were coexpressed in pCEr15 cells. neo+ clones were recovered from pCEr15 cells at a frequency of 0.23 × 10−7 (Table 1). No clones were obtained after expression of either RAG1–RAG2 or I-SceI alone (<0.03 × 10−7; P <0.001). Fluorescence in situ hybridization (FISH) using chromosomes 17 and 14 paints confirmed the expected der(17) in all 11 neo+ clones examined (Fig. 1d). Nine clones had the reciprocal der(14) chromosome, although, interestingly, one clone had a der(6)t(6;17) instead (data not shown).
Translocations were ~1000-fold less frequent in the pCEr15 cells after RAG–I-SceI cleavage relative to pCr15 cells after I-SceI cleavage (Table 1). As RAG-generated DSBs are estimated to occur only a few-fold less efficiently than I-SceI-generated DSBs (Fig. 1c and data not shown), reduced DSB formation can only partially account for the significantly lower translocation frequency. Possibly, RAG recombinase facilitates the joining step of V(D)J recombination to prevent the misuse of DNA ends in other pathways6. An alternative explanation is that the hairpin DNA ends on chromosome 17 are protected from participating in translocations. However, experiments using a signal-end reporter that is similar in design to pCEr15 demonstrated that blunt signal ends also rarely participate in translocations (D.M.W. and M.J., unpublished observations). We also examined two reporters similar to pCEr15 that contain a single 12-RSS in either orientation in place of the paired RSSs. Translocations were not recovered from either reporter (<0.06 × 10−7, P = 0.01; data not shown), indicating infrequent use of a single RSS.
Der(17) and der(14) junctions were sequenced from a number of neo+ clones derived from the pCEr15 reporter. As previously observed for the pCr15 reporter and for oncogenic translocations in patients5, translocation junctions demonstrated typical features of NHEJ, including deletions, insertions and microhomology use (Fig. 1e). A novel feature observed in some pCEr15 junctions was nucleotide additions consistent with hairpin opening (P nucleotides). Overall, the RAG ends were more likely to remain intact than the I-SceI ends (14 out of 25 (56%) versus one out of 15 (4%), P = 0.0001).
As V(D)J recombination is highly dependent on the Ku70–Ku80-dependent NHEJ pathway, we examined whether translocations involving RAG1–RAG2 also use this pathway. The pCEr15 reporter was introduced into Ku70−/− cells7, and RAG1–RAG2 and I-SceI were coexpressed in these cells. Surprisingly, rather than being reduced in frequency, neo+ clones were recovered at a 2.5-fold higher frequency in Ku70−/− cells (0.58 × 10−7) compared with wild-type cells (P <0.001; Table 1). FISH experiments confirmed the expected der(17) and der(14) chromosomes in all eight neo+ clones examined (Fig. 1d and data not shown). Confirming that Ku70 suppresses translocations, ectopic expression of Ku70 in Ku70−/− cells reduced translocations to wild-type levels (0.22 × 10−7). As expected, V(D)J recombination at the pCE allele was reduced in Ku70−/− cells (Fig. 1c and Table 1), excluding the possibility that Ku70 was not involved in normal joining events at this allele. V(D)J recombination was not reduced to a greater extent in Ku70−/− cells9 than the observed 8–9-fold reduction most likely due to the particular sequence context of the coding ends, which have the potential for end annealing after hairpin opening (see Supplementary Information, Fig. 1a). Overall, the ratio of translocations to V(D)J recombination is >20-fold higher with Ku70 deficiency (Table 1), implying that a portion of ends that are normally channeled into V(D)J recombination participate in translocations when Ku70 is absent.
Ku70 suppression of translocations was also observed in cells containing the pCr15 reporter (7.89 × 10−5 in Ku70−/− cells versus 2.87 × 10−5 in wild-type cells; P <0.001); ectopic Ku70 expression in Ku70−/− cells reduced translocations to wild-type levels (3.23 × 10−5). These results indicate that Ku70, most likely in the Ku70–Ku80 heterodimer, suppresses NHEJ between DNA ends from heterologous chromosomes whether the ends are derived from endonuclease or RAG cleavage.
Ku70−/− pCEr15 breakpoint junctions from a number of neo+ clones were characterized (Fig. 1e, f). Overall, junctions obtained from Ku70−/− clones were similar to those from wild-type cells, with a few identical junctions noted for the two genotypes. Microhomology was present at most, but not all, junctions, as in wild-type cells. Some distinctions were noted that reached statistical significance. Longer deletions (>100 base pairs) were more common at junctions from Ku70−/− clones than from wild-type clones (46.7% versus 21.4%, P = 0.03), and coding ends from Ku70−/− clones were less likely to have remained intact than coding ends from wild-type clones (25% versus 56%, P = 0.03). Thus, Ku70 suppresses translocation formation, but can nevertheless affect end modifications at translocation breakpoints. Interestingly, junctions from four clones retained RSS elements, implicating cleavage at a single RSS, although alternative mechanisms are also possible (see Supplementary Information, Fig. 1b). Breakpoint junctions from Ku70−/− pCr15 clones gave similar results (see Supplementary Information, Fig. 1c).
The Ku-dependent pathway of NHEJ was originally characterized through its dual requirement in V(D)J recombination and radiation resistance. Nevertheless, NHEJ can occur in cells deficient for components of the canonical NHEJ pathway, both to repair DSBs in plasmids8,9 and to generate translocations in B cell precursors of p53-deficient mice10,11. Our results suggest that noncanonical NHEJ pathway(s) are the primary mediators of translocation formation — whether these pathways involve some components of the canonical pathway, or are independent of these components, remains to be determined.
In conclusion, DSBs induced by the RAG recombinase can participate with a concurrent DSB on another chromosome to result in a reciprocal translocation, supporting a model proposed for translocation formation in some lymphoid malignancies4. Despite the fact that translocations involve an NHEJ pathway of DSB repair, the canonical NHEJ pathway component Ku70 is not required, but instead suppresses translocation formation. A recent study has provided evidence that Ku70–Ku80 may limit the mobility of DNA ends12, suggesting that DNA ends that would normally be repaired intrachromosomally become mobile in Ku-deficient cells and are able to participate in interchromosomal repair. Different repair-factor usage between intrachromosomal and interchromosomal repair provides an explanation for distinctions that have been noted between junctions found in translocations and those derived from repair of a single DSB5.
Acknowledgments
We thank M. Leversha at the Memorial Sloan-Kettering Cancer Centre, Molecular Cytogenetics Core Facility for performing the FISH analysis, and B. Elliott, J. Stark, V. Lee and members of the Jasin laboratory for helpful discussions. D.M.W. was supported by the Leukemia and Lymphoma Society (5415-05) and a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund. This work was supported by the Lehman Brothers Foundation and National Institutes of Health (NIH) R0154688 (M.J.).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Note: Supplementary Information (including Methods) is available on the Nature Cell Biology website.
References
- 1.Bassing CH, Swat W, Alt FW. Cell. 2002;109:S45–S55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- 2.Lieber MR, Ma Y, Pannicke U, Schwarz K. Nature Rev Mol Cell Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 3.Lieber MR, Yu K, Raghavan SC. DNA Repair. 2006;5:1234–1245. doi: 10.1016/j.dnarep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 4.Marculescu R, et al. DNA Repair. 2006;5:1246–1258. doi: 10.1016/j.dnarep.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 5.Weinstock DM, Elliott B, Jasin M. Blood. 2006;107:777–780. doi: 10.1182/blood-2005-06-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roth DB. Nature Rev Immunol. 2003;3:656–666. doi: 10.1038/nri1152. [DOI] [PubMed] [Google Scholar]
- 7.Gu Y, Jin S, Gao Y, Weaver DT, Alt FW. Proc Natl Acad Sci USA. 1997;94:8076–8081. doi: 10.1073/pnas.94.15.8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang F, Jasin M. J Biol Chem. 1996;271:14405–14411. doi: 10.1074/jbc.271.24.14405. [DOI] [PubMed] [Google Scholar]
- 9.Verkaik NS, et al. Eur J Immunol. 2002;32:701–709. doi: 10.1002/1521-4141(200203)32:3<701::AID-IMMU701>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 10.Difilippantonio MJ, et al. J Exp Med. 2002;196:469–480. doi: 10.1084/jem.20020851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu C, et al. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
- 12.Soutoglou E, et al. Nature Cell Biol. 2007;9:675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]