Abstract
The MRN (Mre11-Rad50-Nbs1)-ATM (ataxia-telangiectasia mutated) pathway is essential for sensing and signaling from DNA double-strand breaks. The MRN complex acts as a DNA damage sensor, maintains genome stability during DNA replication, promotes homology-dependent DNA repair and activates ATM. MRN is essential for cell viability, which has limited functional studies of the complex. Small-molecule inhibitors of MRN could circumvent this experimental limitation and could also be used as cellular radio- and chemosensitization compounds. Using cell-free systems that recapitulate faithfully the MRN-ATM signaling pathway, we designed a forward chemical genetic screen to identify inhibitors of the pathway, and we isolated Z-5-(4-hydroxybenzylidene)-2-imino-1,3-thiazolidin-4-one (mirin, 1) as an inhibitor of MRN. Mirin prevents MRN-dependent activation of ATM without affecting ATM protein kinase activity, and it inhibits Mre11-associated exonuclease activity. Consistent with its ability to target the MRN complex, mirin abolishes the G2/M checkpoint and homology-dependent repair in mammalian cells.
The cellular response to DNA damage coordinates DNA repair with cell cycle progression and/or apoptosis. This response is essential to maintain the integrity of the genome. Defects in the DNA damage response can lead to genomic instability, a mutagenic condition leading to cancer predisposition1,2. Inherited mutations in the ATM, MRE11A and NBN genes are responsible for the cancer-prone syndromes ataxia-telangiectasia, ataxia telangiectasia–like disorder and Nijmegen breakage syndrome (NBS), respectively. These syndromes share clinical and cellular phenotypes including hypersensitivity to ionizing radiation (IR), radio-resistant DNA synthesis, checkpoint deficiencies and chromosomal instability.
ATM belongs to the phosphatidylinositol-3′ kinase-related kinase (PIKK) family and has a critical role in the maintenance of genome integrity3. In response to agents that generate DNA double-strand breaks (DSBs), inactive dimeric ATM protein dissociates into phosphorylated monomers4. The MRN complex is critical for this activation process and also recruits ATM to damaged DNA5–15. Upon activation, ATM phosphorylates several substrates, including p53, Chk2, Nbs1 and BRCA1, that together coordinate cell cycle arrest and DNA repair16.
The MRN complex displays DNA binding and tethering activities and functions as both a single- and double-stranded DNA endonuclease as well as a 3′-5′ double-stranded exonuclease17,18. MRN is required for the maintenance of genome stability during DNA replication, for telomere length maintenance and for DNA repair19. As a result, the MRE11A, RAD50 and NBN genes are each essential for cell viability, whereas ATM is not.
We designed a forward chemical genetic screen to identify inhibitors of the MRN-ATM pathway, and we isolated mirin as an inhibitor of MRN. This inhibitor prevents ATM activation in response to DSBs and blocks homology-directed repair (HDR) in mammalian cells.
RESULTS
Identification of mirin
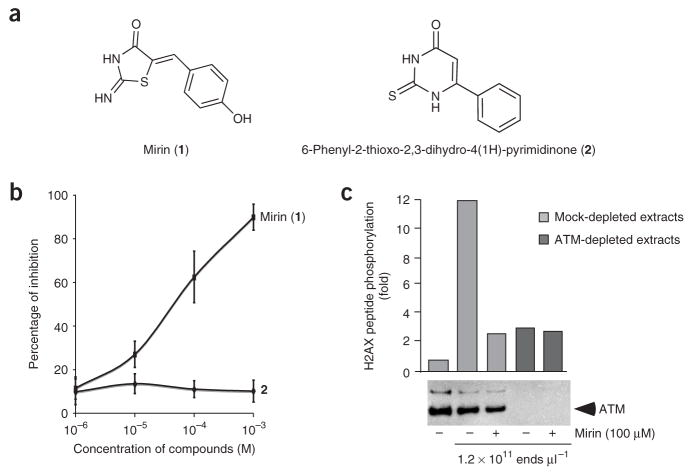
We designed a screen to identify small molecules with potential radio-and/or chemosensitizing properties2,20 that inhibit the MRN-ATM pathway in response to DSBs. The screen was performed in cell-free extracts derived from Xenopus laevis eggs that recapitulate many aspects of the MRN- and ATM-dependent responses to DNA DSBs10,21,22 and some of the aberrant phenotypes, such as radio-resistant DNA synthesis, which is observed in cancer-prone syndromes21,22. X. laevis extracts have been used previously to identify small-molecule inhibitors of actin assembly, spindle assembly and cell cycle progression23–25. In our screen, the DNA damage response was triggered by incubating cell-free extracts with HaeIII-digested plasmid DNA, which mimics DNA DSBs. The readout assay for ATM activity was phosphorylation of a peptide derived from histone H2AX (refs. 21,26); the assay was carried out in a 96-well plate format (Supplementary Fig. 1 online and Methods). In this forward chemical genetic screen, a small molecule could potentially target any step in the pathway leading to H2AX phosphorylation. Here, we used a 10,000-compound DIVERSet library (Chembridge Corporation) that had previously led to the identification of small molecules that inhibit p53 activity or interfere with mitosis and spindle dynamics27,28. The robustness of the assay was determined by calculating the Z′ value for each 96-well plate. The average Z′ value was 0.57 (Supplementary Fig. 2 online). We identified mirin as an inhibitor of DSB-induced ATM activation (Fig. 1a). Purity of mirin was confirmed by NMR (Supplementary Fig. 3 online). The half-maximal inhibitory concentration (IC50) for inhibition of H2AX phosphorylation by mirin was estimated to be 66 μM (Fig. 1b). A similar compound, 6-phenyl-2-thioxo-2,3-dihydro-4(1H)-pyrimidinone (2; Fig. 1a), did not inhibit DSB-induced ATM activation as seen by H2AX peptide phosphorylation (Fig. 1b). To confirm the specificity of the assay for ATM kinase activity, ATM was immunodepleted from extracts before addition of DSB-containing DNA and mirin. H2AX phosphorylation was substantially reduced in ATM-depleted extracts, and the residual H2AX phosphorylation was insensitive to mirin, which demonstrates that the inhibitory effect of mirin on H2AX phosphorylation is entirely ATM dependent (Fig. 1c). Mirin also inhibited the ATM-dependent phosphorylation of the downstream targets Nbs1 and Chk2 (ref. 29) (Fig. 2a) and the MRN-dependent autophosphorylation of ATM at Ser1981 in response to DSBs (Fig. 2b).
Figure 1.
Identification of mirin as an inhibitor of ATM activation by DSBs in X. laevis extracts. (a) Structure of mirin (1) and of 6-phenyl-2- thioxo-2,3-dihydro-4(1H)-pyrimidinone (2). (b) Titration curve of mirin and 2 on H2AX peptide phosphorylation assay. X. laevis extracts were incubated with increasing concentrations of either mirin or 2 in the presence of DSBcontaining DNA (5 ng μl−1). ATM activity was assayed as described in Supplementary Figure 1, and percentages of inhibition were calculated as described in Methods. Each bar represents the average of three different experiments with s.d. shown. (c) Mock-depleted extracts (lanes 1 to 3) or ATM-depleted extracts (lanes 4 and 5) were incubated with streptavidin-bound biotinylated DNA (DNA, lanes 2 to 5) in the presence of 100 μM mirin (lanes 3 and 5) or DMSO (lanes 2, 4, 6 and 8). After DNA pull-down, ATM activation was assayed in soluble fractions by measuring H2AX peptide phosphorylation. The western blot below shows the extent of ATM depletion.
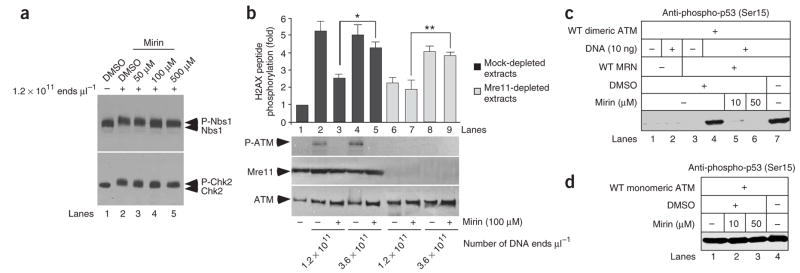
Figure 2.
Mirin inhibits MRN-dependent activation of ATM. (a) Mirin inhibits ATM-dependent phosphorylation of Nbs1 and Chk2. X. laevis extracts were incubated with DMSO or mirin as indicated and then treated with DSBs. Nbs1 (upper panel) and Chk2 (lower panel) electrophoretic mobility were monitored by western blot. (b) Mirin inhibits MRN-dependent activation of ATM. Mock-treated (dark gray) or Mre11-depleted (light gray) extracts were incubated with 100 μM of mirin (lanes 3, 5, 7 and 9). ATM activation was triggered by addition of DSB-containing DNA at the indicated concentrations. Following DNA pull-down, ATM activation was assayed in the soluble fractions by H2AX peptide phosphorylation, or by western blot for phospho-Ser1981 of ATM (P-ATM). Each bar represents the average of four different experiments with s.d. shown. Values marked with asterisks are significantly different (P < 0.003). Mre11 depletion is shown below the graph. (c) Mirin prevents MRN- and DNA-dependent activation of dimeric ATM in vitro. Dimeric ATM activity was assayed in the presence of DNA (lane 2), MRN (lane 3), MRN and DNA (lanes 4 to 7), and with or without mirin (lanes 5 and 6). ATM activation was monitored by western analysis of Ser15-phosphorylated p53. (d) Mirin does not inhibit the activity of monomeric ATM in vitro. Monomeric ATM activity was assayed as in c in the presence of mirin, as indicated. WT, wild type.
Mirin inhibits the MRN-dependent activation of ATM
To identify the target of mirin, extracts were depleted of the MRN complex using an antibody specific for Mre11 and incubated with two different amounts of DSB-containing DNA corresponding to 1.2 × 1011 ends μl−1 and 3.6 × 1011 ends μl−1 (Fig. 2b). Biotinylated DNA was generated by PCR using one biotinylated primer, then purified and bound to streptavidin beads as described10. After removal of streptavidin-bound DNA, ATM activation was monitored in the resulting soluble fractions (Fig. 2b). H2AX peptide was phosphorylated to similar extents in mock-depleted extracts at both DNA concentrations (Fig. 2b). MRN depletion prevented ATM activation at 1.2 ×1011 ends μl−1, but partial ATM activation was observed at 3.6 × 1011 ends μl−1 (Fig. 2b). This confirms that MRN’s requirement for ATM activation can be partially bypassed when the number of DNA breaks is increased, in agreement with results in mammalian NBS cells30 and our own previous data10. Notably, mirin partially inhibited MRN-dependent ATM activation in mock-depleted extracts (Fig. 2b). In contrast, mirin had no effect on MRN-independent ATM activity triggered by 3.6 × 1011 ends μl−1 (Fig. 2b). This result strongly suggests that mirin inhibits MRN-dependent activation of ATM kinase but not ATM kinase activity.
To confirm this finding we turned to an in vitro assay with purified recombinant proteins and DNA. ATM was purified from mammalian cells as a catalytically inactive dimer (Fig. 2c)7 or as a partly active monomer (Fig. 2d)6, as previously described31. Activation of dimeric ATM by MRN and DNA was monitored using phosphorylation of recombinant p53 as a readout with a phosphospecific antibody directed against Ser15 (ref. 7). ATM activation was inhibited by mirin with an IC50 of 12 μM (Fig.(2c and Supplementary Fig. 4 online). In contrast, the catalytic activity of monomeric ATM, which does not require MRN or DNA6, was not inhibited by 50 μM mirin (Fig. 2d).
Mirin inhibits the nuclease activity of Mre11
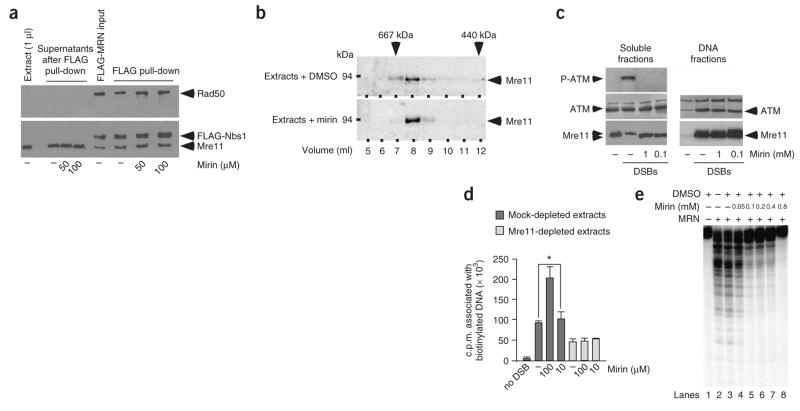
Next, we tested whether mirin triggers MRN complex dissociation. Extracts were incubated with recombinant tagged complex (Mre11–Rad50–FLAG-Nbs1) with or without 100 μM mirin. The MRN complexes were immunoprecipitated with FLAG antibodies, and then the complexes and the resultant soluble fractions were analyzed by western blot for the presence of Mre11, Rad50 and FLAG-Nbs1 (Fig. 3a). We did not observe the release of MRN subunits in soluble fractions from mirin-treated extracts, which indicates that MRN complexes were not dissociated (Fig. 3a). To test the possibility that mirin could modulate the association of the MRN complex with other proteins, extracts treated or not treated with 100 μM mirin were subjected to Superose-6 chromatography9, and the resulting fractions were probed for Mre11 (Fig. 3b). The elution profile of Mre11-containing fractions was not affected by mirin, further demonstrating that endogenous Mre11-containing complexes are not dissociated by mirin.
Figure 3.
Mirin inhibits the nuclease activity of Mre11. (a) Mirin does not trigger MRN complex dissociation in extracts. Extracts were incubated with FLAG-tagged MRN complex and then incubated with DMSO (−) or mirin. Recombinant MRN was isolated, and supernatants or FLAG-resin were probed using antibodies against FLAG, Mre11 or Rad50. X. laevis Rad50 protein is not recognized by the Rad50 antibody. (b) Mirin does not dissociate endogenous Mre11-associated complexes. Extracts were treated with DMSO or 100 μM mirin and fractionated on a Superose 6 gel-filtration column. Fractions were probed by western blot for Mre11. Molecular weight markers are indicated on top. (c) Mirin does not inhibit ATM and Mre11 binding to DNA. Extracts were treated with DMSO (−) or with mirin before incubation with DSB-containing DNA (1.2 × 1011 ends μl−1). Following DNA pull-down, phosphorylation of ATM Ser1981 (upper panel), ATM (middle panel) and Mre11 (lower panel) were monitored by western blotting in soluble fractions and DNA fractions. (d) Mirin does not inhibit MRN-associated DNA tethering activity. Mock-treated (dark gray) or Mre11-depleted (light gray) extracts were incubated with DMSO (−) or mirin at the indicated concentrations. Extracts were then incubated with streptavidin-bound DNA and free radioactive DNA. DNA-tethering activity was assayed by measuring the radioactivity associated with streptavidin-bound DNA. Each bar represents an average of six independent experiments with s.d. shown. Values marked with asterisks are significantly different (P = 0.001). (e) Mirin inhibits the nuclease activity of MRN. Purified human MRN was incubated with a 5′-labeled double-strand oligonucleotide in the presence of DMSO or mirin, as indicated. Digested DNA products were analyzed by denaturing PAGE followed by autoradiography
The MRN complex exhibits DNA-binding and DNA-tethering activities, both of which are required for ATM activation5,10,14,26. Given that MRN promotes ATM recruitment to damaged DNA5,10,14, we sought to determine whether this step is inhibited by mirin. We monitored the association of ATM and Mre11 with DNA by western blot analysis of the DNA-bound fractions isolated from extracts incubated with streptavidin-bound biotinylated DNA (Fig. 3c). Mirin, at concentrations of up to 1 mM, did not affect ATM or Mre11 binding to DNA (Fig. 3c). Next, we measured the MRN-mediated DNA-tethering activity of cell-free extracts in the presence of mirin. We had previously shown that 50% of DNA tethering observed in cell-free extracts is MRN dependent9. DNA tethering was assessed by monitoring the ability of biotinylated DNA bound to streptavidin to associate with nonbiotinylated, radioactive DNA (Fig. 3d). Mirin did not inhibit DNA tethering at 10 μM (Fig. 3d). In fact, we reproducibly observed that 100 μM mirin stimulates MRN-dependent DNA tethering (Fig. 3d). This could reflect a slow turnover of MRN complex on DNA in the presence of mirin18. As anticipated, Mre11 depletion reduced DNA tethering by 50%, and the residual, MRN-independent, DNA-tethering activity was not affected by mirin (Fig. 3d). We recently showed that Rad50-associated adenylate kinase activity is required for MRN-dependent DNA tethering32; nevertheless, this activity was not inhibited by mirin (Supplementary Fig. 5 online). Finally, we tested whether mirin affects the 3′ to 5′ exonuclease activity of MRN (ref. 18) by monitoring the appearance of discretely digested DNA products on polyacrylamide gels (Fig. 3e). We found that mirin inhibits Mre11 nuclease activity at 100 μM (Fig. 3e). Moreover, mirin did not inhibit ExoIII nuclease activity (Supplementary Fig. 6 online). This result strongly suggests that Mre11 is a target for mirin, which is consistent with the experiments in cell-free extracts (Fig. 2a) and the in vitro experiments using recombinant proteins (Fig. 2c).
Mirin inhibits the G2 checkpoint in mammalian cells
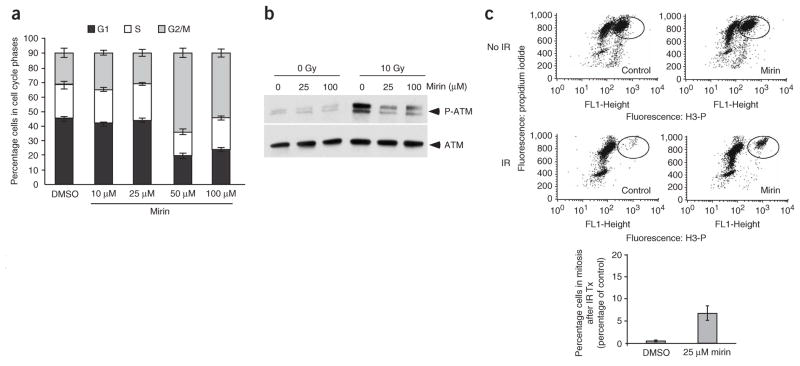
Next we sought to determine the consequences of exposing mammalian cells to mirin (Fig. 4). Exponentially growing TOSA4 (Fig. 4a) cells were treated with increasing concentrations of mirin. Mirin induced a substantial G2 arrest at concentrations of 50 μM and 100 μM (Fig. 4a). This observation suggests that mirin triggers a DNA damage checkpoint in G2, which is consistent with the role for MRN in the maintenance of genome stability during S phase26,33. To investigate the nature of this arrest, we treated U2OS cells with 100 mM mirin or 100 μM mirin in combination with 10 μM of KU-55933 (3), a specific ATM inhibitor34. A similar G2 arrest was observed under these conditions (Supplementary Fig. 7 online), which establishes that this G2 arrest was ATM independent. However, an inhibitor of the MRN-ATM pathway is predicted to abolish the IR-induced G2/M checkpoint. To circumvent mirin-induced G2 arrest observed in asynchronously growing cells, we assessed the consequences of exposing cells in G2 to mirin. U2OS cells were synchronized in G1 by double thymidine block. After release, cells progressed synchronously through S phase and reached G2 at 8.5 h (Supplementary Fig. 8 online). G2 cells were treated with 25 μM mirin, irradiated with 10 Gy, treated with nocodazole (4) and stained for phosphorylated histone H3, a marker of mitotic cells. Treatment with mirin resulted in a ten-fold increase in cells entering mitosis after IR (Fig. 4c). This establishes that mirin can abolish the IR-induced G2/M checkpoint, which is consistent with its ability to inhibit ATM signaling.
Figure 4.
Mirin abolishes the G2/M checkpoint. (a) Cell cycle distribution of TOSA4 cells. TOSA4 cells were treated with DMSO or mirin at the indicated concentrations, and cells were processed for flow cytometric analysis. The cell cycle distribution (G1, S and G2/M) is shown. Each bar represents an average of three independent experiments with s.d. shown. (b) Mirin prevents ATM autophosphorylation on Ser1981. U2OS cells synchronized in G1 were incubated with the indicated concentrations of mirin and then mock-irradiated or irradiated with 10 Gy. Cells were harvested 30 min after irradiation and processed for western blot with antibodies against phosphorylated Ser1981 of ATM (top panel) or total ATM (bottom panel). (c) Mirin abolishes the IR-induced G2/M checkpoint in U2OS cells. U2OS cells were synchronized in G2 (see Methods), treated with DMSO (control) or 25 μM mirin, and irradiated with 10 Gy, and mitotic cells were trapped with nocodazole. A typical FACS profile for each sample is shown on top. The average percentage of phospho-H3-positive cells (in mitosis) following irradiation is shown in control and mirin-treated cells in the graph below. The average of three independent experiments with s.d. is shown.
Mirin inhibits HDR in mammalian cells
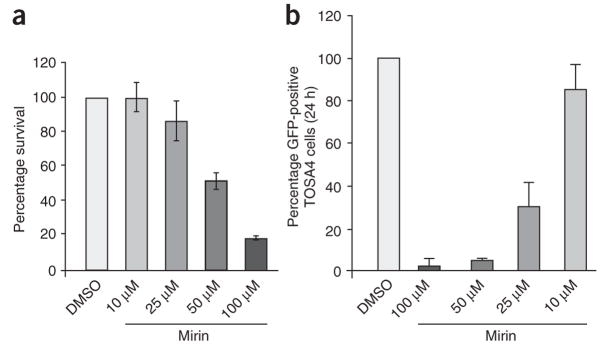
Next, we examined the effects of mirin on DNA repair in mammalian cells. We first analyzed the cellular cytotoxicity of mirin (Fig. 5a). Human embryonic kidney (HEK) 293 cells were exposed to increasing concentrations of mirin for 24 h, and cell survival was analyzed 10 d later. Mirin had little effect on cell survival at 25 μM and showed 50% cytotoxicity at 50 μM. We then investigated the effect of mirin on HDR of DSBs in HEK293-derived TOSA4 cells that harbor a single chromosomally integrated copy of the DR-GFP reporter substrate (Supplementary Fig. 9 online)35,36. The DR-GFP reporter harbors two nonfunctional copies of the gene encoding green fluorescent protein (GFP)—one (SceGFP) that is disrupted by the recognition site for the rare-cutting endonuclease I-SceI and another (iGFP) that only encodes an internal region of the GFP polypeptide. Expression of I-SceI in TOSA4 cells results in a chromosomal DSB at the I-SceI site of SceGFP, and repair of the induced DSB by gene conversion with iGFP yields cells expressing a functional GFP+ gene that can be scored by flow cytometry (Supplementary Fig. 9). Treating these cells for 24 h with 25 μM of mirin, a dose that does not affect cell survival significantly (Fig. 5a), decreased the appearance of GFP-positive cells by 70% (Fig. 5b). This result demonstrates that mirin inhibits HDR.
Figure 5.
Mirin inhibits homology-dependent DNA repair in human cells. (a) Cytotoxicity of mirin in HEK293 cells. HEK293 cells were treated with the indicated concentrations of mirin, then fixed and stained 10 d later (see Methods). Stained plates were counted for colonies. Percentage survival is expressed as the average number of colonies on treated plates divided by the average number of colonies on control plates (0 μM mirin). Each bar represents an average of three independent experiments with s.d. shown. (b) Gene conversion assay. TOSA4 cells treated with DMSO or mirin at the indicated concentrations were transfected with I-Sce1–expressing plasmid. After 24 h, GFP-expressing cells were then scored by cell sorting. The average of four independent experiments is shown. Each bar represents an average of five independent experiments with s.d. shown.
DISCUSSION
Using cell-free extracts derived from X. laevis eggs to screen for compounds that affect the response to DSBs, we have identified and characterized a new inhibitor of the MRN-ATM pathway. This further validates X. laevis cell-free extracts as a powerful system for performing forward chemical genetics screens and for identifying small molecules that modulate the DNA damage response. The readout for the screen was phosphorylation of a peptide derived from γ-H2AX by ATM. In this assay, the pathway leading to ATM activation requires the MRN complex at 1.2 × 1011 ends μl−1 but not at 3.6 × 1011 ends μl−1. Our experiments establish that mirin targets the MRN complex. Mirin inhibits MRN-dependent activation of ATM but does not affect MRN-independent activation of ATM in cell-free extracts (Fig. 2b). Furthermore, using purified, recombinant proteins, we show that mirin does not directly inhibit ATM catalytic activity in vitro, whereas it prevents the activation of ATM in the presence of damaged DNA and MRN recombinant protein complexes (Fig. 2c,d). Finally, experiments in mammalian cells strengthen the idea that mirin targets MRN rather than ATM alone. Mirin triggers a G2/M checkpoint and strongly inhibits HDR—phenotypes that are not expected from an inhibitor of ATM alone.
Mirin inhibits the 3′ to 5′ exonuclease activity associated with the Mre11 subunit of the MRN complex18. Consistent with studies of a nuclease-deficient Mre11 (ref. 37), we found that mirin, and thus the nuclease activity of Mre11, does not affect the ability of Mre11 to interact with Rad50, Nbs1 and damaged DNA. Given that mirin inhibits ATM activation in both X. laevis cell-free extracts and mammalian cells, this suggests that the nuclease and DNA end-processing functions of Mre11 are required for proper ATM activation, which is consistent with experiments in A-TLD cells partially deficient for Mre11 function15. The contribution of Mre11 nuclease activity toward MRN functions has been difficult to assess in vertebrate cells because MRN is essential for cell growth38. Furthermore, nuclease-deficient human MRN complexes are unstable, which hampers biochemical analysis of the function of the nuclease activity in vitro. Together, these results establish mirin as a new tool for dissecting nuclease-dependent and nuclease-independent functions of the Mre11 complex while leaving intact other MRN functions.
Notably, we find that mirin inhibits MRN in mammalian cells. Consistent with the known and predicted functions of the complex, mirin inhibits ATM activation, the G2/M checkpoint and HDR in mammalian cells. Mirin is a powerful tool for the acute inhibition of MRN activity in mammalian cells, thus circumventing the lethality associated with genetic inactivation of the complex. Our data suggest that Mre11 exonuclease activity might be critical for HDR in mammalian cells. Though Mre11 nuclease activity is important for meiotic recombination in yeast, its role in mitotic recombination is more controversial (see ref. 38 for review). However, because mirin is a relatively weak inhibitor, it is difficult to establish unambiguously that its sole target is Mre11. The design of more potent derivatives should help clarify this issue in the future.
METHODS
Chemicals
Caffeine (5) was purchased from Sigma (C-0750) and dissolved in 10 mM PIPES at pH 8.0. [α-32P]ddATP, [α-32P]dCTP and [γ-32P]ATP (10 μCi μl−1) were purchased from GE Healthcare, and M13 single-stranded DNA was from New England Biolabs. The inhibitor KU-55933 was a gift (see Acknowledgments).
X. laevis extracts
Membrane-free egg cytosols were prepared in the presence of cycloheximide (6) as described39. All incubations were performed at 22 °C. X. laevis were handled in accordance to guidelines provided by the Institutional Animal Care and Use Committee at Columbia University, protocol AA5192.
Small-molecule library
The small-molecule library was obtained from Chembridge Corporation. This 10,000 compound DIVERSet library is available in a 96-well format, with 80 compounds per plate. The library was replated and the compounds diluted to achieve 100 μM concentration for screening. We used high compound concentrations because of the high lipid and protein content of X. laevis extracts, which results in the nonspecific sequestration of the small molecules.
Screening procedure
Extracts were incubated with 5 ng μl−1 of DSB-containing DNA (HaeIII-digested pBlueScript DNA) for 20 min at 22 °C. 2 μl aliquots of extracts were transferred to 96-well V-bottom plates containing the small-molecule library on ice. 20 μl of kinase buffer was added to each well (2 μl 1 mMATP, 1 μl of H2AX (AVGKKASQASQEY) or 10 mg ml−1 control peptide (AVGKKAAQAAQEY), 0.4 μl [γ-32P]ATP at 10 μCi μl−1, 16.6 μl Extract Buffer (EB; 80 mM β-glycerophosphate, 10 mM MgCl2, 20 mM HEPES, 1 mM DTT). The samples were mixed thoroughly and incubated for 10 min at 30 °C.
Processing of 96-well P81 plates (Millipore) was performed on a Flusher plate washer (Flushtec). The P81 phosphocellulose 96-well plates were pre-washed with 100 μl of wash buffer containing 0.75% phosphoric acid. The kinase reactions were stopped by placing the plates on ice and adding 20 μl of 0.75% phosphoric acid. The reactions were then transferred to the P81 plates. Binding was allowed for 30 s and the vacuum was applied. The plates were then washed 5 times in 300 μl of 0.75% phosphoric acid and dried with vacuum. The P81 plate backing was removed, excess liquid was absorbed, and the P81 paper was exposed to a storage phosphor screen for 24 h, then processed for image analysis with a PhosphorImager (Molecular Dynamics, GE Healthcare). The phosphorylation of H2AX peptide was calculated for each sample according to the following formula: (value of sample − average of value of negative controls with control peptide)/(average value of 80 samples − average value of negative control). Percentages of inhibition of H2AX peptide phosphorylation were calculated according to the following formula: (1− ((average of sample value − average of value of negative control)/(average value of positive control − average value of negative control))) × 100.
In all the other experiments monitoring ATM activation in extracts, ATM was activated by incubating extracts with biotinylated DNA previously bound to streptavidin beads as previously described10. The reaction was spotted on P81 phosphocellulose filter papers (Upstate Biotechnology), and the radio-activity incorporated was quantified by scintillation. The number of c.p.m. incorporated into control peptide was then subtracted from the number of c.p.m. incorporated into the H2AX peptide, and the values were normalized to untreated extracts.
ATM protein kinase assays with recombinant proteins
Monomeric ATM, dimeric ATM and MRN were purified as described31. Monomeric and dimeric ATM activity were assayed in vitro as previously described6,7.
Nuclease assay
The nuclease activity of Mre11 was assayed as described18.
Cell synchronization
U2OS cells were grown to ~40% confluency in 6-well plates. 24 h after plating, 2 mMthymidine (Sigma) in DMEM was added to the cells for the first block. After 16 h, thymidine was removed by washing the cells twice with 1× phosphate-buffered saline (PBS). Fresh medium was added for 9 h, then 2 mM thymidine was added to cells for the second block for 16 h. Thymidine was then removed followed by 2 washes with 1× PBS, and fresh medium was added.
G2/M checkpoint assay
8.5 h following the second release, either 25 μM mirin, 10 μM KU-55933 (ATM inhibitor) or DMSO was added to the cells. 30 min later (9 h postrelease), the cells were irradiated with 10 Gy of γ-radiation from a 137Cs source or mock-treated, and then incubated at 37 °C for 1.5 h. Each culture was then treated with 1 μg ml−1 nocodazole to trap cells in mitosis. Approximately 15 h later, the cells were harvested and fixed with 70% ethanol and placed at −20 °C overnight. Cells were then resuspended in 1 ml of 0.25% Triton X-100 in PBS and incubated on ice for 15 min. After centrifugation, the cell pellet was suspended in 100 μl of 1% bovine serum albumin (BSA)-PBS with 1.5 μg of a polyclonal antibody that specifically recognizes the Ser10-phosphorylated form of histone H3 (Upstate Biotechnology). The cells were then rinsed with 1% BSA-PBS three times, incubated for 30 min with fluorescein isothiocyanate–conjugated goat anti-rabbit IgG antibody (1:15 in 1% BSA-PBS, Jackson ImmunoResearch Laboratories, Inc.), rinsed three times with 1% BSA-PBS and stained with propidium iodide (Sigma) for 1 h. Cellular fluorescence was then measured by flow cytometry.
HDR assays
HDR assays were conducted essentially as described35 using TOSA4, a subclone of 293T cells that contain a single integrated copy of the DR-GFP reporter. TOSA4 cells were seeded onto 60-mm plates at ~6 × 105 cells per plate in 3 ml of nonselective medium. 24 h later, the cells were transfected with 1 μg per plate of pCBASce, an expression vector encoding I-SceI, using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. At 2 h post-transfection, cells were treated with the indicated concentrations of mirin (in DMSO) or with DMSO alone. At 24 h after transfection, the cells were washed once with PBS and harvested. To measure HDR of an induced DSB, the fraction of GFP-positive cells was quantitated by flow cytometry. To evaluate cell cycle distribution, the cells were fixed in 70% ethanol at −20 °C, washed in 5 ml PBS containing 1% BSA, stained in 800 μl propidium iodide (Sigma) for 1 h at room temperature (18–22 °C) and analyzed on a FACScalibur flow cytometer (Becton Dickinson).
Other methods
Preparation of U2OS cell lysates, immunodepletions in X. laevis extracts, antibodies, production and purification of recombinant FLAG-MRN complex, experiments using biotinylated DNA pull-down, NMR and cytotoxicity assays are described in Supplementary Methods online.
Supplementary Material
Acknowledgments
We thank H. Lindsay for Chk2 antibody and B. Stockwell, O. Haccard and the members of the Gautier laboratory for helpful discussions. We thank G. Smith (KuDOS Pharmaceuticals) for the inhibitor KU-55933. We are indebted to J. Decatur and Y. Itagaki of the Chemistry Department for providing NMR and mass spectral analysis of mirin. This work was supported by US National Institutes of Health grants CA95866 and CA92245 (J. Gautier), CA97403 (R. Baer) and GM54668 (M. Jasin).
Footnotes
Note: Supplementary information and chemical compound information is available on the Nature Chemical Biology website.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 2.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 3.Lavin MF, et al. ATM signaling and genomic stability in response to DNA damage. Mutat Res. 2005;569:123–132. doi: 10.1016/j.mrfmmm.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 4.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 5.You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol. 2005;25:5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JH, Paull TT. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- 7.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 8.Carson CT, et al. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 2003;22:6610–6620. doi: 10.1093/emboj/cdg630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Difilippantonio S, et al. Role of Nbs1 in the activation of the Atm kinase revealed in humanized mouse models. Nat Cell Biol. 2005;7:675–685. doi: 10.1038/ncb1270. [DOI] [PubMed] [Google Scholar]
- 10.Dupre A, Boyer-Chatenet L, Gautier J. Two-step activation of ATM by DNA and the Mre11-Rad50-Nbs1 complex. Nat Struct Mol Biol. 2006;13:451–457. doi: 10.1038/nsmb1090. [DOI] [PubMed] [Google Scholar]
- 11.Kozlov SV, et al. Involvement of novel autophosphorylation sites in ATM activation. EMBO J. 2006;25:3504–3514. doi: 10.1038/sj.emboj.7601231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gatei M, et al. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat Genet. 2000;25:115–119. doi: 10.1038/75508. [DOI] [PubMed] [Google Scholar]
- 13.Paull TT, Lee JH. The Mre11/Rad50/Nbs1 complex and its role as a DNA doublestrand break sensor for ATM. Cell Cycle. 2005;4:737–740. doi: 10.4161/cc.4.6.1715. [DOI] [PubMed] [Google Scholar]
- 14.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 15.Uziel T, et al. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 2003;22:5612–5621. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 17.D’Amours D, Jackson SP. The Mre11 complex: at the crossroads of dna repair and checkpoint signalling. Nat Rev Mol Cell Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- 18.Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 19.Haber JE. The many interfaces of Mre11. Cell. 1998;95:583–586. doi: 10.1016/s0092-8674(00)81626-8. [DOI] [PubMed] [Google Scholar]
- 20.Zhou BB, Bartek J. Targeting the checkpoint kinases: chemosensitization versus chemoprotection. Nat Rev Cancer. 2004;4:216–225. doi: 10.1038/nrc1296. [DOI] [PubMed] [Google Scholar]
- 21.Costanzo V, et al. Reconstitution of an ATM-dependent checkpoint that inhibits chromosomal DNA replication following DNA damage. Mol Cell. 2000;6:649–659. doi: 10.1016/s1097-2765(00)00063-0. [DOI] [PubMed] [Google Scholar]
- 22.Costanzo V, et al. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol Cell. 2001;8:137–147. doi: 10.1016/s1097-2765(01)00294-5. [DOI] [PubMed] [Google Scholar]
- 23.Peterson JR, Lokey RS, Mitchison TJ, Kirschner MW. A chemical inhibitor of N-WASP reveals a new mechanism for targeting protein interactions. Proc Natl Acad Sci USA. 2001;98:10624–10629. doi: 10.1073/pnas.201393198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verma R, et al. Ubistatins inhibit proteasome-dependent degradation by binding the ubiquitin chain. Science. 2004;306:117–120. doi: 10.1126/science.1100946. [DOI] [PubMed] [Google Scholar]
- 25.Wignall SM, et al. Identification of a novel protein regulating microtubule stability through a chemical approach. Chem Biol. 2004;11:135–146. [PubMed] [Google Scholar]
- 26.Costanzo V, Paull T, Gottesman M, Gautier J. Mre11 assembles linear DNA fragments into DNA damage signaling complexes. PLoS Biol. 2004;2:E110. doi: 10.1371/journal.pbio.0020110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komarov PG, et al. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science. 1999;285:1733–1737. doi: 10.1126/science.285.5434.1733. [DOI] [PubMed] [Google Scholar]
- 28.Mayer TU. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. comment 286, 913–914, 1999. [DOI] [PubMed] [Google Scholar]
- 29.Cimprich KA, Shin TB, Keith CT, Schreiber SL. cDNA cloning and gene mapping of a candidate human cell cycle checkpoint protein. Proc Natl Acad Sci USA. 1996;93:2850–2855. doi: 10.1073/pnas.93.7.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitagawa R, Bakkenist CJ, McKinnon PJ, Kastan MB. Phosphorylation of SMC1 is a critical downstream event in the ATM-NBS1-BRCA1 pathway. Genes Dev. 2004;18:1423–1438. doi: 10.1101/gad.1200304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee JH, Paull TT. Purification and biochemical characterization of ataxia-telangiectasia mutated and Mre11/Rad50/Nbs1. Methods Enzymol. 2006;408:529–539. doi: 10.1016/S0076-6879(06)08033-5. [DOI] [PubMed] [Google Scholar]
- 32.Bhaskara V, et al. Rad50 adenylate kinase activity regulates DNA tethering activities of Mre11/Rad50 complexes. Mol Cell. 2007;25:647–661. doi: 10.1016/j.molcel.2007.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trenz K, Smith E, Smith S, Costanzo V. ATM and ATR promote Mre11 dependent restart of collapsed replication forks and prevent accumulation of DNA breaks. EMBO J. 2006;25:1764–1774. doi: 10.1038/sj.emboj.7601045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hickson I, et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 35.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Esashi F, et al. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- 37.Arthur LM, et al. Structural and functional analysis of Mre11–3. Nucleic Acids Res. 2004;32:1886–1893. doi: 10.1093/nar/gkh343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smythe C, Newport JW. Systems for the study of nuclear assembly, DNA replication, and nuclear breakdown in Xenopus laevis egg extracts. Methods Cell Biol. 1991;35:449–468. doi: 10.1016/s0091-679x(08)60583-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.