Abstract
Sizn1 (Zcchc12) is a transcriptional co-activator that positively modulates BMP (Bone Morphogenic Protein) signaling through its interaction with Smad family members and CBP. We have demonstrated a role for Sizn1 in basal forebrain cholinergic neuron specific gene expression. Furthermore, mutations in SIZN1 have been associated with X-linked mental retardation. Given the defined role of SIZN1 in mental retardation, knowing its complete forebrain expression pattern is essential to further elucidating its role in cognition. To better define the dynamic expression pattern of Sizn1 during forebrain development, we investigated its expression in mouse brain development from embryonic day 8.0 (E8.0) to adult. We found that Sizn1 is primarily restricted to the ventral forebrain including the medial ganglionic eminence, the septum, amygdala, and striatum. In addition, Sizn1 expression is detected in the cortical hem and Pallial-subpallial boundary (PSB; anti-hem); both sources of Cajal-Retzius cells. Sizn1 expression in the dorsal forebrain is restricted to a subset of cells in the marginal zone that also express Reln, indicative of Cajal-Retzius cells. These data provide novel information on brain regions and cell types that express Sizn1, facilitating further investigations into the function of Sizn1 in both development and the pathogenesis of mental retardation.
Introduction
Over the past decade mutations in numerous genes have been associated with mental retardation, yet the pathogenesis remains poorly understood in all but a few disorders. SIZN1(ZCCHC12) is one example of such a gene that, when mutated, results in mental retardation in males (Cho et al., 2008a). Biochemical studies indicate that Sizn1 is a positive modulator of BMP signaling and is necessary for normal basal forebrain cholinergic neuron gene expression (Cho et al., 2008b).
Basal forebrain cholinergic neurons are known to be the major cholinergic input to the cerebral cortex and hippocampus (Bigl et al., 1982; Mesulam et al., 1983a; Mesulam et al., 1983b; Woolf et al., 1983, 1984). Loss of this input correlates with cognitive decline in Alzheimer’s disease, implicating an important role of basal forebrain cholinergic neurons in cognition (Baxter and Chiba, 1999; Granholm et al., 2000). In addition, the embryonic septum is not only the origin for many basal forebrain cholinergic neurons, but also a subset of Cajal-Retzius (CR) cell that expresses Reln and are important for normal neocortex lamination. Cajal-Retzius neurons are born from several sites including the septum, the pallial-subpallial boundary, and the cortical hem (Bielle et al., 2005; Meyer et al., 2002; Shinozaki et al., 2002; Takiguchi-Hayashi et al., 2004). From these sites they migrate to the marginal zone to form layer I of the cerebral cortex.
We have hypothesized that mutations in SIZN1 lead to basal forebrain cholinergic neuron deficiencies and therefore intellectual disabilities. Knowing the expression pattern for SIZN1/Sizn1 through development is necessary to gain an understanding of the pathogenesis of SIZN1-associated mental retardation. Therefore, we undertook a series of studies to determine the cell types that express Sizn1 within and beyond the basal forebrain cholinergic neurons. Using both in situ hybridization and immunohistochemistry, we found Sizn1 is expressed in ventral forebrain cell populations in addition to the cholinergic neuron. Furthermore, we find Sizn1 is expressed by Cajal-Retzius neurons of the dorsal forebrain, beginning in their progenitor zones. These data implicate additional sites where Sizn1 is potentially functioning and contributing to a mental retardation phenotype.
1. Results and Discussion
1.1 Genomic structure of Sizn1 and sequence comparison
Sizn1 is located on the X-chromosome (Cho et al., 2008b) and is composed of 4 exons with the entire coding sequence residing in the fourth exon. Sequence comparison using the blast algorithm at Ensembl and UCSC genome browser shows that mammalian orthologs are present only in mammals including humans, cows, chimpanzee, rat and mice.
1.2 Sizn1 transcripts are detected in a dynamic pattern
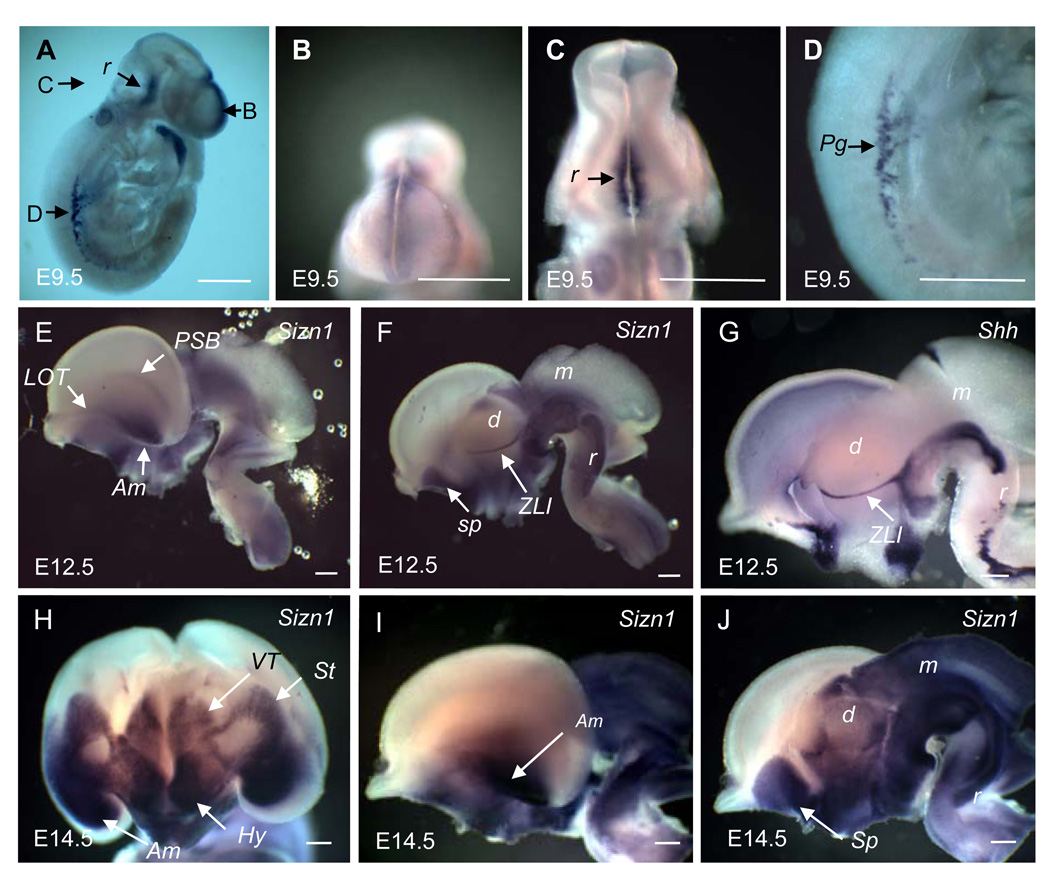
Sizn1 mRNA expression was first analyzed by whole-mount in situ hybridization. At E9.5, Sizn1 was detected mainly in the rostral neural tube and in primordial germ cells (Fig. 1A). In the brain, Sizn1 was detected in the dorsal midline of the telencephalon in the same region where Bmp and Wnt family members are also expressed (Fig. 1A and 1B) (Grove et al., 1998; Shimogori et al., 2004). This dorsal midline expression was limited to the telencephalon; no extension to the dorsal midline of the midbrain or hindbrain was observed (Fig. 1A). In the hindbrain (rhombencephalon), Sizn1 expression was found along the ventral midline anteriorly (Fig. 1A and 1C). Outside the CNS (central nervous system), Sizn1 expression was detected in the migrating primordial germ cells (Fig 1D), which are precursors of germ cells known to originate from the primitive streak before E7.0. These cells migrate to the hindgut to settle in the genital ridges and form adult germ cells (Tres et al., 2004). Expression of Sizn1 in testis occurs at E14.5 (data not shown) and in adult as described previously (Cho et al., 2008b).
Fig. 1.
In situ hybridization for Sizn1 at E9.5, E12.5 and E14.5 whole mouse embryos. In E9.5 stage, Sizn1 is expressed in dorsal midline of the forebrain (A, B), the ventral rhombencephalon (A, C) and in primodial germ cells (A, D). In E12.5 stage, the expression of Sizn1 is now restricted to the ventral neural tube from the telencephalon through the rhombencephalon (E, lateral view and F, medial view). Expression is found in the lateral olfactory tract (LOT), pallium-subpallium boundary (PSB), amygdala, septum and zona limitans intrathalamica (ZLI). (G) The ZLI expression was confirmed by comparison with Shh-in situ hybridization, which is known to be expressed in the ZLI (Puelles and Rubenstein, 2003). While the pattern of E14.5 is generally similar to that at E12.5, there has been considerable expansion of the expression domains. In a mid coronal section of the whole brain (E14.5) shows strong expression in the amygdala, hypothalamus, ventral thalamus and striatum (H). A lateral (I) and medial (J) mid-sagittal view also shows extensive expression nearly circumferentially in the mesencephalon and rhombencephalon. Am=amygdala, d=diencepahlon, Hy=hypothalamus, m=mesencephalon, Pg=primodial germ cell, r=rhombencephalon, Sp=Septum, St=striatum and VT=ventral thalamus. Scale bar is adjusted to 500 µm in each image.
By E12.5, Sizn1 was no longer detected along the expanded dorsal midline of the telencephalon (Fig 1E–F), however, expression in the cortical hem (hippocampus) was detected (see Fig. 2C, 2G and 2H). In addition, it was detected in the lateral olfactory bulb (LOT), amygdala and pallial-subpallial boundary (PSB) (Fig 1E). Prominent expression was found in the septum as previously described (Cho et al., 2008b) and zona limitans intrathalamica (ZLI) (Fig 1F). ZLI expression was confirmed by comparing expression with the known ZLI marker, Shh (Fig 1G) (Puelles and Rubenstein, 2003). In the midbrain (mesencephalon) and hindbrain (rhombencephalona), Sizn1 is mostly restricted ventrally, although the expressions extended more laterally when compared to the earlier stages (Fig 1F and 1C).
Fig. 2.
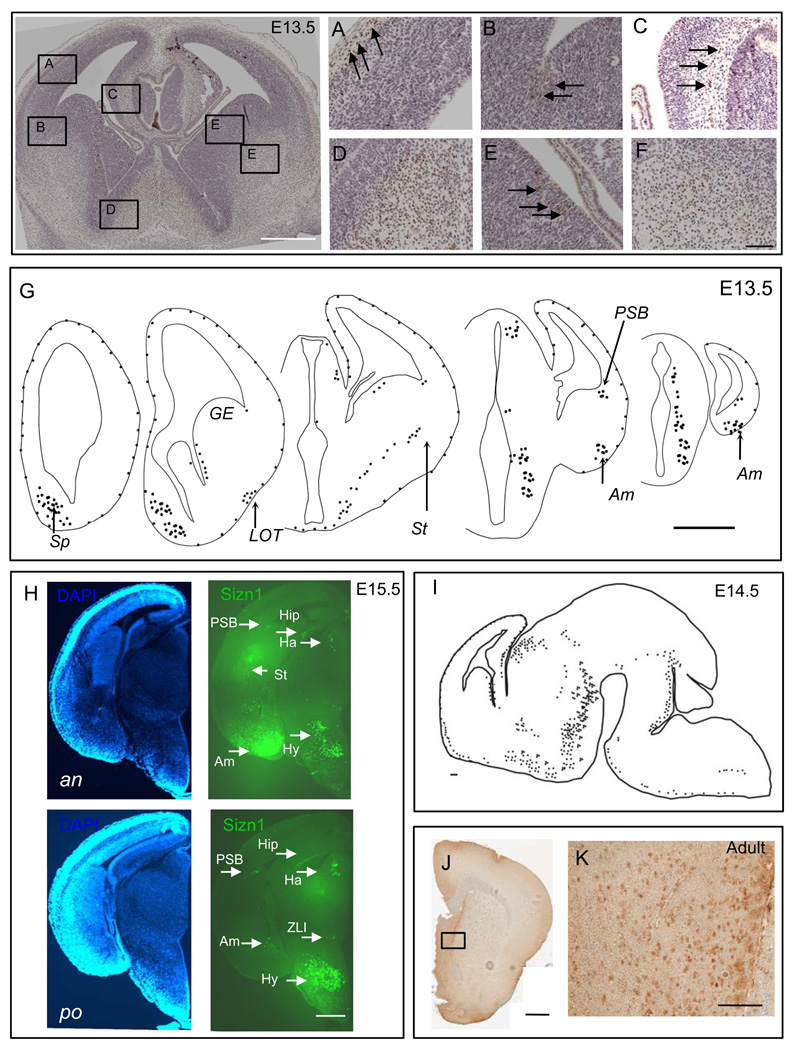
Immunohistochemistry for Sizn1 at E13.5, E14.5, E15.5 and adult brains. Top left panel is low magnification image of A–F (Scale bar is 500 µm). (A) Sizn1 expression is found in the molecular layer of the developing cortical plate corresponding to Cajal-Reitzius cells (arrows and see Figure 3). (B) Scattered labeled cells are found at the pallial-subpallial boundary (PSB, arrows). (C) Scattered Sizn1 labeled cells are present in the cortical hem (arrows). Numerous labeled cells are found in the developing septal region (D) and striatum (F). (E) Labeled cells in the medial ganglionic eminence (MGE, arrows) are primarily found in the ventricular zone. Scale bar in image F is 100 µm and corresponded to images A–F. Schematic diagrams of E13.5 brain coronal section (G; Scale bar=500 µm)) and of E14.5 sagittal section (I; Scale bar=100µm) for Sizn1 expression pattern. (H) Coronal section of E15.5 embryonic caudal forebrain. Anterior (an) and posterior (po) DAPI labeled sections on the left and Sizn1 immunofluorescence on the right (green). Sizn1 expression is localized in the amygdala, habenula, hippocampus, hypothalamus, pallium-subpallium boundary and zona limitans intrathalamica (scale bar = 500 µm). (J and K) Immunostaining for Sizn1 in the adult brain with emphasis on the septal region (Scale bar=500 µm (I) and 100 µm (J)). Am=amygdala, Ha=habenula, Hip=hippocampus, Hy=hypothalamus, Sp=Septum, St=striatum, LOT=lateral olfactory tract, GE=gangilonic eminence, PSB=pallium-subpallium boundary and ZLI= zona limitans intrathalamica.
At E14.5, Sizn1 expression in the brain primarily mirrors that seen at E12.5. Its expression in the telencephalon was mainly restricted to ventral areas including the striatum, amygdala (Fig. 1H and 1I) and septum (Fig. 1J), the ventral diencephalon and the hypothalamic nuclei (Fig. 1H and 1J). Interestingly, expression in the midbrain spans the entire neural tube from ventral to dorsal (Fig 1I and 1J). Although previous reports suggested that Sizn1 (Zcchc12) is expressed broadly within the neuroepithelium of the forebrain, diencephalon, midbrain and hindbrain of E10.5 mouse embryos (Li et al., 2009), our data indicate there is spatially and temporally restricted Sizn1 gene expression in the developing embryonic mouse brain.
1.3 Sizn1 protein is detected in the nucleus of the specific neuronal populations
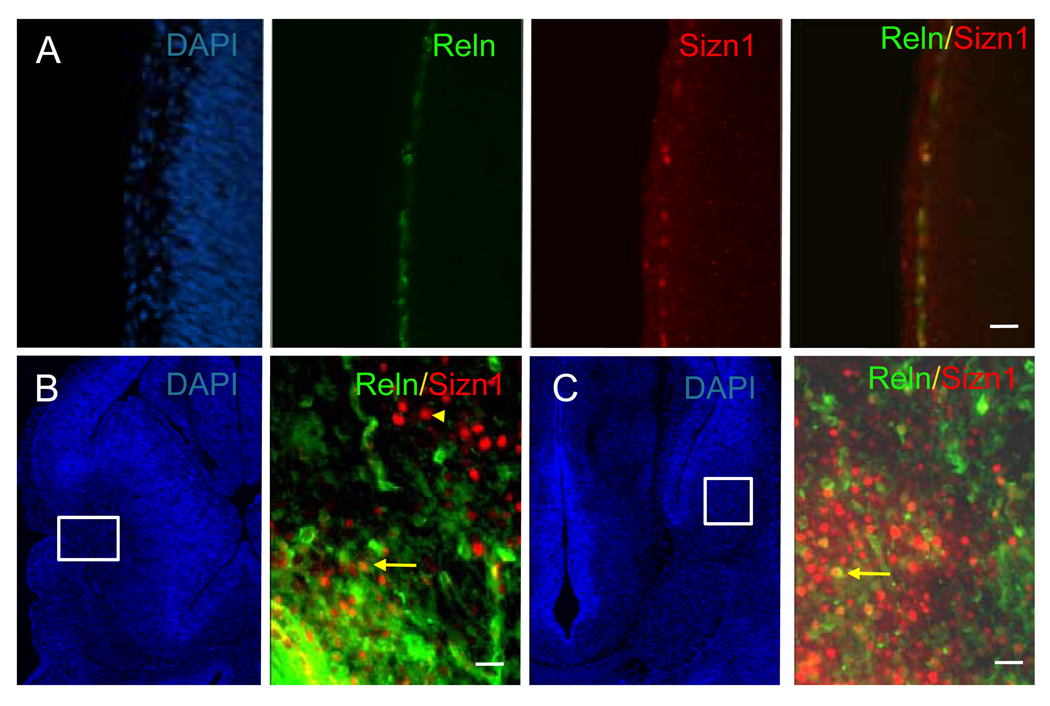
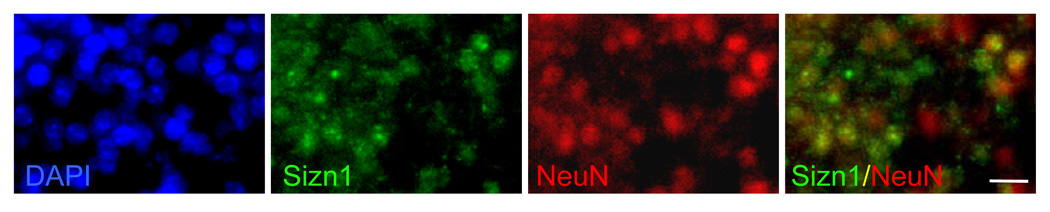
To characterize Sizn1 expression pattern, we performed immunostaining with an anti-Sizn1 antibody on E8.0 (data not shown), E13.5 (Fig. 2A–G), E14.5 (Fig. 2I), E15.5 (Fig. 2H) and adult sections (Fig. 2J–K). The immunolabeling pattern closely mirrored that observed for Sizn1 mRNA expression (Fig. 2). Although Sizn1 expression was not detected at E8.0 stage (data not shown), we cannot exclude the possibility that a few cell have expression of Sizn1. In the telencephalon of the E13.5 and E15.5 embryos, Sizn1 is expressed in a subpopulation of cells in the marginal zone of the neocortex (Fig. 2A). We also observed Sizn1 expression in the pallial-subpallial boundary (PSB) (Fig. 2B and Fig. 2H), the cortical hem (Fig. 2C) and the septum (Fig. 2D); all known sources of CR neurons (Bielle et al., 2005). In addition, the medial ganglionic eminence shows scattered Sizn1 positive cells (Fig. 2E). Sizn1 was also detected in numerous striatal cells (Fig 2F and Fig. 2H) and in the amygdala at E13.5 and E15.5 (Fig. 2G, Fig. 3C and Fig. 2H). In the diencephalon, Sizn1 is expressed in a subset of cells in the habenula, hypothalamus and ZLI (Fig. 2G and 2H). In adult brain tissue, the expression of Sizn1 remains similar to that observed at E13.5–E15.5 (Fig. 2J as example). Co-labeling experiments with anti-Sizn1 and anti-Reln antibodies confirmed that Sizn1 is expressed in Cajal-Retzius neurons (Fig. 3A). In the LOT and amygdala region, Sizn1 also was found to co-label with Reln but not in the striatum (Fig. 3B–C). To determine if the cell types that express Sizn1 are neurons, sections were labeled with anti-Sizn1 and the neuronal specific antibody, NeuN (Mullen et al., 1992). These were a near complete overlap of with these two antibodies (Fig. 4), however Sizn1 and GFAP showed virtually no overlap (data not shown). These data indicate the majority of cells that express Sizn1 are neurons.
Fig. 3.
Sizn1 is expressed by Cajal-Retzius cell in the cerebral cortical molecular layer at E14.5 (A). Immunofluorescence for Reln, a marker of early Cajal-Retzius cells, decorates a row of cells in the molecular layer. Labeling with an anti-Sizn1 antibody shows a similar layer of cells and merging of these labels shows complete overlap. DAPI labeling highlights the tissue structure (scale bar = 20 µm). (B and C) Sizn1 is also co-labeled with Reln in the LOT and amygdala. Arrows and arrowhead highlight examples of co-labeled and non co-labeled cell, respectively.
Fig. 4.
Sizn1 is expressed in neurons. Sizn1-expressing cell also express NeuN (scale bar = 20 µm).
In summary, Sizn1 shows a spatially and temporally dynamic expression pattern through development. We have confirmed the subcellular localization in the nucleus of cells (Cho et al., 2009). As reported previously, Sizn1 is expressed in ventral forebrain including the septum, striatum and amygdala (Cho et al., 2008b). In addition, we have found Sizn1 is expressed in all known sources of CR neurons including the septum, the PSB, the MGE and cortical hem (Bielle et al., 2005; Meyer et al., 2002; Shinozaki et al., 2002; Takiguchi-Hayashi et al., 2004). Their expression of Sizn1 continues as they migrate out over the surface of the brain. This expression pattern will facilitate further studies aimed at understanding the pathology of SIZN1 related X-linked mental retardation.
2. Experimental procedures
2.1. Whole-mount mRNA in situ hybridization
In situ hybridization on whole embryos was performed as previously described (Lim et al., 2005). Digoxigenin (Dig)-labeled anti-sense Sizn1 was prepared from full-length Sizn1 cDNA cloned into pBluescript (Stratagene) and digested with EcoRI (Cho et al., 2008b)(AY466375). The Shh probe was prepared as previously described (Lim and Golden, 2002). The probes were generated by using T7 RNA polymerase in the presence of Dig-UTP. Mouse embryos were harvested from a time-mated female; whole embryos (E9.5 and E10.5) and embryonic brains (E12.5 and E14.5) were dissected and fixed overnight at 4°C in 4% paraformaldehyde. In situ hybridization was performed as previously described (Lim and Golden, 2002).
2.2. Immunostaining
Immunohistochemistry on cryostat and paraffin sections was carried out as previously described (Cho et al., 2008b). After paraffin removal by Xylene, the sections were microwaved in EDTA solution (Invitrogen) for 10 mins. After treatment of 1% H2O2 for 10 min, sections were blocked with TBS (20mM Tris-HCl pH 7.5, 150mM NaCl) containing 10% goat serum (Sigma) and 0.1% Triton X-100 for 1 hr. Then sections were incubated with primary antibody against Sizn1 (1:50) (Cho et al., 2008b), Reln (A10; Chemicon), NeuN (A60; Chemicon) and GFAP (rat monoclonal antibody, kindly provided by Dr. V. Lee, University of Pennsylvania) for 2hrs. For immunofluorescence staining sections were incubated for 1 hr in polymer-HRP-conjugated-secondary antibody against rabbit (Dako, Envision). The signal was detected by TSA-Cy3 (Perkin Elmer). For chromogenic detection, the ABC elite kit (Vector Lab) was used. Protocols for the TSA-Cy3/TSA-fluorescein and ABC elite kit were according to the manufacturers instructions. The detail protocol about TSA and ABC elite kits were followed by the suggestion that the manufacture provided.
Acknowledgements
This work is supported by National Institutes of Health Grant, NS45034 (JAG) and the Alavi-Dabiri postdoctoral fellowship at the Children’s Hospital of Philadelphia (GC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baxter MG, Chiba AA. Cognitive functions of the basal forebrain. Curr Opin Neurobiol. 1999;9:178–183. doi: 10.1016/s0959-4388(99)80024-5. [DOI] [PubMed] [Google Scholar]
- Bigl V, Woolf NJ, Butcher LL. Cholinergic projections from the basal forebrain to frontal, parietal, temporal, occipital, and cingulate cortices: a combined fluorescent tracer and acetylcholinesterase analysis. Brain Res Bull. 1982;8:727–749. doi: 10.1016/0361-9230(82)90101-0. [DOI] [PubMed] [Google Scholar]
- Cho G, Bhat SS, Gao J, Collins JS, Rogers RC, Simensen RJ, Schwartz CE, Golden JA, Srivastava AK. Evidence that SIZN1 is a candidate X-linked mental retardation gene. Am J Med Genet A. 2008a;146A:2644–2650. doi: 10.1002/ajmg.a.32472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho G, Lim Y, Golden JA. SUMO interaction motifs in Sizn1 are required for promyelocytic leukemia protein nuclear body localization and for transcriptional activation. J Biol Chem. 2009;284:19592–19600. doi: 10.1074/jbc.M109.010181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho G, Lim Y, Zand D, Golden JA. Sizn1 is a novel protein that functions as a transcriptional coactivator of bone morphogenic protein signaling. Mol Cell Biol. 2008b;28:1565–1572. doi: 10.1128/MCB.01038-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granholm AC, Sanders LA, Crnic LS. Loss of cholinergic phenotype in basal forebrain coincides with cognitive decline in a mouse model of Down's syndrome. Exp Neurol. 2000;161:647–663. doi: 10.1006/exnr.1999.7289. [DOI] [PubMed] [Google Scholar]
- Grove EA, Tole S, Limon J, Yip L, Ragsdale CW. The hem of the embryonic cerebral cortex is defined by the expression of multiple Wnt genes and is compromised in Gli3-deficient mice. Development. 1998;125:2315–2325. doi: 10.1242/dev.125.12.2315. [DOI] [PubMed] [Google Scholar]
- Li H, Liu Q, Hu X, Feng D, Xiang S, He Z, Zhou J, Ding X, Zhou C, Zhang J. Human ZCCHC12 activates AP-1 and CREB signaling as a transcriptional co-activator. Acta Biochim Biophys Sin (Shanghai) 2009;41:535–544. doi: 10.1093/abbs/gmp042. [DOI] [PubMed] [Google Scholar]
- Lim Y, Cho G, Minarcik J, Golden J. Altered BMP signaling disrupts chick diencephalic development. Mech Dev. 2005;122:603–620. doi: 10.1016/j.mod.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Lim Y, Golden JA. Expression pattern of cLhx2b, cZic1 and cZic3 in the developing chick diencephalon. Mech Dev. 2002;115:147–150. doi: 10.1016/s0925-4773(02)00091-6. [DOI] [PubMed] [Google Scholar]
- Mesulam MM, Mufson EJ, Levey AI, Wainer BH. Cholinergic innervation of cortex by the basal forebrain: cytochemistry and cortical connections of the septal area, diagonal band nuclei, nucleus basalis (substantia innominata), and hypothalamus in the rhesus monkey. J Comp Neurol. 1983a;214:170–197. doi: 10.1002/cne.902140206. [DOI] [PubMed] [Google Scholar]
- Mesulam MM, Mufson EJ, Wainer BH, Levey AI. Central cholinergic pathways in the rat: an overview based on an alternative nomenclature (Ch1–Ch6) Neuroscience. 1983b;10:1185–1201. doi: 10.1016/0306-4522(83)90108-2. [DOI] [PubMed] [Google Scholar]
- Mullen RJ, Buck CR, Smith AM. NeuN, a neuronal specific nuclear protein in vertebrates. Development. 1992;116:201–211. doi: 10.1242/dev.116.1.201. [DOI] [PubMed] [Google Scholar]
- Puelles L, Rubenstein JLR. Forebrain gene expression domains and the evolving prosomeric model. Trends Neurosci. 2003;26:469–476. doi: 10.1016/S0166-2236(03)00234-0. [DOI] [PubMed] [Google Scholar]
- Shimogori T, Banuchi V, Ng HY, Strauss JB, Grove EA. Embryonic signaling centers expressing BMP, WNT and FGF proteins interact to pattern the cerebral cortex. Development. 2004;131:5639–5647. doi: 10.1242/dev.01428. [DOI] [PubMed] [Google Scholar]
- Tres LL, Rosselot C, Kierszenbaum AL. Primordial germ cells: what does it take to be alive? Mol Reprod Dev. 2004;68:1–4. doi: 10.1002/mrd.20056. [DOI] [PubMed] [Google Scholar]
- Woolf NJ, Eckenstein F, Butcher LL. Cholinergic projections from the basal forebrain to the frontal cortex: a combined fluorescent tracer and immunohistochemical analysis in the rat. Neurosci Lett. 1983;40:93–98. doi: 10.1016/0304-3940(83)90285-9. [DOI] [PubMed] [Google Scholar]
- Woolf NJ, Eckenstein F, Butcher LL. Cholinergic systems in the rat brain: I. projections to the limbic telencephalon. Brain Res Bull. 1984;13:751–784. doi: 10.1016/0361-9230(84)90236-3. [DOI] [PubMed] [Google Scholar]