Summary
The transcription factor MYB was recently proposed to be a promising oncogene candidate in salivary gland adenoid cystic carcinoma (ACC). However, the up-regulation of MYB in ACC could not be explained solely by deletion of its 3′ end. It is widely accepted that the promoter methylation status can regulate the transcription of genes, especially in human cancers. Therefore, it is important to know whether MYB promoter demethylation could explain the over-expression of MYB in ACC. By using the Methprimer program, we identified nine CpG islands in the promoter of MYB. All of these CpG islands were located within the −864 to +2,082 nt region relative to the transcription start site of MYB. We then used bisulfite genomic sequencing to evaluate the methylation levels of the CpG islands of MYB in 18 primary ACC tumors, 13 normal salivary gland tissues and nine cancer cell lines. Using cell lines, we also determined the relative MYB expression levels and correlated these with the methylation levels. With bisulfite genomic sequencing, we found no detectable methylation in the CpG islands of MYB in either ACC or normal salivary gland tissues. There was a variable degree of MYB expression in the cell lines tested, but none of these cell lines demonstrated promoter methylation. Promoter hypomethylation does not appear to explain the differential expression of MYB in ACC. An alternative mechanism needs to be proposed for the transcriptional control of MYB in ACC.
Keywords: adenoid cystic carcinoma, MYB, DNA methylation
Introduction
Salivary gland adenoid cystic carcinoma (ACC) is an aggressive, pernicious malignancy known for its persistent slow growth, proclivity for perineural invasion, high rate of recurrence, and formation of distant metastasis. While the five-year survival is favorable at approximately 75%, the long-term survival is poor, with only 39.6% of patients alive after 15 years 1. Histopathologically, ACC is characterized by three patterns: solid, cribriform, and tubular. All three patterns may be found within the same tumor. The solid pattern is rare but typically has a worse clinical course 1. Surgical therapy with adjuvant radiation remains the main treatment for ACC and typically provides excellent locoregional control. Chemotherapy has been considered as a treatment for ACC due to its frequent distant metastasis, though only limited response has been found thus far. The basis for these disappointing results is a lack of understanding of the basic biologic mechanisms involved in ACC carcinogenesis 2.
Several oncogene candidates have been proposed for ACC. Among them, the transcription factor MYB seems to be a very promising one. MYB was recently shown to be overexpressed in 100% of ACC samples tested by a novel mechanism of forming a t(6;9)(q22–23;p23–24) translocation, which fuses the MYB and NFIB genes 3. In an independent study with a larger primary ACC cohort, the overexpression of MYB was confirmed in the majority of salivary ACC cases. However, the formation of the fusion MYB-NFIB product was found in only 28% of primary and 35% of metastatic ACCs 4. Therefore, fusion MYB-NFIB formation must not be the only mechanism to explain the overexpression of MYB in ACC.
DNA methylation in the promoter region is a key mechanism in regulating gene transcription. Many tumor suppressor genes are found to be silenced by methylation, and oncogenes re-activated by demethylation in various human cancers 5. In fact, the methylation status of some genes can be used as biomarkers for tumorigenesis 6. In salivary gland ACC, promoter methylation has been reported in several candidate tumor suppressor genes 7–13. Furthermore, promoter methylation of these candidates was correlated with clinical and pathologic parameters, such as tumor histological grade or subtype 10,11,13. However, DNA methylation status in the MYB promoter in ACC has remained unexplored. In our current study, we investigated the methylation status of the MYB promoter region in ACC in hopes of elucidating an alternative mechanism to explain MYB overexpression in ACC.
Materials and methods
Clinical samples
Eighteen primary ACC tissues were obtained via the Johns Hopkins Pathology Department under the Johns Hopkins Institutional Review Board approved protocol IRB#92-07-21-01. Written informed consent was obtained from all those patients. Normal salivary gland samples were collected through a Johns Hopkins IRB-approved waste tissue protocol through the Department of Pathology that waived patient consent, and no patient identifiers were maintained. Tumor tissues from fresh surgical specimens were snap frozen in liquid nitrogen immediately after surgical resection and stored at −196°C until use. Tumor blocks with at least 80% tumor cells were used for DNA isolation. A cohort of normal parotid tissues obtained from separate patients with benign or inflammatory disease were also freshly frozen and utilized for this study as controls. The normal tissue used from benign tumors was selected to be distant from the tumor and confirmed histologically that no tumor or inflammation was found within the sample block before proceeding to DNA isolation.
DNA and RNA extraction
Samples were digested in 1% SDS and 50 μg/mL proteinase K (Boehringer Mannheim) at 48°C overnight, for removal of proteins bound to DNA. DNA was then purified by phenol-chloroform extraction and ethanol precipitation as described previously 14. The DNA was subsequently resuspended in 500 μL of LoTE (EDTA 2.5 mmol/L and Tris-HCl 10 mmol/L) and stored at −80°C until use.
Total cellular RNA was isolated using the RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions.
Cell strain and cell lines
The normal salivary gland epithelial cell strain HPAF1 was obtained from ATCC (with the kind permission of Dr. Dharam Chopra) and grown according to ATCC instructions. Cells were grown in collagen IV coated flasks (BD Bioscience, San Jose, CA). The medium used was Keratinocyte Basal Medium (Lonza, Walkersville, MD) supplemented with KGM Single Quots (Lonza).
The ACC83 cell line was a kind gift from Dr. Osamu Tetsu (personal communication), who performed the genotyping of the available ACC cell lines. While ACC83 is thought to be of epithelial origin, its genotype could not necessarily be proven to be of ACC origin, though it did not overlap with other known cell lines. ACC83 was cultured in RPMI1640 medium with 10% FBS and 1% penicillin/streptomycin in a 5% CO2, 37°C incubator.
Human head and neck squamous cell carcinoma (HNSCC) cell lines JHU-O22, and JHU-O28 were from the Johns Hopkins University Department of Otolaryngology-Head and Neck Surgery 15. UM22A and UM22B cell lines were provided by Ajay Verma (Merck & Co., North Wales, PA). FaDu cells originated from the American Type Culture Collection.
Three non-small cell lung cancer (NSCLC) cell lines, H1703, H1299 and H1437 were maintained in RPMI 1640 medium with 10% fetal bovine serum 16.
5-Aza dC/TSA treatment
In order to evaluate whether MYB could be under the control of promoter methylation, we treated the ACC83 cell line in triplicate with 5-aza-deoxycytidine (5-Aza dC) and trichostatin (TSA) as described previously 17. Briefly, cells were split to low density (5×105/100 mm dish) 24 hours before treatment. Stock solutions of 5-Aza dC (Sigma, St. Louis, MO) and TSA (Sigma, St. Louis, MO) were dissolved in 50% acetic acid and 100% ethanol, respectively. Cells were treated with 5 uM 5-Aza dC for 96 hours and 300 nmol/L TSA for the last 24 hours. Baseline expression was established by mock-treated cells with the same volume of acetic acid or ethanol in triplicate. Total cellular RNA was isolated using the RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions.
Bisulfite treatment and bisulfite genomic sequencing
The EpiTect Bisulfite kit (Qiagen, Valencia, CA) was used to convert unmethylated cytosines to uracils in DNA according to the manufacturer’s instructions. Converted DNA was stored at −80°C. Subsequently, bisulfite-treated DNA was amplified using primers designed by MethPrimer to span areas of CpG islands in the promoter of MYB. Primer sequences were designed to contain no CG dinucleotides and are listed in Table 1. Touch-down PCR was performed as follows: 95°C for 5 min, followed by cycles consisting of a 30 second denaturation step at 95°C, holding at an annealing temperature for 30 sec, initial extension at 72°C for 1 min, with a final extension at 72°C for 5 min. The annealing temperature was gradually decreased, e.g. two cycles at 64°C, 62°C, 60°C, 58°C and then 35 cycles at 56°C. The PCR products were purified using the QIAquick 96 PCR Purification Kit (Qiagen, Valencia, CA), according to the manufacturer’s instructions. Purified PCR products were then subjected to direct sequencing (Genewiz Inc, Germantown, MD).
Table 1.
Clinical and pathologic characteristics of patient populations
| Category | Subcategory | Normal | ACC |
|---|---|---|---|
| Patients, n | 13 | 18 | |
| Age, yr, median (range) | 54 (29–81) | 47 (26–68) | |
| Sex, n (%) | Male | 10 (76.9%) | 9 (50%) |
| Female | 3 (23.1%) | 9 (50%) | |
| Smoking status, n (%) | Never | 6 (46.2%) | 7 (38.9%) |
| Former | 4 (30.8%) | 5 (27.8%) | |
| Current | 3 (23.1%) | 5 (27.8%) | |
| Unknown | 0 (0%) | 1 (5.6%) | |
| Tumor location, n (%) | Parotid | - | 4 (22.2%) |
| Submandibular | - | 5 (27.8%) | |
| Minor | - | 9 (50%) | |
| Stage, n (%) | I | - | 6 (33.3%) |
| II | - | 3 (16.7%) | |
| III | - | 2 (11.1%) | |
| IV | - | 4 (22.2%) | |
| Unknown | - | 3 (16.7%) | |
| Perineural invasion, n (%) | Yes | - | 6 (33.3%) |
| No | - | 6 (33.3%) | |
| Unknown | - | 6 (33.3%) | |
| Local recurrence, n (%) | Yes | - | 6 (33.3%) |
| No | - | 11 (61.1%) | |
| Unknown | - | 1 (5.6%) | |
| Metastasis, n (%) | Yes | 4 (22.2%) |
ACC - Adenoid Cystic Carcinoma
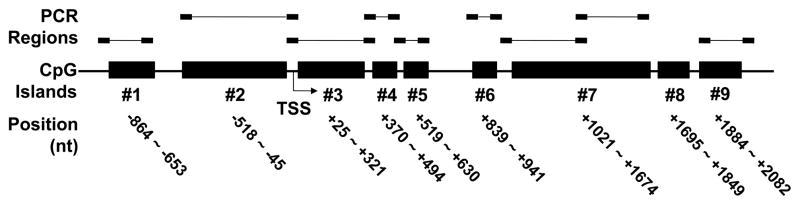
By using Methprimer 18, the criteria for CpG islands are: 1) an Observed/Expected CpG ratio over 0.6, 2) the percentage of G plus C over 50%, and 3) a window size of at least 100 bp. By these criteria, there are nine clustered CpG islands within −864 to +2,082 nt region relative to the transcription start site of MYB. We designed bisulfite sequencing primers in each of the CpG islands (Table 2). All islands, except for island number 8, were amplified successfully.
Table 2.
Bisulfite genomic sequencing primers for MYB
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| Isd 1a | TTTGTAGGTGTGTAATTTAGGGTG | TCTCTAATTTAACTTATTAATAACAAAAAA |
| Isd 2 | TAGAGGGTATAGTTGTAAATTTTGA | ACAAAAATACCATCAAACAAAAAACTT |
| Isd 3 | GGGGAGTGTTTAAAGTTTTTTGTT | AAATCTAACAACCCCTCAAATAATC |
| Isd 4 | GGATTATTTGAGGGGTTGTTAGATT | TCCCCATACAACTTACAAAATAAAC |
| Isd 5 | TAAGTTGTATGGGGATATATTTTTTTT | ACCATATACTCTACACTTCCAAATCC |
| Isd 6 | AAAGGGGTTTTTATTTTTGTAAATTGT | ATAAAAAAACTACACCCATCTTCCC |
| Isd 7-1 | GGGTGTAGTTTTTTTATATTTTTGGT | AATTTACTTTACCTCCATAAACTTC |
| Isd 7-2 | TTTATGGAGGTAAAGTAAATTTATT | AACTATCCTATAAAATCCTAAAAAC |
| Isd 9 | TTTTGAGGTTTTAGGTAGTTTAGAGG | TAACCTAATTAATATAATCCCTACC |
Isd 1, CpG island #1, the PCR amplified regions are in Figure 1.
Quantitative reverse transcription PCR (qRT-PCR)
Total RNA was measured and adjusted to the same amount for each cell line, and then cDNA synthesis was performed using the iScript™cDNA Synthesis Kit (Bio-Rad, Hercules, CA). The final cDNA products were used as the templates for subsequent qRT-PCR with SYBR green and primers designed spanning Exon 5 and Exon 6 of MYB. The MYB RT primers are: forward. 5′-GCT GGC TTT TGA AGA CTC CT -3′ and reverse, 5′-AAA TGA CTG TTC TTC TGG AAG -3′. PCR conditions were 95°C for 5min, 45 cycles of 95°C for 15 sec, and 64°C for 30 sec. β-actin was examined to ensure accurate relative quantitation in qRT-PCR. The β-actin RT primers are: forward, 5′-AGT CCT GTG GCA TCC ACG AAA CTA-3′ and reverse, 5′-ACT GTG TTG GCG TAC AGG TCT TTG-3′. PCR conditions were the same as MYB RT primers. All reactions were performed in triplicate with water controls, and relative quantity was calculated after normalizing to β-actin expression.
Results
Clinical samples
The demographic characteristics of the patients are summarized in Table 1. Patients with ACC ranged in age from 26 to 68 years, whereas the controls ranged in age from 29 to 81 years. The ACC groups had similar male and female patients, while the normal groups had more males than females: 76.9% vs 23.1%. The smoking status of patients with ACC and subject with controls were similar.
DNA methylation in CpG islands of MYB in primary ACCs, normal salivary glands, and cancer cell lines
In the UCSC human genome browser (http://genome.ucsc.edu/), Feb. 2009 (GRCh37/hg19) assembly, there are 33 splice variants for MYB. Twenty-nine of these transcripts spanned a ~38 kb region within chromosome 6 (chr6:135,502,453–135,540,313). We first used Methprimer 18 to screen for CpG islands in the MYB region plus a 5 kb region upstream of MYB. The default parameters for defining CpG islands are, 1) an Observed/Expected CpG ratio over 0.6, 2) the percentage of G plus C over 50%, and 3) a window size of at least 100 bp. Methprimer located a total of 11 CpG islands, the sizes of which ranged from 112 bp to 654 bp. Nine of total 11 CpG islands were clustered in the −864 ~ +2,082 nt region relative to the MYB transcription start site (TSS) (Figure 1). We then used Methprimer to design primers that did not contain CpG dinucleotide sequences (Figure 1, Table 2). The other two CpG islands were short (<150 bp) and located in the introns of MYB at +21,096 nt and +24,613 nt relative to TSS. Their short length and long distance from the TSS suggest that they might have little effect on the transcriptional control of MYB. Therefore, we did not analyze the methylation status of these two CpG islands.
Figure 1. CpG islands of MYB and regions analyzed by bisulfite genomic sequencing.
This cartoon shows the location of the 9 CpG islands of MYB (indicated by solid black boxes), which are numbered #1 to #9. Subregions analyzed by bisulfite genomic sequencing (not to scale) are indicated above each CpG island. Two subregions in CpG island #7 were analyzed. The transcription start site (TSS), was assigned as nt 0 in this cartoon. All positions of the CpG islands are relative to the TSS: the positions upstream are assigned negative values, and the positions downstream are positive. CpG islands were clustered in a region from −864~+2,082 nt region. The relative position of each CpG island is indicated below each CpG island.
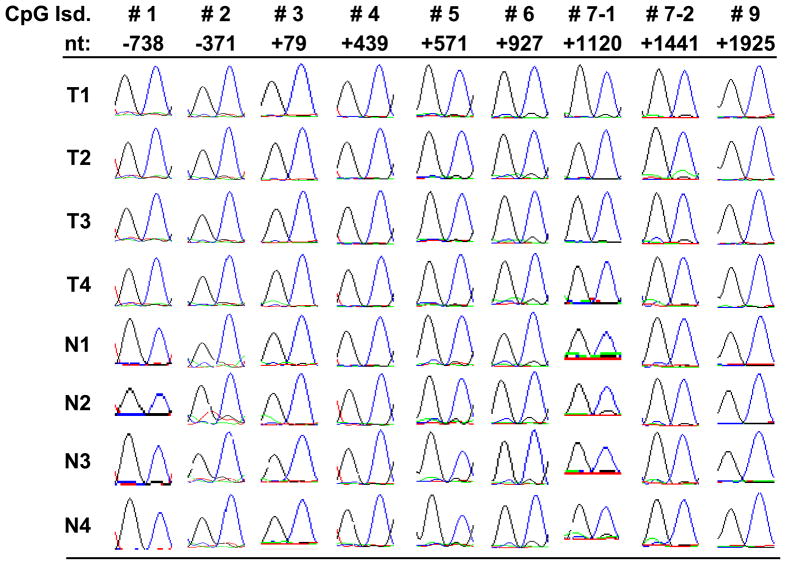
From bisulfite genomic sequencing, our results showed that none of the analyzed CpG islands demonstrated detectable methylation in the 18 primary salivary adenoid cystic carcinomas or in the 13 normal salivary gland tissues. The representative chromatography sequencing results are shown in Figure 2. The methylation status of all CpG sites were clearly unmethylated as demonstrated by singly predominant T peaks seen on sequencing chromatographs.
Figure 2. Representative sequencing results in ACC and normal salivary gland tissues.
We used bisulfite genomic sequencing to evaluate the DNA methylation levels of MYB CpG islands. Here we are showing the sequencing results (in chromatograph form) from randomly selected CpG sites within MYB CpG islands in four salivary ACC and four normal salivary gland tissues. Numbered CpG islands (CpG Isd) from #1 to #9 correspond to those shown in Figure 1. Nt indicates the position of selected CpG dinucleotides relative to the TSS of MYB. The blue peak stands for G; black, T; green, C; red, A. All CpG dinucleotides deomstrated dominant peaks of TG, which indicate unmethylated CpG. There was no discernible difference in methylation between ACC and normal salivary tissue.
We further analyzed one normal salivary gland epithelial cell strain, HPAF1, several head and neck squamous cell carcinoma (HNSCC), and non-small cell lung carcinoma (NSCLC) cell lines by bisulfite genomic sequencing. Similar to the primary tissue samples, we did not find detectable methylation in any of the CpG islands.
Expression levels of MYB
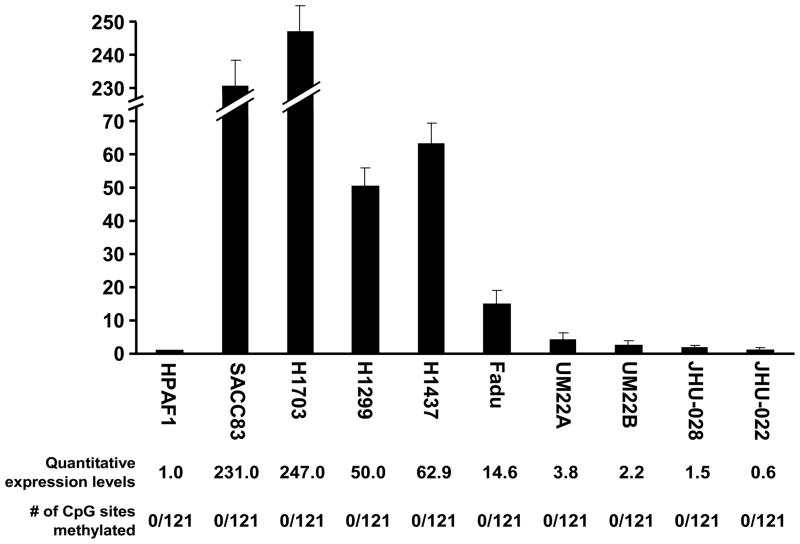
Since the all cell strain/lines were similarly unmethylated in most CpG islands in the MYB promoter, we then wanted to know the expression status of MYB. We quantified the exon 5/6 junction of MYB by qRT-PCR in those cell lines. The expression level of MYB in the normal cell strain HPAF1 was assigned as 1. The MYB levels in cell lines were quite variable despite having no detectable promoter methylation. The NSCLC cell lines overall showed higher expression (H1703 was 247.0, H1299 was 50.0, H1437 was 62.9) than the HNSCC cell lines (Fadu was 14.6, UM22A was 3.8, UM22B was 2.2, JHU-O28 was 1.5, and JHU-O22 was 0.6). SACC83 also had relatively high expression level of 231.0 (Figure 3). The methylation levels of the MYB promoter region were low in all examined cells lines: none of the cytosines were methylated in a total of 121 interpretable CpG sites (Figure 3). Our results confirm that the variable expression levels of MYB were not correlated with promoter methylation status.
Figure 3. Variable MYB expression in cancer cell lines.
We used qRT-PCR with SYBR green to investigate the expression levels of MYB in various head and neck squamous cell carcinoma cell lines (Fadu, UM22A, UM22B, JHU-O28 and JHU-O22), non-small cell lung cancer cell lines (H1703, H1299, and H1437) and SACC83. HPAF1, which is a normal salivary gland epithelial cell strain, was used as a control and assigned a relative value of “1” for its expression level of MYB. Other cell lines were then compared to HPAF1. The quantitative expression levels and the number of CpG sites methylated of MYB are shown below each indicated cell line. Error bars indicate the standard deviation. The methylation level of MYB was determined by counting the number of methylated CpG in total 121 interpretable CpG sites in all examined CpG islands.
Validation of the effects of 5-Aza dC/TSA treatment in MYB re-expression
5-Aza dC/TSA treatment can induce global demethylation in the genome, which may reactivate the transcription of genes that were silenced via promoter methylation. A cell line with methylated MYB would be very helpful to investigate whether MYB were subjected to methylation control. However, since none of the screened cancer cell lines, including SACC83, showed detectable methylation, we did a tentative 5-Aza dC/TSA treatment in the only available ACC cell line, SACC83. After 5-Aza dC/TSA treatment, the expression levels of MYB were actually decreased from 231.0 to 127.7. Therefore, there was no significant reactivation of MYB expression (data not shown).
Discussion
The transcription factor, MYB, plays a critical role in regulating stem and progenitor cells in the bone marrow, colonic crypts and neurogenic tissues 19. MYB has been implicated in various human malignancies including colon cancer, breast cancer, melanoma, pancreatic cancer, esophageal cancer, and more recently, salivary ACC 19,20. MYB target genes were summarized into three categories by Ramsay, et al, including housekeeping genes (MAT2A, GSTM1), differentiation genes (ELA2, MIM1, CD4 and PTCA), and genes with oncogenic potential (MYC, KIT, BCL2, GATA3) 20. MYB has been studied as a therapeutic target in colon cancer and breast cancer 21,22. For example, a DNA vaccine against MYB has been shown to suppress tumor growth in colon cancer 21.
The mechanism of MYB overexpression in salivary ACC is postulated to be related to creation of a fusion gene product formed by a chromosomal translocation, t(6;9)(q22–23;p23–24), which leads to a loss of the 3′ untranslated region (UTR) harboring miR-15a/16 and miR-150 binding sites 3. In a separate study with a larger cohort of salivary ACC’s, the overexpression of MYB was found in, but not limited to, the primary ACC with such chromosomal translocations 4. This discrepancy suggested that the overexpression of MYB in salivary ACC has an alternative regulatory mechanism in addition to miRNA. The mechanism of regulating MYB expression has been proposed to be through external signals, such as mitogens, signal pathway through extracellular signal-regulated kinases, and Wnt 20. Promoter methylation control is an important epigenetic mechanism for tumor suppressor genes or oncogenes; this has been frequently demonstrated in human cancers. For the MYB gene, its TSS is surrounded by nine clustered CpG islands. Therefore, we hypothesized that MYB transcription might be under the control of promoter methylation.
In this study, we investigated the methylation levels in almost all CpG islands overlapping the promoter region of MYB in primary ACC and normal salivary gland tissues. No detectable methylation was observed in either primary ACC or normal salivary gland tissue. While we would expect that promoter hypomethylation would lead to overexpression, this was not an ACC tumor specific event, but occurred in the normal samples as well. We further examined several HNSCC and NSCLC cell lines, none of which demonstrated detectable methylation in the MYB promoter. These cell lines also demonstrated variable expression levels, further suggesting that MYB is not under the influence of promoter methylation. Our final conclusion is that the variable expression levels of MYB could not be explained by differential methylation in the promoter.
There are very few studies investigating the relationship of MYB methylation and its transcription. One study in 1998 suggested MYB promoter methylation might be linked to the transformation in MNNG-exposed Bloom Syndrome (BS) B-lymphoblastoid cell populations, which have been reported to be highly susceptible to malignant transformation after carcinogen treatment 23. The authors showed unexpected hypermethylation of the c-myb locus in MNNG-BS cell populations compared to control cell lines. The DNA methylation analysis was performed by using methylation sensitive enzymes, HpaII and MspI followed by Southern blotting. However, only a limited number of CpG sites were studied, and MYB transcription levels were not determined. In our study, we were unable to identify a cell line with a methylated MYB promoter, nor were there an existing, reliable ACC cell line model; therefore, there was no way to investigate whether MYB could be re-expressed after inducing promoter demethylation with 5-Aza dC/TSA. This study could be carried out when appropriate cells lines are available.
Additionally, the MYB promoter harbors many GGA sequences which might potentially form G-quadruplex structures 24. A G-quadruplex is a four-stranded DNA or RNA structure stabilized by G-quartets, in which four guanines interact via Hoogsteen hydrogen bonds to form a planar ring 25,26. The biological implications of these sequences in promoter region are suggestive of their involvement in the regulation of MYB gene expression 27.
In summary, we found no methylation in the promoter regions of MYB in ACC or normal controls, and therefore, promoter demethylation does not appear to explain the over-expression of MYB in ACC tumors. An alternative mechanism needs to be proposed for the transcriptional control of MYB in ACC, as growing evidence shows that this may be an important event in ACC carcinogenesis.
Acknowledgments
This paper/analysis is based on a web database application provided by Research Information Technology Systems (RITS) - https://www.rits.onc.jhmi.edu/
Footnotes
Conflict of Interests Statement
None declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fordice J, Kershaw C, El-Naggar A, Goepfert H. Adenoid cystic carcinoma of the head and neck: predictors of morbidity and mortality. Arch Otolaryngol Head Neck Surg. 1999;125:149–152. doi: 10.1001/archotol.125.2.149. [DOI] [PubMed] [Google Scholar]
- 2.Mithani SK, Shao C, Tan M, et al. Mitochondrial mutations in adenoid cystic carcinoma of the salivary glands. PLoS One. 2009;4:e8493. doi: 10.1371/journal.pone.0008493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Persson M, Andren Y, Mark J, Horlings HM, Persson F, Stenman G. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci U S A. 2009;106:18740–18744. doi: 10.1073/pnas.0909114106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitani Y, Li J, Rao PH, et al. Comprehensive Analysis of the MYB-NFIB Gene Fusion in Salivary Adenoid Cystic Carcinoma: Incidence, Variability and Clinicopathological Significance. Clin Cancer Res. 2010 doi: 10.1158/1078-0432.CCR-10-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 6.Sidransky D. Emerging molecular markers of cancer. Nat Rev Cancer. 2002;2:210–219. doi: 10.1038/nrc755. [DOI] [PubMed] [Google Scholar]
- 7.Uchida D, Begum NM, Almofti A, Kawamata H, Yoshida H, Sato M. Frequent downregulation of 14-3-3 sigma protein and hypermethylation of 14-3-3 sigma gene in salivary gland adenoid cystic carcinoma. Br J Cancer. 2004;91:1131–1138. doi: 10.1038/sj.bjc.6602004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maruya S, Kurotaki H, Shimoyama N, Kaimori M, Shinkawa H, Yagihashi S. Expression of p16 protein and hypermethylation status of its promoter gene in adenoid cystic carcinoma of the head and neck. ORL J Otorhinolaryngol Relat Spec. 2003;65:26–32. doi: 10.1159/000068658. [DOI] [PubMed] [Google Scholar]
- 9.Maruya S, Kurotaki H, Wada R, Saku T, Shinkawa H, Yagihashi S. Promoter methylation and protein expression of the E-cadherin gene in the clinicopathologic assessment of adenoid cystic carcinoma. Mod Pathol. 2004;17:637–645. doi: 10.1038/modpathol.3800104. [DOI] [PubMed] [Google Scholar]
- 10.Li J, El-Naggar A, Mao L. Promoter methylation of p16INK4a, RASSF1A, and DAPK is frequent in salivary adenoid cystic carcinoma. Cancer. 2005;104:771–776. doi: 10.1002/cncr.21215. [DOI] [PubMed] [Google Scholar]
- 11.Zhang CY, Mao L, Li L, et al. Promoter methylation as a common mechanism for inactivating E-cadherin in human salivary gland adenoid cystic carcinoma. Cancer. 2007;110:87–95. doi: 10.1002/cncr.22758. [DOI] [PubMed] [Google Scholar]
- 12.Williams MD, Chakravarti N, Kies MS, et al. Implications of methylation patterns of cancer genes in salivary gland tumors. Clin Cancer Res. 2006;12:7353–7358. doi: 10.1158/1078-0432.CCR-06-1272. [DOI] [PubMed] [Google Scholar]
- 13.Durr ML, Mydlarz WK, Shao C, et al. Quantitative methylation profiles for multiple tumor suppressor gene promoters in salivary gland tumors. PLoS One. 2010;5:e10828. doi: 10.1371/journal.pone.0010828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoque MO, Lee CC, Cairns P, Schoenberg M, Sidransky D. Genome-wide genetic characterization of bladder cancer: a comparison of high-density single-nucleotide polymorphism arrays and PCR-based microsatellite analysis. Cancer Res. 2003;63:2216–2222. [PubMed] [Google Scholar]
- 15.Sun W, Liu Y, Glazer CA, et al. TKTL1 is activated by promoter hypomethylation and contributes to head and neck squamous cell carcinoma carcinogenesis through increased aerobic glycolysis and HIF1alpha stabilization. Clin Cancer Res. 2010;16:857–866. doi: 10.1158/1078-0432.CCR-09-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoque MO, Soria JC, Woo J, et al. Aquaporin 1 is overexpressed in lung cancer and stimulates NIH-3T3 cell proliferation and anchorage-independent growth. Am J Pathol. 2006;168:1345–1353. doi: 10.2353/ajpath.2006.050596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamashita K, Upadhyay S, Osada M, et al. Pharmacologic unmasking of epigenetically silenced tumor suppressor genes in esophageal squamous cell carcinoma. Cancer Cell. 2002;2:485–495. doi: 10.1016/s1535-6108(02)00215-5. [DOI] [PubMed] [Google Scholar]
- 18.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 19.Stenman G, Andersson MK, Andren Y. New tricks from an old oncogene: gene fusion and copy number alterations of MYB in human cancer. Cell Cycle. 2010;9:2986–2995. doi: 10.4161/cc.9.15.12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nat Rev Cancer. 2008;8:523–534. doi: 10.1038/nrc2439. [DOI] [PubMed] [Google Scholar]
- 21.Williams BB, Wall M, Miao RY, et al. Induction of T cell-mediated immunity using a c-Myb DNA vaccine in a mouse model of colon cancer. Cancer Immunol Immunother. 2008;57:1635–1645. doi: 10.1007/s00262-008-0497-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonda TJ, Leo P, Ramsay RG. Estrogen and MYB in breast cancer: potential for new therapies. Expert Opin Biol Ther. 2008;8:713–717. doi: 10.1517/14712598.8.6.713. [DOI] [PubMed] [Google Scholar]
- 23.Bhalla A, Sachdeva G, Bamezai R. T-cell receptor-gamma rearrangement and c-myb methylation in MNNG-exposed Bloom syndrome B-lymphoblastoid cells. Cancer Lett. 1998;126:1–6. doi: 10.1016/s0304-3835(97)00529-6. [DOI] [PubMed] [Google Scholar]
- 24.Palumbo SL, Memmott RM, Uribe DJ, Krotova-Khan Y, Hurley LH, Ebbinghaus SW. A novel G-quadruplex-forming GGA repeat region in the c-myb promoter is a critical regulator of promoter activity. Nucleic Acids Res. 2008;36:1755–1769. doi: 10.1093/nar/gkm1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc Natl Acad Sci U S A. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsumagari K, Qi L, Jackson K, et al. Epigenetics of a tandem DNA repeat: chromatin DNaseI sensitivity and opposite methylation changes in cancers. Nucleic Acids Res. 2008;36:2196–2207. doi: 10.1093/nar/gkn055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci U S A. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]