Abstract
Recent studies provide a glimpse of future potential therapeutic applications of custom-designed zinc finger proteins in achieving highly specific genomic manipulation. Custom-design of zinc finger proteins with tailor-made specificity is currently limited by the availability of information on recognition helices for all possible DNA targets. However, recent advances suggest that a combination of design and selection method is best suited to identify custom zinc finger DNA-binding proteins for known genome target sites. Design of functionally self-contained zinc finger proteins can be achieved by (a) modular protein engineering and (b) computational prediction. Here, we explore the novel functionality obtained by engineered zinc finger proteins and the computational approaches for prediction of recognition helices of zinc finger proteins that can raise our ability to re-program zinc finger proteins with desired novel DNA-binding specificities.
Keywords: Zinc finger proteins, Genome targeting
Introduction
Efficient design of biological systems has been the ‘holy grail’ of synthetic biology. With the advent of genomics over the past decade, the genome of many plants and mammalian genomes including the human genome have been completely sequenced and annotated. Genes and control elements are the two basic types of information coded by the genomes which are responsible for carrying out developmental and physiological function; and modulation of gene expression levels, respectively. Gene expression in all organisms is controlled by DNA–protein interactions, thus constituting the central theme in the area of molecular recognition. The easy availability of complete genomes has provided a novel platform for developing potent therapeutic strategies by modulation of the genetic information. A long term goal of molecular biologists has been the ability to manipulate the human genome at specific sites. Advancements in design and engineering of DNA-binding proteins have led to the use of zinc finger proteins (ZFP) in genome targeting by fusing them to other functional domains for several therapeutic purposes. Different approaches have been reported to create ZFP modules that are functionally self-contained and which can be recombined to provide novel DNA-binding sequence-specificity (Durai et al. 2005). Here, we discuss the essential elements of DNA-ZFP interactions for designing synthetic ZFP and strategies incorporating the computational and informatic expertise for prediction of DNA recognition by zinc finger proteins.
Zinc finger proteins (ZFP)
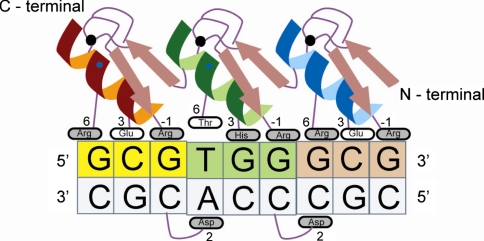
ZFP belong to the class of DNA binding proteins. They bind to DNA and sometimes even to RNA and other proteins, through a finger-shaped fold, which is stabilized by zinc ions coordinated to a combination of cysteine and histidine residues. Most of the cells nuclei carry ZFP where they regulate gene activity by binding to a specific nucleotide sequence and hence turning them on or off. The very structure of ZFP and their DNA binding domain make them the most efficient systems for development of artificial DNA binding proteins (Pabo et al. 2001; Beerli and Barbas 2002). Each ZFP motif consists of 30 amino acids, which fold into a ββα-structure, in which the Zn ion stabilizes the conserved Cys2His2 residues. Binding of ZFP to DNA is characterized by the insertion of an α-helix of ZFP into the major grove of DNA double helix (Pavletich and Pabo 1991). The recognition site for ZFP motifs primarily consists of a specific three nucleotide sequence (triplet) within a DNA substrate (Fig. 1). Their recognition specificity is due to the amino acids located at positions −1, +1, +2, +3, +4. +5 and +6 relative to the start of α- helix. Any change in these amino acids can bring about a change in its specificity. The remaining amino acids, which can remain unchanged, constitute the consensus backbone of ZFP. A number of such ZFP motifs can be linked in tandem to form ZFP, which can recognize longer DNA sequences (Fig. 1) (Liu et al. 1997; Beerli et al. 1998; Kim and Pabo 1998; Alwin et al. 2005). The designed ZFP technology provides a very powerful technique through which different functional domains like transcription activator (A), transcription repressor (R), methylases (M) and non specific FokI cleavage (N) can be fused to specific ZFP motifs to form zinc finger transcription activators (ZFA), zinc finger transcription repressors (ZFR), zinc finger methylases (ZFM) and zinc finger nucleases (ZFN), respectively (Durai et al. 2005).
Fig. 1.
Representation of a zinc finger protein and its specific target DNA site. a ββα—three zinc finger domain protein (Zif268) wrapping around the target DNA; the zinc finger amino acid side chains at the alpha helical positions −1, 3 and 6 makes sequence-specific hydrogen bond interactions with the nucleotides on the top DNA strand; a cross-strand interaction to the complementary DNA strand is also depicted (amino acid side chain at position 2 makes the cross-strand interaction)
Applications of engineered ZFP
Engineered ZFP fused to different functional domains have led to the advent of engineered chimeric DNA modifying enzyme technology. A number of such functional domains capable of acting directly on DNA, like DNA methyltransferase, integrase (Tan et al. 2004), resolvase and nuclease, fused with custom designed ZFP can be used to modify DNA at specific loci on the chromosomes (Urnov et al. 2005).
Zinc finger nucleases (ZFN) formed by fusing zinc finger motif with a nuclease domain, are used to create a double strand break at the specific genomic location. This finds application in the area of targeted gene therapy and for engineering plant and mammalian genomes. Over the last few years, ZFN have been instrumental in modifying the genomes of a number of higher organisms like Drosophila melanogaster, tobacco, zebra fish (Doyon et al. 2008) apart from various mammalian cell lines (Carroll 2008). Evaluation of ZFNs application in the potential treatment of HIV/AIDS is in the clinical trial stage. The proposed treatment consists of disruption of CCR5 gene in CD4+ human T cells using ZFNs. ZFNs are also being studied as a means to carry out homologous recombination to replace the gene copy with an extra chromosomal donor in patients suffering from monogenic disorders like X linked severe combined immune deficiency (SCID) (Urnov et al. 2005), sickle cell anemia (SCA), haemophilia etc. This correction of one particular faulty gene would be possible by employing ZFN at or near the site of mutation.
Another major area of application of ZFP is gene silencing using targeted DNA methylation. This uses the unique property of DNA methylases of methylating a particular base in a specific DNA sequence, thus making the gene (of which this DNA sequence is a part) non-functional (gene silencing). Fusion of the methylase domain of DNA methylases and ZFP (comprising of the designed DNA binding motif), thus, can be used to silence any gene. There are reports wherein such zinc finger-methylase fused proteins proposed to bind to the p53 site from p21WAF1/CIP1 gene have been used to methylate the target oligonucleotides in vitro within the gene sequence (Xu and Bestor 1997). Further, methylation of a large number of sequences using designer ZFP has also been reported in vivo. Though this area presents huge potential in genetic engineering, the toxicity of methylase domain has always acted to restrict the fast growth of this field.
Gene regulation is another major application field of ZFPs. A target gene can be regulated by engineering the proteins responsible for gene regulation e.g. designer transcription factors (an activator domain fused to custom designed ZFP). Such designer transcription factors find widespread use in drug therapy for formalizing genes as the drug target and in human therapeutics (Jamieson et al. 2003). Literature is filled with the explorations of the effects of such artificial transcription factors on promoter regions to regulate endogenous gene expression. Some of these include regulation of human erythropoietin gene (EPO1) (Zhang et al. 2000), the erbB-3 protoncogene as well as silencing of the multidrug resistance 1 gene (MDR1) (also known as ABCB1), the erbB-2 protoncogene (Beerli et al. 2000), the peroxisome proliferator activated receptor-γ gene (PPARγ) (Ren et al. 2002) and the checkpoint kinase 2 (CHK2). Regulation of specific gene expression in mammalian cell lines using ZFP artificial transcription factors were the first reported successful experiments (Beerli and Barbas 2002). Artificial transcription factors have also been formed by fusing ZFP to the independent repressor domains e.g. Krupel associated box (KRAB), ERF repressor domain (ERD), Mad SID or even parts of TATA box binding protein (TBP) (Li et al. 2008; Tian et al. 2009). Active research is also taking place in development of treatment of human peripheral arterial disease using vascular ZFPs. The specific ZFPs activate endothelial growth factor (VEGF) and thus stimulate vascular growth (Klug 2005). The creation of the above mentioned zinc finger chimeras, be it zinc finger- nucleases or zinc finger-transcription factors, depend on the reliable creation of zinc finger proteins (ZFP) that can specifically recognize a target sequence. Availability of highly reliable design and selection approaches to evolve novel ZFP will help realize the full potential of the above technology in therapeutic applications in the future.
Computational design of ZFP
Modular protein engineering has been successful for ZFP design or selection. These methods are, however, time consuming, expensive and have techniques not accessible to all laboratories (Beerli and Barbas 2002). For example, some of them involve several rounds of phage display, a laborious process (Greisman and Pabo 1997). In contrast, strategies encompassing computational and informatics elements can offer advantages of being faster, universally accessible and cheaper. ZFPs are attractive systems that benefit most by computational design, owing to the ready amount of experimental data available for them relative to other types of DNA binding proteins. The regularity in structures of zinc fingers which recognize and bind to nucleotide base triplets, where only the few amino acids at fixed positions interacting with the DNA strand vary, offer the possibility of devising and using a prediction algorithm. Computational design, in principle, can be divided into prediction tools of two types. The first is based on experimental data, which involve a statistical approach, to look for patterns and extrapolate to give a set of results that have higher relative likelihood than all the other possibilities in the library. Even in this category there are tools which are based purely on sequential data (in this case of the sequence of residues in the recognition helices and the DNA binding site sequence) and those based on structural data. Each prediction tool is limited to a specific context and is only as good as the variety and amount of the data used to train the algorithm. Even very good algorithms must rely on waiting for and incorporating sequences and structures into their datasets as they are discovered with time. The second type of computational tools strives to computationally simulate the fundamental science behind the phenomenon—in this case biochemical binding and specificity. It does good to remember that the science behind phenomena is based on models created by scientists, which labour under certain approximations and assumptions themselves. Furthermore, just like an experimental technique is more relevant when accessible to most laboratories, a prediction tool would be more relevant if it would run smoothly on PCs (or better, on a web platform) rather than needing more advanced computational resources like supercomputers. In order to cater to this utility, the algorithms are simplified using heuristic techniques and mathematical approximations. In many cases, exactitude isn’t even desired, because the task at hand doesn’t need it per se for fulfillment. One could take the example of the protein tertiary structure prediction problem. Computational chemistry involved is simplified to a model represented by balls and sticks, because a structural biologist probably doesn’t need to probe further to the quantum physics level. Therefore, no prediction tool is expected to give a single exact answer because the computer does not simulate the fundamental science behind the binding process perfectly, but instead only up to a level to perhaps cut down 90% of the absolutely arbitrary possibilities and to give a fairly good head-start. Finally, as the binding model of the zinc finger motif is more fundamentally understood, these types of prediction tools would have an opportunity to improve themselves. Hence in both cases, computational tools rely on experimental data and research.
Several databases pertaining to zinc finger proteins, storing various types of data have been developed in the past, giving computational biologists various approaches to design prediction tools. One such example is SysZNF which is an online, freely available database of C2H2 zinc finger genes. Each entry in the database is presented as a card containing details of gene physical location on the chromosome, gene model, Affymetrix gene expression probes, protein domain structures, homologs, PubMed references and links to other databases. A specialty about this database is that it arranges C2H2 gene information based on the physical location in the respective genome. It also has a graphical gene model/protein viewing system to visually represent the sequences and helps in determining ortholog and paralog relationships across species. A helpful feature is its allowing the user data access in complex ways using MySQL’s “select” statement (Ding et al. 2009). Another database ZNDB classifies the various zinc binding motifs according to the structure of the fold around the zinc atom (Krishna et al. 2003). The authors classify all known zinc binding structures into eight fold groups according to inferred homology between them. They also correlate this structural information with the function of the protein. Of these eight groups, the members of C2H2-like, treble clef and Zn2/Cys6 interact and complex with DNA. A third database, ZifDB (Fu et al. 2009) is a part of the Zinc Finger Consortium (http://www.zincfingers.org). It houses sequence, recognition helix and binding site information of 716 individual zinc fingers and engineered zinc-finger arrays. Each entry in the database additionally specifies at which position (in a 3-finger array) it binds most specifically, because the binding sites for the same 3 fingers would be different for proteins having a different order of the fingers, due to synergistic interactions between fingers. A program called ZifiT (Sander et al. 2007) is also interfaced to the database, which, given a nucleotide sequence, returns binding regions on the stretch, linking out to ZifDB’s entries of available proteins, and a set of Consortium reagents needed to engineer the protein. Prof. Akinori Sarai of Kyushu Institute of Technology, Japan, who has substantial work dedicated to understanding protein-DNA interactions in general has assisted in developing a useful collection of databases containing a wealth of structural information about protein-DNA complexes (http://gibk26.bse.kyutech.ac.jp/jouhou/jouhoubank.html). Finally, we come to a database developed by our own team, ZifBASE (http://web.iitd.ac.in/~sundar/zifbase) a manually curated repository of information containing various natural and engineered zinc finger proteins collected from the Protein Data Bank, Mod Base, Protein Model Portal and from literature (Jayakanthan et al. 2009). For natural zinc finger proteins, the name of the source organism for an entry is also stored. Each entry links out to popular databases available on the web. Searches can be made by giving a raw gene/DNA sequence as input, wherein if a zinc finger binding site is encountered, it is highlighted and information about the zinc finger protein provided. Each entry also lists the recognition helices (here the amino acid residues at positions −1, +1 to +6 relative to the alpha-helix) for each finger in the protein. The current version of ZifBASE contains 139 entries of which 89 are engineered ZFPs, containing 3-7F totaling to 296 fingers. There are 50 natural zinc finger protein entries ranging from 2-13F, totaling to 307 fingers.
The first attempts towards prediction were to find a recognition code, a simple table which matches the 64 nucleotide triplets to their corresponding amino acids at the positions −1, +3 and +6 in the helix (Desjarlais and Berg 1993; Choo and Klug 1994). Later it was found that zinc fingers have more complex interactions, which may not be modular and also involve interactions with the secondary strand. Like experimental techniques, computational tools also have a bias due to the underlying assumption about the binding model. Zinc Finger Tools (Mandell and Barbas 2006) is a web-based utility which employs a non-redundant set of 49 helices to target as many DNA triplets. This set includes zinc fingers that recognize 16 GNN, 15 ANN, 15 CNN and 3 TNN triplets. A search tool identifies all target sites (contiguous or separated) comprising of any of the 49 DNA triplets on both strands. Each site consists of results of a multi-target ELISA specificity assay for the appropriate zinc finger which predicts overall specificity of the ZFP containing these domains. The default backbone and linker provided can be altered by the user. It also includes a reverse engineering utility to predict the binding site for a ZFP of known sequence. Another tool available can conduct a search within a long DNA sequence for a shorter sequence that exactly or closely matches a specified sequence. The user can specify a number of allowed mismatches permitted in any position of the target site. A computational method based on data obtained from crystal structures of protein-DNA complexes to get a score indicating the compatibility between a DNA site and a protein sequence has also been reported (Mandel-Gutfreund et al. 2001). However, such an approach assumes that the set of contacts between protein and DNA doesn’t change upon substitution of the sequences. A simple linear predictor to discern DNA-binding proteins from non-binding ones by calculating their net charge, electric dipole moment and quadrupole moment has been reported (Ahmad and Sarai 2004). This method was able to achieve prediction accuracies of 82.6, 77.4 and 73.7% using these quantities, respectively. However, the cut-off values selected might be rendered inaccurate as more and more data becomes available. A method to predict target sites of transcription factors using a family-wise approach is also available (Kaplan et al. 2005). This involves collection of statistical information about the DNA-binding preferences of amino acid residues under different contexts from available information on transcription factors of a family binding to DNA. This information is used to predict context-specific DNA recognition preferences of other transcription factors from same family for which DNA targets have not been identified. In addition to information on zinc finger protein-DNA interactions, information about those protein-DNA pairs which bind weakly or not at all have also been included (Persikov et al. 2009). They demonstrate the potential of information on weakly or non-binding protein- DNA pairs to more accurately predict protein-DNA interactions. Their approach is based on support vector machines. An advantage is that it also affords the possibility to include higher order interactions not covered by the canonical model by using polynomial SVMs. Approaches assuming that binding preferences of one amino acid is independent of those in its neighborhood are not satisfactory because such dependence is observed. Hence, a new scheme was proposed to include this interdependence of amino acid interactions with DNA into a prediction algorithm (Cho et al. 2008). Here, to model this interdependence and for a given amino acid in a zinc finger, the binding probability to all 64 different triplets was calculated using a Hidden Markov Model. The top 5% of predicted sequences were fed into Match from the TRANSFAC database (http://www.gene-regulation.com/cgi-bin/pub/databases/transfac). The 2.1 kb upstream promoter regions of all human genes were scanned and the top 5% sequences obtained using which ZIFIBI was built. However, this study also leaves out structural parameters that determine protein-DNA interactions. A detailed review on several structure based strategies towards prediction of protein-DNA interactions is reported elsewhere (Sarai and Kono 2005). One for example, is purely based on the spatial probability of finding a certain amino acid in a particular point in space around the DNA and subsequently calculating the z-score, which gives a measure of specificity. More recently, a structure-based geometrical approach has been developed for prediction of C2H2 zinc finger binding specificity (Siggers and Honig 2007). The principle was to calculate a score based purely on the spatial distances between relevant amino acids residues and local nucleotides for two complexes called the interfacial alignment score (IAS). This score was found to give a good measure of specificity. In the template structure, the best conformations for the bases and residues were calculated by choosing the lowest-energy set using a Monte Carlo procedure. The minimisation was performed by the minimize program in the TINKER package. Our lab has developed a web tool, Zif-Predict, which is a sequence-based prediction tool for zinc-finger proteins, which, given a nucleotide sequence, finds the possible binding sites of zinc-fingers and returns the amino acid sequences of their recognition helices (in this case, amino acids at position −1, +1 to +6 in the alpha helix of the zinc finger motif). It can also predict the sequences of zinc finger nucleases which are useful in genome editing. It can account for synergistic interactions between consecutive fingers interacting with a stretch of several nucleotide triplets, and the interface also offers the user the choice to predict based on an assumption of absence of these interactions (i.e. modular design). The algorithm is trained purely by sequential data and that too, only of the 7-residue recognition helices and their matching nucleotide triplets. These sequences are primarily derived from the above mentioned Zif-BASE. The prediction algorithm is based on artificial neural networks. The network consists of layers, characteristic to neural networks, in this case: an input layer, two hidden layers and a single output neuron. The algorithm makes use of adaptive control wherein its parameters keep improving with every iteration according to a learning rate. The algorithm can become even more meaningful if the data on which it was trained included the sequences of recognition helices for all 64 nucleotide triplet possibilities. Hence, as more and more sequences of zinc finger proteins and the DNA sequences that they bind to are unearthed, the algorithm can be improved. Zif Predict is available as a software tool on the web platform at http://web.iitd.ac.in/~sundar/zifpredict (Molparia et al. 2010).
Theoretically a zinc finger protein with the backbone of Zif268 and with specific amino acids residues at the recognition helix will bind to its respective binding sites. What is important is that the conserved hydrophobic residues and the residues interacting with the zinc ion remain intact. Zif268 is a three fingered protein and hence binds to a 9-bp stretch of DNA. An experimental method to determine the sequence of the engineered zinc-finger protein based on Zif268 is the inspiration for our approach towards a computational prediction system (Greisman and Pabo 1997). In said experimental technique, one begins with a library of zinc finger proteins, which is fully randomized at the base contacting residues. One finger is selected at a time, while keeping the other two same as in Zif268. Initially a library containing randomizations only at the first finger is created. When the most competent structure for the first finger is selected by phage display, the second finger is randomized and so on. In our approach, we begin with the protein-DNA crystal structure of Zif268 with its natural binding site (PDB structure, 1AAY). The user submits a nucleotide sequence of arbitrary length. The program walks a 9-bp window across it. Using computational tools available (such as DeepView, SwissProt; http://spdbv.vital-it.ch/) one can very easily mutate the structure at specific residues and obtain resulting mutant structures. So initially the first three nucleotides of the DNA sequence in the crystal structure are replaced with the first triplet of the 9-bp window. The first finger is mutated at seven key positions in the α-helix (−1, +1 to +6) which interact with the DNA. For each permutation, the resulting structure of the mutant complex is obtained. Then one calculates a parameter or a combination of these which characterises the number of hydrogen bonds, hydrogen bond distances between residues and bases, the interaction energy of the complex and so on. This parametric function is still being investigated by our team. For sensitive binding, this parameter is expected to fall in a favourable range of values (determined from a set of 31 available zinc finger protein-DNA PDB structures), and its value is the basis for ranking it against other possibilities. Even if the absolute value of the parameter is inaccurate, if the values are reliable enough to compare possibilities with each other and eliminate the ones falling blatantly out of range, it would suffice. After the best possibility for the first finger is found, corresponding changes are made in the original structure, and now the program moves to the second finger, and so on. Computationally, the program must deal with 207 possibilities for the sequences for each finger, but this number can be reduced in various ways. First, by eliminating those amino acids which have never been found in zinc finger proteins. Then, by pre-filtering those combinations which would disrupt the structure of the α-helix and hence the integrity of the zinc finger protein. Also the fact that one is dealing with a narrow context of proteins based on the backbone of Zif268 (C2H2 type, which bind according to canonical model) would significantly simplify the problem. We have found some insights thus far which can help exactly there: two-thirds of hydrogen bonds with bases are involved in bidentate and complex interactions, and provide the greatest specificity (Suzuki 1994; Mandelgutfreund et al. 1995). The hydrogen bond distribution clearly demonstrates that particular amino acid–base pairs are favoured: arginine, lysine, serine and histidine with guanine; and asparagine and glutamine with adenine. Furthermore, a technique to predict DNA binding property by using theoretically calculated values of dipole moment from protein structure has been developed (Ahmad and Sarai 2004). This can function as a preliminary low-stringency filter to eliminate a large number of initial possibilities. This approach on the whole would ensure that the prediction system relies purely on the structural context.
Conclusion
Exploiting biology to our requirements has been the major part of synthetic biology research. Methods exploiting the various essential properties of ZFP including positively charged electrostatic patches, DNA-binding structural motifs, sequence composition, solvent accessibility and secondary structure, net charge, dipole and quadrapole moments, and size of largest positive surface patch and amino acid composition, etc. need to be unraveled and adopted to evolve an accurate prediction protocol, which could be routinely used by genome scientists before they proceed with designing proteins to target a specific site on a genome. It is envisioned that in the long run, computational models and tools to re-program DNA-binding specificity in ZFP will have great promise to engineer novel ZFP for several therapeutic applications.
Acknowledgments
KC and KG were recipients of the Summer Undergraduate Research Award (SURA) from IIT Delhi and DS was supported by a grant from the Department of Biotechnology (DBT), Govt. of India under the IYBA scheme. The authors of this paper would like to acknowledge the Bioinformatics Facility at the DBT-funded Distributed Information Sub Centre at IIT Delhi.
Abbreviations
- ZFP
Zinc finger proteins
Footnotes
A. Grover, A. Pande, K. Choudhary, K. Gupta authors contributed equally to this work.
References
- Ahmad S, Sarai A. Moment-based prediction of DNA-binding proteins. J Mol Biol. 2004;341:65–71. doi: 10.1016/j.jmb.2004.05.058. [DOI] [PubMed] [Google Scholar]
- Alwin S, Gere MB, Guhl E, Effertz K, Barbas CF, III, Segal DJ, Weitzman MD, Cathomen T. Custom zinc-finger nucleases for use in human cells. Mol Ther. 2005;12:610–617. doi: 10.1016/j.ymthe.2005.06.094. [DOI] [PubMed] [Google Scholar]
- Beerli RR, Barbas CF., III Engineering polydactyl zinc-finger transcription factors. Nat Biotechnol. 2002;20:135–141. doi: 10.1038/nbt0202-135. [DOI] [PubMed] [Google Scholar]
- Beerli RR, Segal DJ, Dreier B, Barbas CF. Toward controlling gene expression at will: Specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc Natl Acad Sci USA. 1998;95:14628–14633. doi: 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli RR, Dreier B, Barbas CF., III Positive and negative regulation of endogenous genes by designed transcription factors. Proc Natl Acad Sci USA. 2000;97:1495–1500. doi: 10.1073/pnas.040552697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D. Progress and prospects: zinc-finger nucleases as gene therapy agents. Gene Ther. 2008;15:1463–1468. doi: 10.1038/gt.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SY, Chung M, Park M, Park S, Lee YS. ZIFIBI: prediction of DNA binding sites for zinc finger proteins. Biochem Biophys Res Commun. 2008;369:845–848. doi: 10.1016/j.bbrc.2008.02.106. [DOI] [PubMed] [Google Scholar]
- Choo Y, Klug A. Selection of DNA binding sites for zinc fingers using rationally randomized DNA reveals coded interactions. Proc Natl Acad Sci USA. 1994;91:11168–11172. doi: 10.1073/pnas.91.23.11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjarlais JR, Berg JM. Use of a zinc-finger consensus sequence framework and specificity rules to design specific DNA binding proteins. Proc Natl Acad Sci USA. 1993;90:2256–2260. doi: 10.1073/pnas.90.6.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding G, Lorenz P, Kreutzer M, Li Y, Thiesen HJ. SysZNF: the C2H2 zinc finger gene database. Nucleic Acids Res. 2009;37:D267–D273. doi: 10.1093/nar/gkn782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Amacher SL. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durai S, Mani M, Kandavelou K, Wu J, Porteus MH, Chandrasegaran S. Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005;33:5978–5990. doi: 10.1093/nar/gki912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu F, Sander JD, Maeder M, Thibodeau-Beganny S, Joung JK, Dobbs D, Miller L, Voytas DF. Zinc Finger Database (ZiFDB): a repository for information on C2H2 zinc fingers and engineered zinc-finger arrays. Nucleic Acids Res. 2009;37:D279–D283. doi: 10.1093/nar/gkn606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greisman HA, Pabo CO. A general strategy for selecting high-affinity zinc finger proteins for diverse DNA target sites. Science. 1997;275:657–661. doi: 10.1126/science.275.5300.657. [DOI] [PubMed] [Google Scholar]
- Jamieson AC, Miller JC, Pabo CO. Drug discovery with engineered zinc-finger proteins. Nat Rev Drug Discov. 2003;2:361–368. doi: 10.1038/nrd1087. [DOI] [PubMed] [Google Scholar]
- Jayakanthan M, Muthukumaran J, Chandrasekar S, Chawla K, Punetha A, Sundar D. ZifBASE: a database of zinc finger proteins and associated resources. Bmc Genomics. 2009;10:421. doi: 10.1186/1471-2164-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan T, Friedman N, Margalit H. Ab initio prediction of transcription factor targets using structural knowledge. PLoS Comput Biol. 2005;1:5–13. doi: 10.1371/journal.pcbi.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Pabo CO. Getting a handhold on DNA: design of poly-zinc finger proteins with femtomolar dissociation constants. Proc Natl Acad Sci USA. 1998;95:2812–2817. doi: 10.1073/pnas.95.6.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug A. Towards therapeutic applications of engineered zinc finger proteins. FEBS Lett. 2005;579:892–894. doi: 10.1016/j.febslet.2004.10.104. [DOI] [PubMed] [Google Scholar]
- Krishna SS, Majumdar I, Grishin NV. Structural classification of zinc fingers. Nucleic Acids Res. 2003;31:532–550. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Yang D, Bai Y, Mo X, Huang W, Yuan W, Yin Z, Deng Y, Murashko O, Wang Y, Fan X, Zhu C, Ocorr K, Bodmer R, Wu X. ZNF418, a novel human KRAB/C2H2 zinc finger protein, suppresses MAPK signaling pathway. Mol Cell Biochem. 2008;310:141–151. doi: 10.1007/s11010-007-9674-4. [DOI] [PubMed] [Google Scholar]
- Liu Q, Segal DJ, Ghiara JB, Barbas CF., III Design of polydactyl zinc-finger proteins for unique addressing within complex genomes. Proc Natl Acad Sci USA. 1997;94:5525–5530. doi: 10.1073/pnas.94.11.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelgutfreund Y, Schueler O, Margalit H. Comprehensive analysis of hydrogen-bonds in regulatory protein DNA-complexes—in search of common principles. J Mol Biol. 1995;253:370–382. doi: 10.1006/jmbi.1995.0559. [DOI] [PubMed] [Google Scholar]
- Mandel-Gutfreund Y, Baron A, Margalit H. A structure-based approach for prediction of protein binding sites in gene upstream regions. Pac Symp Biocomput. 2001;6:139–150. doi: 10.1142/9789814447362_0015. [DOI] [PubMed] [Google Scholar]
- Mandell JG, Barbas CF., III Zinc Finger Tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 2006;34:W516–W523. doi: 10.1093/nar/gkl209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molparia B, Goyal K, Sarkar A, Kumar S, Sundar D. ZiF-Predict: a web tool for predicting DNA-binding specificity in C2H2 zinc finger proteins. Genomics Proteomics Bioinf. 2010;8:122–126. doi: 10.1016/S1672-0229(10)60013-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo CO, Peisach E, Grant RA. Design and selection of novel Cys(2)His(2) zinc finger proteins. Annu Rev Biochem. 2001;70:313–340. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Persikov AV, Osada R, Singh M. Predicting DNA recognition by Cys(2) His(2) zinc finger proteins. Bioinformatics. 2009;25:22–29. doi: 10.1093/bioinformatics/btn580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren DL, Collingwood TN, Rebar EJ, Wolffe AP, Camp HS. PPAR gamma knockdown by engineered transcription factors: exogenous PPAR gamma 2 but not PPAR gamma 1 reactivates adipogenesis. Genes Dev. 2002;16:27–32. doi: 10.1101/gad.953802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Zaback P, Joung JK, Voytas DF, Dobbs D. Zinc finger targeter (ZiFiT): an engineered zinc finger/target site design tool. Nucleic Acids Res. 2007;35:W599–W605. doi: 10.1093/nar/gkm349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarai A, Kono H. PROTEIN-DNA recognition patterns and predictions. Annu Rev Biophys Biomol Struct. 2005;34:379–398. doi: 10.1146/annurev.biophys.34.040204.144537. [DOI] [PubMed] [Google Scholar]
- Siggers TW, Honig B. Structure-based prediction of C2H2 zinc-finger binding specificity: sensitivity to docking geometry. Nucleic Acids Res. 2007;35:1085–1097. doi: 10.1093/nar/gkl1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M. A framework for the DNA-protein recognition code of the probe helix in transcription factors—the chemical and stereochemical rules. Structure. 1994;2:317–326. doi: 10.1016/S0969-2126(00)00033-2. [DOI] [PubMed] [Google Scholar]
- Tan WJ, Zhu K, Segal DJ, Barbas CF, Chow SA. Fusion proteins consisting of human immunodeficiency virus type 1 integrase and the designed polydactyl zinc finger protein E2C direct integration of viral DNA into specific sites. J Virol. 2004;78:1301–1313. doi: 10.1128/JVI.78.3.1301-1313.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Xing GC, Xie P, Lu KF, Nie J, Wang J, Li L, Gao M, Zhang LQ, He FC. KRAB-type zinc-finger protein Apak specifically regulates p53-dependent apoptosis. Nat Cell Biol. 2009;11:580–591. doi: 10.1038/ncb1864. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Xu GL, Bestor TH. Cytosine methylation targetted to pre-determined sequences. Nat Genet. 1997;17:376–378. doi: 10.1038/ng1297-376. [DOI] [PubMed] [Google Scholar]
- Zhang L, Spratt SK, Liu Q, Johnstone B, Qi H, Raschke EE, Jamieson AC, Rebar EJ, Wolffe AP, Case CC. Synthetic zinc finger transcription factor action at an endogenous chromosomal site. Activation of the human erythropoietin gene. J Biol Chem. 2000;275:33850–33860. doi: 10.1074/jbc.M005341200. [DOI] [PubMed] [Google Scholar]