Abstract
In the backdrop of accelerated efforts for creating synthetic organisms, the nature and scope of an ideal programming language for scripting synthetic organism in-silico has been receiving increasing attention. A few programming languages for synthetic biology capable of defining, constructing, networking, editing and delivering genome scale models of cellular processes have been recently attempted. All these represent important points in a spectrum of possibilities. This paper introduces Kera, a state of the art programming language for synthetic biology which is arguably ahead of similar languages or tools such as GEC, Antimony and GenoCAD. Kera is a full-fledged object oriented programming language which is tempered by biopart rule library named Samhita which captures the knowledge regarding the interaction of genome components and catalytic molecules. Prominent feature of the language are demonstrated through a toy example and the road map for the future development of Kera is also presented.
Keywords: Synthetic biology, Programming language, Design and construction of organisms, Computational biology
Introduction
The paradigm shift from traditional biology to the new biology can be most succinctly referred as an evolution from low throughput analysis to high throughput analysis and synthesis. Computational means to put forward strong hypotheses for physically feasible genome modifications is increasingly being demanded in the space of synthetic biology (Endler et al. 2009). This process is partially enabled by GUI based web servers such as GenoCAD (Pedersen et al. 2009); Tinkercell (Chandran et al. 2009) and Biojade (Goler 2004) etc. However, for a greater control over modeling leading to synthesis, programming languages are currently being developed. Antimony (Smith et al. 2009) and GEC (Pedersen et al. 2009), developed by Microsoft, are prominent examples.
In our opinion, the language that will survive the rigors of biological engineering shall be the one that can handle widest variety of contextual interactions. Versatile programming constructs can make programming languages a powerful tool at the hand of synthetic biologists. Considering that the rules of biological composition are unclear, we have attempted to develop a new language that covers functionalities of existing languages and adds a rule library named samhita.
In this paper we present a brief overview of the field of computational synthetic biology, including programming languages. We point out some lacunae in the present languages and describe the basic structure of our new language Kera and the associated evolving rule libraries named Samhita and demonstrate its flavor by a few toy examples.
Kera in Sanskrit means coconut. The most innocent reason for choosing this name is that our state is known for the abundance of coconut trees. From a scientific standpoint coconut represents one of the finest biological engineering of nature due to its multi level and modular natural construction.
A brief introduction to computational synthetic biology
Computational support to synthetic biology first emerged in the form of repository of biological parts. These repositories which have been evolving for more than a decade by now, consists of sequences of promoter, ribosome binding sites, coding sequences and terminators. The Biobrick registry of standard biological parts (http://bbf.openwetware.org/) is one of the most widely used repository in the field. Scientist have attempted to build composite devices using these building blocks (Young and Alper 2010). These building blocks along with associated information enable stitching of parts into composite devices. One of the earliest successful demonstration of synthesizing genetic devices was provided by Elowitz and Leibler, Gardner who constructed a repressilator with a stand-alone capability in the host that housed the constructs (Elowitz and Leibler 2000; Gardner et al. 2000). This involved building in silico model of the circuit, determining the right parameters, modifying the binding co-efficient of molecules experimentally and generating oscillations in vivo with an accuracy of a computational model (Bashor et al. 2010). Following the successful implementation of these genetic devices, a new hope of designing genetic circuits ground up emerged in the community. Bacterial toggle switch and oscillator, programmed pattern formation, bacteria designed to detect cancer, Artemisinin produced in yeast, RNA- based devices (Purnick and Weiss 2009) are some of the significant achievements of evolving synthetic biology the community. All these efforts involved some amount of computational work in the form of databases, sequence analysis and molecular interaction modeling (Khalil and Collins 2010).
A review of the existing synthetic biology programming languages
With the popularity of Biobrick repository and the emerging non-biobrick repositories, there is a concomitant increase in complimentary computational tools to support mathematical and computational modeling of parts devices and circuits. Systems Biology Markup Language (SBML, Hucka et al. 2003) is a community standard of exchanging information on part and interactions. Biojade was the first computational tool designed to meet the emerging requirements of synthetic community and was lanched at the synthetic biology meeting 1.0. Subsequently more tools were developed that offer more functionalities and user friendliness (Goler 2004). While tools are emerging recently, new approaches have emerged. These are in the form of full-fledged or non-full-fledged programming languages, aimed at giving finer control over representation and modeling. GenoCAD is a web based tool with some features of programming language (Czar et al. 2009). GenoCAD has a GUI and a built-in database containing Biobricks. GenoCAD provides some rules for assembling systems S. cerevisiae and E. coli. It uses mass action rates for simulation (Pedersen et al. 2009).
Antimony (Smith et al. 2009) is closer to a programming language than GenoCAD and built on the concept of modularity (Cai et al. 2007). User can define models, store, and simulate antimony code of the model. Antimony uses polymerase per second (PoPS) and ribosomes per second (RiPS) rates for simulating instead of mass action rates in GenoCAD.
The latest in the series of programming language is GEC (Genetic Engineering of living Cells) released by Microsoft foundation. GEC is a rule based language. It has associated part database and reaction database. GEC uses Prolog based engine to choose compatible parts from database. In its current form, GEC comes with some performance issues. Example: retrieving data from database is time consuming. GEC does not give condition for optimal expression and GEC data model is not robust yet. Some researchers in the field feels that, model storing and distribution of genetic material is losing its relevance and new computational strategies are to be developed to correlate sequence and functions through the programs and models (Clancy and Voigt 2010).
In the next section, we describe our effort to develop another programming language for synthetic biology, which address many of these lacunae pointed out above and also tries to feature a powerful rule library named Samhita integrated into language.
The Kera programming language
Kera is an object oriented, Knowledge based programming language for synthetic biology which enables users to create, edit, combine, and display in-silico simulation run of experimental synthetic genomes. It provides C like data types and control structures, and is more object oriented than its counter parts, thus delivering more of the traditional advantages of object oriented thinking, viz, clarity in managing complex thoughts. We feel that this aspect is crucial as the language evolves to cover multiple levels within organisms and absorb the technological developments in biology through synthesis.
In the v1.0 of Kera we have focused on structure and functionality rather than language efficiency. Kera 1.0 is based on the C# platform. We plan to make Kera standalone only after it matures, attracts a critical mass of users and proves its relevance sufficiently. For now the intent is to capture biological knowledgebase into programming language formats.
Given that biologists will be the end users of this language, not computer scientists or technologists, we have sacrificed brevity for readability, aimed at biologists. However, we have provided ‘cryptic’ and ‘verbose’ options of the language for those who might think otherwise. To aid rapid prototype development and experimentation, Kera works in the interpreter mode by default. An efficient compilation facility is also available.
Kera has three basic classes to capture genomic knowledge- the cell, the genome and the proteins. The cell class carries with itself a large user-edited library named Samhita. Samhita will check compatibility of bioparts and suggests optimal bioparts for required design. Samhita has a rule library and a part library which are dynamic, with permission to each user to add non-persistent parts or rules to Samhita.
The syntax of Kera Language
Kera program starts with a definition of cell and compartment of interest. In the following toy Kera code related to an E. coli cell, a cell object is created and stored in X. The compartment of interest can be nucleus, mitochondria, cytoplasm and so on.
Cell X = new Cell(“E. Coli”);
X.compartment(“nucleus”);
The above Kera code works with one gene at a time selected by the ‘selectgene’ method in the cell object. Once the gene is selected, various actions can be performed on the gene upstream and downstream. For instance, the promoter can be replaced with another one of our choice as in the following example.
X.selectgene(“g1”);
X.promoter(“replace”, “ACTGCTA”);
The replacement invokes the Samhita library and would return a compatibility indicator, which can be verified using conditional statements of Kera for instance. In addition to the program level checking, Kera also supports a type ahead dropdown menu of rule-confirming options. The following Kera code attempts a replacement with “ACTGCTA” and if it fails, attempts another replacement with “TTGATGCTA”.
boolean compatibility;
compatibility = X.promoter(“replace”, “ACTGCTA”);
if compatibility = 0
{
X.promoter(“replace”, “TTGATGCTA”);
}
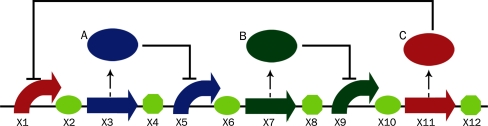
Let’s consider a simple synthetic genetic regulatory network—repressilator [] which was implemented in Escherichia coli. It acts as an electrical oscillator system with fixed time periods. The repressilator has three genes which are connected in a feedback loop and each gene represses the other in the loop. The network diagram is given bellow (Fig. 1).
Fig. 1.
The repressilator
For simulating model of a repressilator in Kera, we begin by creating a E. Coli cell variable X and setting the desired compartment as nucleus defined by the following Kera code
cell X = new cell(“E. Coli”);
X.compartment(“nucleus”);
Each segment including promoter, RBS and terminator in the network are defined by the following code, which related to the first part of the repressilator, consisting of X1, X2, X3, and X4. The remaining part of the network is defined in similar manner
X.seg(1) = new prom();
X.seg(2) = new rbs();
X.seg(3) = new pcr();
X.seg(4) = new ter();
Proteins produced by each of the three coding sequences are programmed as follows:
Protein A = new protein ();
X.seg(3).codes(A);
Protein B = new protein ();
X.seg(7).codes(B);
Protein C = new protein();
X.seg(11).codes(B);
Now the regulatory information can be coded as follows
X.seg(1).regulates(C,−1);
X.seg(5).regulates(A,−1);
X.seg(9).regulates(B,−1);
This completes the formal representation of a repressilator. Now to display the synthetic sequence, the following code can be added:
X.display();
Kera converts the defined model into corresponding sequence and the user can save this file using the code:
X.writedna(“filename.txt”);
Samhita–Biorule library
Samhita bio-rule library is the most value adding feature of Kera language. It attempts to distill coarse biological rules that apply to the bioparts that Kera aims to alter for manipulation through the programming language. Consequently the rule library is based on the part library. The part library is a relational database, consisting of the part id, part type and the sequence. The part ids are adopted from Biobrick ids. However, the user can define addition to the part library which will be non-persisting. The types included in the Kera are promoter, ribosome binding site, coding sequence, and terminator. Sample part library extract is shown in the Table 1.
Table 1.
An extract from part database
| Part id | Part type | Sequence |
|---|---|---|
| BBa_I760005 | Promoter | atgacaaaattgtcat |
| BBa_J63003 | Ribosome binding site | cccgccgccaccatggag |
| BBa_B0015 | Terminator | ccaggcatcaaataaaacgaaaggctcagtcgaaagactgggcctttcgttttatctgttgtttgtcggtgaacgctctctactagagtcacactggctcaccttcgggtgggcctttctgcgtttata |
| BBa_G00100 | Primer | tgccacctgacgtctaagaa |
| BBa_J18922 | Linker | ggtagcggcagcggtagcggtagcggcagc |
| ORF83P1 | Promoter | cgagccgctttccatatctattaacgcataaaaaactctgctggcattcacaaatgcgcaggggtaaaacgtttcctgtagcaccgtgagttatactttgt |
| aceB | Promoter | caacaagttatcaagtatttttaattaaaatggaaattgtttttgattttgcattttaaatgagtagtcttagttgtgctgaacgaaaagagcacaacgat |
The rule library has a relational database with an onto relational with the part library. This would mean that every rule in the rule library, which would refer one or more parts in part library, but not necessarily vise versa. The rule library database consists of the rule id, part id, and the regulatory effect. One part may have more than one known regulatory effect. For the purpose of database normalization, these shall be encoded as separate rules. An example of Samhita rule database is shown in Table 2.
Table 2.
The samhita database
| Part | Regulation | Kera code |
|---|---|---|
| BBa_I1051 | LuxR (positive regulation) | BBa_I1051.regulates (LuxR,1) |
| BBa_I1051 | cI dimer (negative regulation) | BBa_I1051.regulates (cI dimer,−1) |
| BBa_I751501 | AHL (positive regulation) | BBa_I751501.regulates (AHL,1) |
| BBa_I751501 | Lambda cI (negative regulation) | BBa_I751501.regulates (Lambda cI,−1) |
| BBa_I739104 | P22 cII (negative regulation) | BBa_I739104.regulates (P22 cII,−1) |
In the rule library also permits non-persistent and a non-contradictory addition of the rule by users as mentioned in the description of the programming language. The methods of various classes of Kera language will call the rule library and return compatibility status as Boolean variables. The library can be used to choose parts based on requirements. As the programming language is capable of invoking the rules during runtime, part addition can be iteratively checked and designed at runtime. The power of the rule library depends on the experimental results available in the public domain and would need to be upgraded much frequently than the language versions itself.
Kera 1.0 is confined to qualitative data management rather than quantitative for obvious reasons. Quantitative characterization of even widely employed parts are lacking (Machisio and Stelling 2009). However, future versions of Kera plan to include a framework where reaction rates and predictions based on dynamic models of biological circuits are incorporated.
Conclusion
Kera 1.0 provided subtle design power to the synthetic biologists through the powerful programming constructs and biorule library. The language is currently in its infancy but designed for a long term evolution into a leading tool for the synthetic biologists. The authors have attempted to compare various features of the closest competitors to Kera. (One must admit that this comparison is unfair, as some of these tools have no resemblance to a formal programming language).
We summarize in the Table 3, the comparison of Kera with GenoCAD, Antimony, Little b, and GEC. It must be pointed out that associated rule base and part base are widely varying in the compared tools. What ultimately matters is whether the language serves the perceived and creative needs of synthetic biologists in a futuristic way. From this standpoint the authors believe that Kera is well positioned. The real power of Kera or similar tools would emerge once the quantitative modeling is seamlessly integrated into these tools.
Table 3.
A comparative study of existing programming languages
| GenoCAD | Antimony | Little b | GEC | Kera | |
|---|---|---|---|---|---|
| GUI | √ | × | × | × | √ |
| BioBrick support | √ | √ | × | √ | √ |
| Part repository | √ | × | × | √ | √ |
| Rule based | √ | × | × | √ | √ |
| Modularity | × | √ | √ | √ | √ |
| User can create models | × | √ | √ | √ | √ |
| Text based | × | √ | √ | √ | √ |
| Import other file formats (like SBML) | × | √ | √ | × | √ |
| Open source | √ | √ | √ | × | √ |
| Object oriented | × | × | × | × | √ |
| Full scale programming construction | × | √ | √ | √ | √ |
References
- Bashor CJ, Horwitz AA, Peisajovich SG, Lim WA (2010) Rewiring cells: synthetic biology as a tool to interrogate the organizational principles of living systems. Rev Lit Arts Am. doi:10.1146/annurev.biophys.050708.133652 [DOI] [PMC free article] [PubMed]
- Cai Y, Hartnett B, Gustafsson C, Peccoud J. A syntactic model to design and verify synthetic genetic constructs derived from standard biological parts. Bioinformatics. 2007;23(20):2760. doi: 10.1093/bioinformatics/btm446. [DOI] [PubMed] [Google Scholar]
- Chandran D, Bergmann FT, Sauro HM. TinkerCell: modular CAD tool for synthetic biology. J Biol Eng. 2009;3:19. doi: 10.1186/1754-1611-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy K, Voigt CA (2010) Programming cells: towards an automated ‘genetic compiler’. Curr Opin Biotechnol 572–581. doi:10.1016/j.copbio.2010.07.005 [DOI] [PMC free article] [PubMed]
- Czar MJ, Cai Y, Peccoud J. Writing DNA with GenoCAD TM. Grammars. 2009;37:40–47. doi: 10.1093/nar/gkp361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403(6767):335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- Endler L, Rodriguez N, Juty N, Chelliah V, Laibe C, Li C et al. (2009) Designing and encoding models for synthetic biology rapid response designing and encoding models for synthetic biology. J R Soc. doi:10.1098/rsif.2009.0035.focus [DOI] [PMC free article] [PubMed]
- Gardner TS, Cantor CR, Collins JJ (2000) Construction of a genetic toggle switch in Escherichia coli. Nature 339–342 [DOI] [PubMed]
- Goler JA (2004) BioJADE: a design and simulation tool for synthetic biological systems. MIT Computer Science and Artificial Intelligence Laboratory AT Technical Report 2004-003
- Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- Khalil AS, Collins JJ (2010) Synthetic biology: applications come of age, 11. doi:10.1038/nrg2775 [DOI] [PMC free article] [PubMed]
- Machisio MA, Stelling J. Computational design tools for synthetic biology. Science Direct. 2009;20(4):479–485. doi: 10.1016/j.copbio.2009.08.007. [DOI] [PubMed] [Google Scholar]
- Pedersen M, Phillips A, Pedersen M, Phillips A (2009) Towards programming languages for genetic engineering of living cells Towards programming languages for genetic engineering of living cells. Interface. doi:10.1098/rsif.2008.0516.focus [DOI] [PMC free article] [PubMed]
- Purnick PE, Weiss R. The second wave of synthetic biology: from modules to systems. Nature Publishing Group. 2009;10(6):410–422. doi: 10.1038/nrm2698. [DOI] [PubMed] [Google Scholar]
- Smith LP, Bergmann FT, Chandran D, Sauro HM. Antimony: a modular model definition language. Bioinformatics. 2009;25(18):2452–2454. doi: 10.1093/bioinformatics/btp401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young E, Alper H (2010) Synthetic biology: tools to design, build, and optimize cellular processes. J Biomed Biotechnol. doi:10.1155/2010/130781 [DOI] [PMC free article] [PubMed]