Abstract
Natural killer (NK) cells are lymphoid effectors that are involved in the innate immune surveillance against infected and/or tumor cells. Their function is under the fine-tuning control of cell surface receptors that display either inhibitory or activating function and in healthy condition, mediate self-tolerance. It is known that inhibitory receptors are characterized by clonal and stochastic distribution and are extremely sensible to any modification, downregulation or loss of MHC class I surface expression that are induced in autologous cells upon viral infection or cancer transformation. This alteration of the MHC class I expression weakens the strength of the inhibitory receptor-induced interaction, thus resulting in a prompt triggering of NK cell function, which ends up in the inhibition of tumor progression and proliferation of pathogen-infected cells. Thus, the inhibitory function of NK cells is only one face of the coin, since NK-cell activation is controlled by different arrays of activating receptors that finally are involved in the induction of cytolysis and/or cytokine release. Interestingly, the inhibitory NK-cell receptors that are involved in dampening NK cell-mediated responses evolved during speciation in different, often structurally unrelated surface-expressed molecules, all using a conserved signaling pathway. In detail, during evolution, the inhibitory receptors that assure the recognition of MHC class I molecules, originate in, at least, three different ways. This ended up in multigene families showing marked structural divergences that coevolved in a convergent way with the availability of appropriate MHC ligand molecules.
Key words: killer inhibitory receptor, natural cytotoxicity receptors, NKG2D, primate, gene loci evolution
The MHC Class I Inhibitory Receptors
While humans1 and higher primates evolved mainly Killer immunoglobulin (Ig)-like receptor (KIR) molecules, rodents and other species used type II transmebrane Ly49 molecules that belong to the C-type lectins receptors.2–8 Recently, lower primates or prosimians have been reported to evolve a third strategy based on the use of different combinations of polymorphic CD94/NKG2 hetero-dimeric receptors.9
In contrast to inhibitory receptors, activating receptors appear to be evolutionary more conserved than inhibitory counterparts. They are organized as multimeric structures with tyrosine-based signal-transducing polypeptides. The genes coding for these signaling polypeptides have been conserved during speciation for more that 400 million years and some are even used for T-cell receptor signaling.
Inhibitory NK receptors evolved to interact with MHC class I molecules in a “lock and key” fashion. The fine molecular mechanism of interaction changed during evolution, since this function has been maintained by completely different receptor structures during speciation. On the other hand, it is evolutionary relevant that all the inhibitory receptors have been found to use a conserved mechanism of signaling based on the presence of immunoreceptor tyrosine-based inhibitory motif (ITIM; V/I/L/SxYxxL/V) in their cytoplasmic tails. Upon MHC class I recognition, ITIM sequences become tyrosine-phosphorylated by src-kinases. The inhibitory pathway ends up with the association with the intracellular Src homology-2 (SH2) domain-containing phosphatases 1 or 2 (SHP1 or SHP2), that are dominantly involved in the dampening of all triggering signaling pathways controlled by the different activating NK cell receptors.10
KIR genes evolved almost 135 million years ago from a common ancient gene precursor to originate the KIR3DL and KIR3Dx gene loci. The encoded KIR inhibitory receptors have been involved only recently in the regulation of NK cell function in higher primates.11
KIR loci that are expressed in primates originate by gene duplication and expansion from the KIR3DL remnant of the common ancient gene precursor that maps telomerically to the other ancestral KIR3Dx locus (Fig. 1) Notably, during KIR loci evolution upon macaque's speciation, there was a consistent expansion of KIR3D loci that encode transmembrane receptors characterized by three extracellular Ig-like domains.12,13 In contrast, the number of KIR2D gene loci that display only two Ig-like domains increased more recently and followed the emergence of MHC-C genes from orangutan speciation. Different KIR2D loci co-evolved with MHC-C alleles in the last 15 million years, and they encode important inhibitory receptors that regulate NK-cell function in humans.14,15
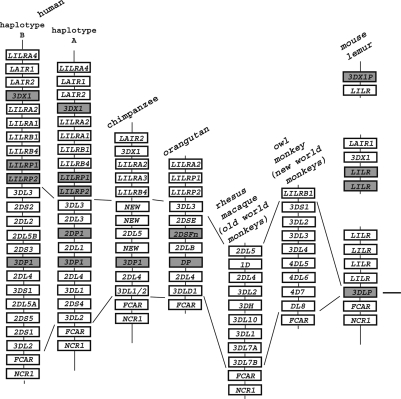
Figure 1.
KIR haplotypes gene loci organization from six primate species. The structure and schematic gene representation of KIR haplotypes from human,16,17 chimpanzee,62 orangutan,15 rhesus macaque,12 owl monkey35 and mouse lemur.9 Grey rectangles denote pseudogenes, the ancestral 3DP locus of mouse lemur is indicated, as well as lines shown the amplification of KIR loci in the different species. The maps have not been drawn to scale.
Homo sapiens KIR genes map on chromosome 19q13.42, in the telomerically region of Leukocyte Receptor complex (LRC), which is close to Leukocyte Ig-like Receptor (LILR) gene loci that also belong to LRC. The KIR multigene family accounts for a total of 17 gene loci (two of them are pseudogenes) that are organized in two possible haplotypes (A and B).16
In contrast to the ancestral KIR3DL locus that evolved by gene duplication and expansion into the presently known primate KIR loci, the KIR3DX1 locus, has been conserved in human, common chimpanzee, gorilla, rhesus macaque and common marmoset, as an unused leftover of the evolution of the KIR multigene family.
In humans, it maps centromerically, close to the LAIR2 gene and is located between the two LILR gene clusters but outside the region encoding KIRs.17
Interestingly, different from primates, cattle is the unique non-primate species known to have expanded KIR loci and originate KIR multigene family through the expansion of the KIR3Dx ancestral locus.11,18
Although, the ancestral KIR genes are older than the rodent/primate split from a common ancestor rising to 87 million years ago,19 mouse, rat and other non-primate species evolved completely different MHC class I-specific inhibitory NK cell receptors. All these species actually use type II integral transmembrane homodimeric glycoproteins that belong to the C-type lectins and are known as Ly49 receptors.5,6,20–22 These type II transmembrane receptors are structurally very different from the Ig-like type I transmembrane molecules used by higher primates.
Thus, mouse and rat expanded NK gene complex (NKC) Ly49 loci (15 and 36 genes, respectively), while only a single copy of Ly49 gene/pseudogene is left in primates. Indeed, a gene related to human Ly49L was found in common in chimpanzee, gorilla, orangutan, gibbon, baboon, African green monkey and other mammalian orders.23 This locus is non-functional both in human24 and chimpanzee due to a premature termination codon.23 This mutation might have arisen in the common ancestor of gorilla, chimpanzee and human, thus affecting the Ly49H function in these primates, while baboon and other lower primates might have a single functional conserved Ly49H gene, whose potential role is unknown.23 Along this line, although mouse and rat use a different NK-cell receptor system to sense MHC-class I expression, their genomes display two KIR loci mapping on chromosomes X and 1, respectively.25,26 More precisely mouse Kirl1 displays 40% amino acid identity with primate KIR family members and its transcript is expressed at high levels on immature mouse thymocytes with unknown function.26
Recently, the analysis of gene loci encoding NK receptor in prosimians (Strepsirrhini), a suborder of primates that split about 78 millions years ago,19 added a novel degree of complexity to the mechanism of co-evolution of MHC class I inhibitory NK receptors and their ligands and made it fundamental to our improved understanding the mechanisms of evolution of self/non-self discrimination.
Interestingly, the NK-related gene loci found in Microcebus murinus (grey mouse lemur) possibly suggests an alternative mechanism to produce polymorphic MHC class I receptors, since these lemurs display only a single copy of both Ly49 and KIR genes. More precisely, the ancestral KIR3DL locus of grey mouse lemur that is normally expanded in higher primates, is a pseudogene (3DLP). The other ancestral KIR3Dx locus that is typically expanded in cattle, has been duplicated generating a single functional gene KIR3DX1 with unknown function and a pseudogene KIR3DX1P.9 This indicates that in lemurs, a different mechanism is used to generate receptors involved in self/nonself discrimination, since a single KIR is clearly insufficient to generate a functional MHC-class I NK-cell receptor system.8 These data have been confirmed by the analysis of a second lemur species (Varecia variegata) that diverged from the other lemur species about 43 million years ago.9,27
Interestingly, in contrast with other species, prosimian lemurs show amplification of the CD94 and NKG2 gene loci that might be involved in the regulation of NK cell functions. Indeed, the CD94/NKG2 receptor evolved in different species to interact with MHC-E (Qa-1 in rodents) molecules, non-classical class I molecules that have in the peptide cleft the signal peptide derived from the various MHC-class I alleles.28–32 This interaction evolved as a second safety check on overall expression of class I molecules.8 Differently, lemurs appear to miss MHC-E orthologue and any other functional homologues. It is intriguing to hypothesize that since CD94/NKG2 evolved in different species to interact with a class I-like molecule, in lemurs, polymorphic CD94/NKG2 heterodimers might use the same buried surface area of interaction with histocompatibility molecules to sense not only non-classical MHC-E molecules but also MHC class I allele variability. Thus, in lemurs, the receptor formed by the combination of different CD94 and NKG2 molecules might be the result of positive selection mechanisms aimed to the maintenance of a critical innate immune function involved in the balance of self/nonself interaction.
MHC Class I/Receptor Interaction
In primates, the major family of HLA class I-inhibitory receptors is structurally characterized by Ig-like molecules with two or three extracellular Ig-like C2-type domains (KIR2DL and KIR3DL), a transmembrane region containing non-polar amino acid residues, and a long cytoplasmic tail containing two ITIM sequences. Phylogenetic analysis of KIRs expressed in catarrhini (hominoid and old world monkeys) revealed that these receptors might be classified in five different lineages. Receptors belonging to the lineage I (KIR2DL4, KIR2DL5) are structurally characterized by two extracellular Ig-like C2-type domains with D0 + D2 Ig-like domain configuration. Lineage II is characterized by the KIR3DL2 and KIR3DL1 receptors sharing D0 + D1 + D2 domains specific for MHC-A and MHC-B alleles, respectively. Lineage III contains MHC-C specific molecules sharing D1 + D2 or D0 + D1 + D2 configuration. Macaque's KIRs essentially belong to lineage IV, with the exception of Mm3DL1 and 3DL10 that have been classified in lineage II, while human KIR3DL3 molecules characterize the lineage V.15,33 Recent data indicated that new world monkeys (platyrrhini), which share with old world monkeys (catarrhini) a common ancestor about 35 million years ago,34 might have evolved gene loci encoding KIR that does not belong to any of the five lineages describe above.35
The number of loci encoding lineage III KIR receptors expanded in hominoids following the appearance of MHC-C allele in orangutan about 15 million years ago.15 This lineage originates in humans the expansion of KIR2DL loci that resulted to be specific for two HLA-C allele groups characterized by the presence of a dimorphism at position 80.36–45 Indeed, KIR2DL1 is specific for HLA-Cw2, -Cw4, -Cw5 and -Cw6 alleles that are characterized by Lys80, while KIR2DL2 and KIR2DL3 are specific for all HLA-C alleles, such as HLA-Cw1, -Cw3, -Cw7 and -Cw8, that display Asn80.8 In humans, recognition of HLA class I alleles is also supported by the KIR3DL1 molecules sensitive to all HLA-B alleles sharing the Bw4-supertypic specificity that is characterized by the presence of a particular amino acid at position 80 in a similar region involved in defining the KIR2DL/HLA-C specificities. Finally, KIR3DL2 is known to be specific for HLA-A3/A11 alleles.8 The interaction between KIR and HLA alleles has been confirmed by crystallographic analysis revealing that these molecules interact with each other approximately orthogonally with involvement of several conserved residues residing on the HLA molecule as well as the residues 7 and 8 of the C-terminal portion of the HLA-bound peptide.46,47
In humans, the LRC-encoded LILRB1 (LIR1, ILT2, CD85j) and LILRB2 (LIR2, ILT4, CD85d) receptors have also been shown to bind a broad range of classical and non-classical MHC and MHC-like molecules, such as the HLA-G molecules that are expressed by placenta trophoblasts and thymic epithelial cells.48–52
LILRB2 and LILRB1 recognize different HLA class I alleles essentially in a β2-microglobulin dependent way, since more than 70% of the binding surface interaction is involving 14 β2-microglobulin residues.53–55 In contrast, the same receptors bind HLA-G more tightly with different α3-domain amino acid residues of HLA-G molecules involved in the interaction.53
The recognition of other non-classical HLA class I molecules, such as HLA-E, has been shown to be dependent on the expression of the CD94/NKG2A receptors.
Crystallographic studies revealed that CD94 contributes for more than 69% in the interaction with HLA-E and the peptide, while the role of NKG2A in the peptide recognition is minimal, but essential to determine the ligand recognition outcome.56,57 It is to be stressed that CD94 by itself, although is critical for the interaction, is unable to bind HLA-E.
More interestingly, in lemurs, unlike other species where CD94 and NKG2 molecules are monomorphic, the same loci are polymorphic indicating a more complex behavior of lemur CD94/NKG2 molecules in interacting with ligand(s). In particular, lemur CD94 loci produce higher sequence variability than NKG2 loci, since 15 amino acids have been positively selected for CD94 compared to only 6 for NKG2 molecules.9 Since the prominent role of CD94 in the MHC interaction is known, these data might indicate that lemurs use the heterodimeric polymorphic CD94/NKG2 receptors to recognize different MHC class I alleles, and not merely the MHC-E molecule.
Along this line, the evolution of CD94 and NKG2 loci probably it might also be present in cattle (Bov taurus), since this species displays, together with an unconventional expansion of KIR3Dx loci, multiple distinct NKG2 and CD94 genes. Some of these loci show minor allelic variation and multiple alternatively spliced forms,58 thus indicating that two different receptor families that are probably involved in sensing different MHC class I alleles might coexist in Bov taurus.
The MHC Class I Activating Receptors
It is interesting to note that in all species analyzed and more importantly, in all gene families known to encode inhibitory MHC class I NK receptors, we always find a variable number of gene loci coding for triggering receptor. This phenomenon is true either for genes encoding type II receptor molecules like Ly 49 and CD94/NKG2 or for genes encoding LLIRs (LILRA) and KIRs Ig-like receptors. In particular, Ly49D, H, K, L, M, N, P, R, U, W7 and CD94/NKG2C59 and CD94/NKG2E heterodimers do not inhibit but rather activate NK mediated cytolysis. The same triggering function is also associated to Ig-like superfamily receptors belonging to the LLIR (LILRAs) and KIRs (KIR2DS and KIR3DS).8
These triggering receptors associate at the cell surface with signal-transducing polypeptides forming a multichain immune recognition receptor, similar, or identical, to those used by T- and B-cell antigen receptors.60
In primates, activating KIRs originate within the last 35 million years (my) from common ancestor gene loci coding for inhibitory receptors. They are apparently continuously acquired and lost during evolution as a response to the rapid evolution of KIR loci.61,62 The unstable presence of activating KIR during evolution might indicate that although they have played a favorable effect in host-resistance to pathogens, they also might have determined a detrimental role, since they might have increased the risk to induce autoimmunity.61 On the contrary, although activating KIR genes appear to be unstably maintained over evolution, the CD94/NKG2C heterodimer appear to be stably expressed in rodents63,64 as well as in macaques (M. fascicularis, M. mulatta) and in common chimpanzee.13,65,66 It is of note that the comparison of the human and chimpanzee sequences for CD94 and NKG2 loci indicates that the former locus is more conserved than the other. In particular, the NKG2C and NKG2E genes resulted to be the most divergent NKG2 loci.66 The sequence comparison between the Homo sapiens and Pan troglodytes genes shows a value of sequence identity different than expected for chimpanzee/human gene sequence divergences under neutral selection, thus suggesting that the evolution of the heterodimeric CD94/NKG2 receptors might be driven by external factors, such as pathogens.
Cynomolgous and rhesus macaque NK cells express functional CD94/NKG2C molecules sharing a high degree of sequence homology with human molecules.65 All these data indicate that NKG2 loci evolved under strong positive selection throughout primate evolution.67 Thus, it is possible that the positive selection mechanism, underlying the NKG2 and CD94 loci evolution in lemurs might have defined a complete novel set of NK receptors using a combination of different CD94/NKG2 encoded molecules that ended up to be specific for different alleles of classical MHC class I molecules.
Natural Cytotoxicity Receptors
The main human NK-cell receptors involved in NK-mediated cell cytotoxicity are the natural cytotoxicity receptors (NKp46, NKp44 and NKp30) that belong to the Ig-like superfamily, and NKG2D (CD314), which is a member of the C-type lecting family of type II transmembrane receptors. Interestingly, while NKp46 and NKG2D have been conserved during speciation since rodents evolution, NKp44 and NKp30 appear to be differently conserved during evolution.
NKp46 (CD335) is encoded by the NCR1 gene,8,68 which is located on the telomeric end of LRC close to the Fcα receptor-enconding gene (FCαR) on chromosome 19 in human, chimpanzee, orangutan and macaque.8,21,60,68,69 This locus is also conserved in lemurs, mice, rats, bovine and the encode protein displays a similar pattern of expression and function.71–77 NKp46 has an aminoterminal extracellular portion organized in two extracellular Ig-like C2-type domains with known structure (PDB: 1OLL78), a transmembrane region containing a positively charged arginine residue and a small cytoplasmic tail of 25 amino acids. In the context of the hydrophobic transmembrane enviroment, the positively charged amino acid is involved in a salt-bridge formation with the negatively charged residue (aspartic acid) present on the transmembrane region of CD3ζ and FcεRγ, two ITAM-bearing signal-transducing polypeptides involved in the NKp46-mediated cell activation.8,69
The NKp46 receptor is expressed both on resting and activated NK cells, being the major receptor involved in NK-mediated cancer cell killing.8,69,79
Differently than NKp46, the NKp44 (NCR2) expression is only induced upon interleukin-2 (IL-2) activation and this receptor is essentially expressed on activated NK and γ/δT cells but not on resting NK cells.80 The NCR2 locus maps on chromosome 6p21.1 and is located telomerically to the human triggering receptor expressed on myeloid cells (TREM) gene cluster.80,81 Interestingly, the murine syntenic region of the human chromosome 6 located on mouse chromosome 17 lacks any NKp44 gene orthologue, although the same region displays different orthologous loci of the TREM gene cluster that are known to be close to the NCR2 gene locus in human genome.81,82 The NKp44 structure (PDB: 1HKF) is characterized by a single extracellular IgV-like domain with an extra disulphide bridge, which is typical of IgV-domains of receptors that belong to the CD300-like multigene family (IREM1, PDB: 2NMS, and mClm1, PDB: 1ZOX).83 The Ig domain is anchored to the transmembrane region through a proline-rich stalk followed by a cytoplasmic tail that contains a single, nonfunctional ITIM sequence.8,80 NKp44 associates with disulfide-linked homodimer of the ITAM-bearing signal-tranducing DNAX-activating protein of 12 kDa (DAP-12) through a positively charged transmembrane lysine residue. Additional pathway of NKp44-mediated activation, which is different than those of other NCR receptors, and uniqueness of NKp44 expression on activated cells might explain higher cytolytic potential of activated NK cells as compared to resting cells.84
NKp44 cellular ligand(s) expressed in cells of different histological origin are apparently sensible to trypsin treatment and their expression seems to be modified along the progression of cell cycle.85 In addition, NKp44 has been reported to bind directly to the bacteria cell wall of mycobacterium-related species apparently using a ligand different to the one involved in the cellular interaction.86 Although the natural cellular ligands for NKp46 and NKp44 are still unknown, surface heparan sulfate proteoglycans (HSPGs) are believed to be involved in the recognition of tumor cells by NKp44.87 In addition, NKp46 and NKp44 are involved in the response against influenza virus, since they appear to recognize the viral-encoded hemagglutin (HA) (IV-HA) and hemagglutinin- neuroaminidase (SV-HN) molecules.88–91 Moreover, NKp44 has been reported to bind influenza virus hemagglutinin of both H1N1 and H5N1 strains.88,92
Similar to NKp46, NKp30 (NCR3) is expressed by both resting and activated human NK cells. The NCR3 locus has been conserved during speciation at least in macaque and chimpanzee, while in mouse, the locus is generally non-functional, being a pseudogene, with the exception of a single Mus caroli mouse strain.72,74,93–95 In humans, the NCR3 locus maps in the HLA class III region on human chromosome 6p21.3.96
NKp30 has a single IgV-like domain, a short stalk region followed by a transmembrane region containing the positively charged arginine residue and by a short cytoplasmic tail without any signaling motifs. NKp30 and NKp46 are expressed at the cell surface associated with CD3ζ and FcεRIγ ITAM-bearing signaling polypeptides through a salt bridge.93,97 Apparently, both receptors are also associated with the glycosylphosphatidylinositol (GPI)-linked CD59 molecule that is involved in tyrosine phosphorylation of the associated CD3ζ-chain.98 Recently, B7-H6, which is involved in the induction of NKp30-mediated cell cytotoxicity and cytokine secretion, has been suggested as a possible cellular ligand for NKp30.99 In addition, the nuclear factor HLA-B-associated transcript 3 has also been associated with NKp30-mediated cytokines release and tumor rejection.100
On the contrary, the human cytomegalovirus molecule pp65 has been proposed to induce a viral-mediated escape to the innate immune recognition by inducing NKp30 uncoupling to the signal transducing CD3ζ subunit and thus blocking any receptor-mediated function.101
NKp30 is not only involved in the control of tumor cell transformation and pathogens by the triggering of innate immunity but also plays an important role in the fine regulation of adaptive immune responses.101 NKp30 is directly involved in the cross-talk between NK lymphocytes and dendritic (DC) cells101 and participates in the selection and maturation of immature DC (iDC),103–106 thus modulatiing and editing the intensity and quality of adaptive immune responses.
The information gained so far on the characterization of NCRs gene loci during phylogenesis highly suggests that a staged appearance of these receptors was probably driven by the pathogen environment. In particular, they might be involved in retrovirus susceptibility, thus providing useful tools for the study of natural immunity correlates of protection in primate SIV/SHIV infection models.
The NKG2D triggering receptor is a type II transmembrane protein encoded by a gene locus mapping on human chromosome 12p13.2, inside the region defined NK gene complex encompassing the NKG2 and CD94 gene loci. NKG2D is expressed in rodents and primates (macaques, chimpanzee), including humans.8,65,76,107–111 It is expressed at the cell surface of NK cells, γ/δ and CD8+ T lymphocytes112 as a homodimer and associates with two DAP10 (KAP10) signaling homodimers, thus forming an hexameric receptor structure.113 Interestingly, NKG2D triggering does not involve ITAM-induced signaling pathways, since DAP10 cytoplasmic tails display the YINM motif that upon phosphorylation, is able to recruit either phosphatidylinositol 3-kinase (PI3K) p85 subunit or the Grb2 adapter protein in a mutual exclusive pattern.114–116 Human NKG2D binds to different molecules, including MICA and MICB, the stress-inducible MHC class I-related molecules that are encoded by loci mapping on MHC complex on human chromosome 6p21.3.117–123 Recently, MICA/MICB-like orthologous genes have also been described in macaques.124 In addition, NKG2D has also been reported to bind to the human cytomegalovirus UL16 binding proteins (ULBP1-5), which are encoded by gene loci present on the long arm on chromosome 6q24.2-q25.3 and are homologous to the murine retinoic acid inducible RAET1 molecules that are known to be NKG2D ligands in mouse.125–128 ULBP1-3 are GPI-linked molecules (RAET1I, RAET1H, RAET1N), while ULBP-4 (RAET1E) and ULBP-5 (RAET1G) are type I transmembrane proteins.129
MICA and MICB are expressed on the cell surface and differently than MHC class I molecules, are not associated with the β2-microglobulin subunit and peptides.117 In contrast to MICA and MICB, all ULBPs miss the extracellular α3 domain typical of class I-related molecules.
Murine NKG2D, like the human homologue, is characterized by the recognition of multiple ligand molecules, such as the H60 minor histocompatibility antigen, the murine ULPB-like transcript 1 MULT1 molecule and the RAEα-ε retinoic acid early inducible proteins.130–136
NKG2D is able to interact with multiple ligands albeit they essentially do not share significant homology (<20%). Although the receptor-ligand interfaces are apparently highly similar, at least for ULBP-3 and MICA, they use different amino acid residues in the interaction.137–139 In humans, some of different MICA (60) and MICB (25) alleles are probably associated with the pathogenesis of different diseases, such as rheumatoid arthritis, multiple sclerosis, celiac disease and diabetes mellitus.140–142 NKG2D ligands are known to be upregulated in response to viral infection, oxidative and genotoxic stresses.143–146 Although MIC molecules have been found conserved in non-human primates, lemur, cattle and pigs, no orthologues have been defined in the mouse MHC region.6,140,141,147
Among the receptors involved in natural cytotoxicity, the NKp46, NKp30, NKp80 and NKG2D molecules expressed in macaques PBMC and in in vitro-derived NK cell populations and clones, were shown to react with monoclonal antibodies (mAbs) originally raised against human NK cells. More interestingly, the same mAbs can be used to analyze the function of these receptors either by redirected killing assay or mAb-mediated masking assay.65,72 In addition to macaques PBMC, chimpanzees PBMC also display variable reactivity with mAbs specific for human NKp46, NKp80 and NKG2D molecules, but major differences apply to NKp30 expression. Probably due to post-transcriptional regulation, NKp30 is not expressed (or has low expression level) on freshly derived PBMC, while it is clearly expressed in activated NK cells. Thus, differently from humans, chimpanzees NK cells express NKp30 upon culturing them in IL-2-rich media.76 It is tempting to speculate that the differential expression of NKp30 might lead to a differential maturation of immature-DC in chimpanzee affecting and modifing the outcome of HIV-1 related lentivirus SIVcpz infection.148 Since chimpanzees are known to have the relative resistance in the development of AIDS (even in the presence of a high viral load), the association of an impaired NKp30-dependent DC maturation and NK activation may in part explain the different behaviour following HIV viral infection. Thus, it is tempting to hypothesize a role of NKp30 as one of the factors implicated in the longer survival of chimpanzees in the case of a long-standing HIV-1 infection.
More recently, NKp44 has been found to be expressed in Pan troglodytes NK cells upon activation, like in human, although the NKp44 transcription appears to increase slightly less compared than in humans.82 Since humans and chimpanzees are the unique species known to express functional NKp44 molecules, it is tempting to speculate that, NKp44 locus has recently emerged during evolution. It is possible that NKp44 locus appeared during macaque speciation, since cynomolgus showed weak transcription levels, occurrence of high-frequency of out-of-frame transcripts, without the surface expressinon of functional receptor molecules.82
Perspective on Antiviral Defenses
According to phylogenetic evidences gathered so far, it appears that patrolling of NK cell receptors for self/nonself recognition evolved in a highly dynamic fashion, and this evolution could be primarily driven by the necessity to cope with adaptation to changing environments with ever changing pathogens. The required extent of flexibility in molecular mechanisms driving this shaping of native and adaptive immune responses is reflected, for example, by the still ongoing battle between host defense responses and escape tricks adopted by human cytomegalovirus to hide and survive.
When adaptation to the environment and evolution is considered, innate immune responses that include NK cells, match those induced by the adaptive immune system. Adaptive immune responses developed to high specificity are tuned to recognize and destroy/control individual pathogens and transformed cells. They operate under considerable molecular constraints and are endowed with more limited evolutionary plasticity as compared to NK cells. Natural Killer cells have been originally believed to be a simpler and ancient effectors defense mechanism compared to the cells responsible for adaptive immunity. On the contrary, molecular studies of NK cell receptor loci development convey a view of an extremely dynamic immune-defense system. These receptor molecules underwent evolution or adaptation together with the evolution MHC molecules in order to optimally tackle the challenges of intracellular pathogen infection or tumor-transformation.
In humans, several lines of evidence link differences in NK cell receptor expression to differential responses to invading pathogens. In contrast, little is known about this link in other animal species with the possible exception of rodents. This discrepancy contributes to a paradox situation: when it comes to our understanding of comparative NK-cell receptor function, animal models that have been established and widely used to study immune responses to pathogens and to evaluate vaccines including macaque models, represent in part black boxes when compared to humans. In addition, differences in NK-cell receptor organization, regulation and function prevent transfer of information from humans to animal models or vice versa.
With regard to clinical responses to invading pathogens, different reports have shown that both activating and inhibitory NK cell receptors may be involved in the shaping of different clinical outcomes.
When considering the role of inhibitory NK-cell receptors, a seminal work based exclusively on molecular biology studies in patients exposed to HCV infection showed that the chance of clearing HCV upon acute infection is five times higher for patients homozygous for KIR2DL3 inhibitory NK receptors and HLA-C1.149 Since the affinity of this receptor for its HLA ligand (HLA-C1) is lower compared to the affinity of KIR2DL2 for HLA-C1 or of KIR2DL1 for HLA-C2, these patients could take advantage of a less inhibited NK cell function when their NK cells sense autologous HCV-infected hepatocytes that are still expressing HLA class I molecules, possibly leading to a more efficient virus clearance. Although no functional proof of this mechanism is provided, this data interpretation might explain why homozygosity for KIR2DL3/HLA-C1 bears a higher probability of success in clearing acute infection with low titer exposure to HCV (injection as opposed to blood transfusions). In line with this finding, the same group has recently reported the similar association in exposed-uninfected subjects and in chronically HCV-infected patients responding to treatment.150 Other studies have associated KIR2D alleles to outcome of liver transplantation and repeat cirrhosis, albeit with discordant results.151,152 With the cautionary note in mind that functional proof of KIR/HLA relevance on control of virus replication in hepatocytes is not available yet, these data suggest that interventions with techniques aimed at modulation of KIR expression (or of their function) on NK cells could influence the outcome of acute HCV infection preventing progression to chronic persistent infection. Unfortunately, correlates of this observation in animal models are lacking so far. Although experimental HCV infection in non-human primates (chimpanzees) has been developed as a model for vaccine purposes,153,154 KIR diversity in chimpanzees155 and lack of correspondence to human KIR/HLA156 prevent the possibility of a direct transfer of this information. This leads to the impossibility to devise and verify tools for the manipulation of KIR/HLA interaction for the treatment or prevention of HCV infection. Given the above discussion on KIR evolution, it will become difficult to embed manipulation of KIR/HLA interaction in established animal models (e.g., chimpanzee for HCV). This could constitute a complicating issue also with regard to the envisaged transition to small animal models for the development of HCV vaccines.157
Results from other human studies independently contribute to the hypothesis that KIR/HLA interaction might represent a relevant parameter to be considered during HCV infection. Indeed, as recently reported from a large study in Europe on chronically infected patients undergoing treatment,158 exploring KIR genotyping (KIRotype) may become a useful tool to evaluate chances of clinical response to standard treatment in patients chronically infected with HCV. In this study, the presence of either KIR2DL2 heterozygosity or KIR2DL2 homozygosity was associated to increased rates (three-fold) of non-response to standard treatment at 6 mo The reverse, i.e., KIR2DL3 heterozygosity or homozygosity in the presence of HLA-C1 haplotype, was correspondingly associated with increased odds (3-fold) of responses to treatment with clearance of virus replication.158 In this case, functional correlates on in vitro NK cells or in animal models are still lacking, as well, and there are so far no answers whether it is feasible to manipulate KIR expression (or interfere with it) to favor response to treatment during chronic HCV infection.
Activating KIRs are also associated to different disease courses in humans infected with pathogens characterized by persistent or latent replication cycles. Severe, invasive, HPV-6/11-associated recurrent respiratory papillomatosis (RRP) has been shown to associate with lack of activating KIRs such as KIR3DS1, KIR2DS2, KIR2DS5, while their presence on NK cells was more frequent in patients with moderate RRP.159 The relevance of short-tailed, activating KIRs during chronic HCV infection has been only partially elucidated in humans. Expression of both KIR3DS1 and HLA Bw4 has been recently reported to associate with liver injury and cirrhosis in HCV infected patients.160 Blocking of triggering KIRs and reducing KIR2DL3 interaction with its natural MHC ligands may therefore represent future options to increase survival of chronically infected patients who would otherwise proceed to liver injury and cirrhosis with end-stage liver disease.
Over recent years, attention to short-tailed isoforms of KIRs during human HIV infection has been also raised. KIR3DS1, in combination with HLA-B alleles that encode molecules with isoleucine at position 80 (HLA-B Bw4-80Ile), has been associated with delayed progression to AIDS in individuals infected with human immunodeficiency virus type 1 (HIV-1).161 Patients with homozygous expression of KIR3DS1 and HLA-B Bw4-80Ile alleles (including HLA-B57) bear a subset of NK cells that display enhanced control of HIV-1 replication in vitro.162 Patients bearing the HLA-B57/B5801 haplotype are over represented in cohorts of elite controller long-term non-progressors163 and are more likely to develop LTNP or slowly progressing disease, when compared to other HLA haplotypes. Depending possibly on the KIR haplotype expressed by NK cells, HIV-infected patients bearing KIR3DS1+/HLA-B Bw4-80Ile+ may respond with improved innate control of viral replication and conserved CD4+ T-cell counts. In these patients, the relationship between a minor fraction of KIR3DL1+ NK cells (≈5–10%), the ability of these cells to directly control HIV-1 replication in all interested districts, and their role in the protection from NCR perturbations and NK cell activation is still undefined. In line with this consideration, recent studies on LTNP cohorts failed to identify enrichment in KIR3DS1+/HLA-B Bw4-80Ile+,164 or improved NK cell function.164,165
So far, animal models for the study of HIV infection are limited to few animal species. The only known natural occurring model of animal HIV-1 infection is chimpanzee,148 while macaque models exploit infection with heterologous or molecularly manipulated Simian Immunodeficiency Virus (SIV). In general, chimpanzee infected with HIV as well as the vast majority of African non-human primates infected with homologous virus behave as human long-term non-progressors.148,166 Contrary to these models, human long-term non-progressors represent a minority of HIV-infected patients (≤2%). Currently available animal AIDS models, including macaques and mice, require artificial manipulation of either the host or of the virus167,168 and bear consistent limitations in their comparability to human HIV infection.169 In addition, as discussed above, differences between macaque and human NK-cell receptor expression support the cautionary notes that have been recently issued.170 Indeed, one can envisage consistent differences in NK cell interaction with HIV in cynomolgus and rhesus macaques compared to humans. In these species, for example, NK cells do not express NKp44 as its transcription is inefficient.82 As a consequence, aspects of NKp44/CD4+ T cells/HIV interaction are missing and cannot be evaluated, preventing exploitation of knowledge derived from human studies for vaccine purposes.170,171 In addition, different from NK cells in HIV-infected patients,172 NCR expression is not decreased in SIV-infected Macaca fascicularis that develop AIDS-like illness.65 This raises questions regarding the appropriateness of some immunological correlates of protection when macaques are employed for vaccine studies.
The observation of a particular regulation of NKp30 expression in chimpanzees (both uninfected and HIV-infected) opens up the possibility that the decreased pathogenicity of HIV in this species could be associated to this finding, preserving chimpanzees from intense NK-DC crosstalk and therefore from intense downstream immune responses with generalized immune activation.76 Although correlates of this regulation in humans are lacking, this observation could raise the possibility that in humans, different responses to chronic infection by intracellular pathogens may reflect, at least in part, differential NCR regulation.
Footnotes
Previously published online: www.landesbioscience.com/journals/selfnonself/article/11717
References
- 1.André P, Biassoni R, Colonna M, Cosman D, Lanier LL, Long EO, et al. New nomenclature for MHC receptors. Nature Immunol. 2001;2:661. doi: 10.1038/90589. [DOI] [PubMed] [Google Scholar]
- 2.Ljunggren HG, Karre K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol Today. 1990;11:20–22. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 3.Takei F, Brennan J, Mager DL. The Ly49 family: genes, proteins and regulation of class I MHC. Immunol Rev. 1997;155:67–77. doi: 10.1111/j.1600-065x.1997.tb00940.x. [DOI] [PubMed] [Google Scholar]
- 4.Yokoyama WM, Plougastel BF. Immune functions encoded by the natural killer gene complex. Nat Rev Immunol. 2003;3:304–316. doi: 10.1038/nri1055. [DOI] [PubMed] [Google Scholar]
- 5.Yokoyama WM, Kim S, French AR. The dynamic life of natural killer cells. Annu Rev Immunol. 2004;22:405–429. doi: 10.1146/annurev.immunol.22.012703.104711. [DOI] [PubMed] [Google Scholar]
- 6.Dimasi N, Moretta L, Biassoni R. Structure of the Ly49 family of natural killer (NK) cell receptors and their interaction with MHC class I molecules. Immunol Res. 2004;30:95–104. doi: 10.1385/IR:30:1:095. [DOI] [PubMed] [Google Scholar]
- 7.Dimasi N, Biassoni R. Structural and functional aspects of the Ly49 natural killer cell receptors. Immunol Cell Biol. 2005;83:1–8. doi: 10.1111/j.1440-1711.2004.01301.x. [DOI] [PubMed] [Google Scholar]
- 8.Biassoni R. Human natural killer receptors and their ligands. Curr Protoc Immunol. 2009;14:10. doi: 10.1002/0471142735.im1410s84. [DOI] [PubMed] [Google Scholar]
- 9.Averdam A, Petersen B, Rosner C, Neff J, Roos C, Eberle M, et al. A novel system of polymorphic and diverse NK cell receptors in primates. PLOS Genet. 2009;5:1000688. doi: 10.1371/journal.pgen.1000688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olcese L, Lang P, Vély F, Cambiaggi A, Marguet D, Bléry M, et al. Human and mouse natural killer cell inhibitory receptors recruit the PTP1C and PTP1D protein tyrosine phosphatases. J Immunol. 1996;156:4531–4534. [PubMed] [Google Scholar]
- 11.Guethlein LA, Abi-Rached L, Hammond JA, Parham P. The expanded cattle KIR genes are orthologous to the conserved single-copy KIR3DX1 gene of primates. Immunogenetics. 2007;59:517–522. doi: 10.1007/s00251-007-0214-x. [DOI] [PubMed] [Google Scholar]
- 12.Bimber BN, Moreland AJ, Wiseman RW, Hughes AL, O'Connor DH. Complete characterization of killer Ig-like receptor (KIR) haplotypes in mauritian cynomolgus macaques: novel insights into nonhuman primate KIR gene content and organization. J Immunol. 2008;181:6301–6308. doi: 10.4049/jimmunol.181.9.6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LaBonte ML, Hershberger KL, Korber B, Letvin NL. The KIR and CD94/NKG2 families of molecules in the rhesus monkey. Immunol Rev. 2001;183:25–40. doi: 10.1034/j.1600-065x.2001.1830103.x. [DOI] [PubMed] [Google Scholar]
- 14.Guethlein LA, Flodin LR, Adams EJ, Parham P. NK cell receptors of the orangutan (Pongo pygmaeus): a pivotal species for tracking the coevolution of killer cell Ig-like receptors with MHC-C. J Immunol. 2002;169:220–229. doi: 10.4049/jimmunol.169.1.220. [DOI] [PubMed] [Google Scholar]
- 15.Guethlein LA, Aguilar A, Abi-Rached L, Parham P. Evolution of killer cell Ig-like receptor (KIR) genes: definition of an orangutan KIR haplotype reveals expansion of lineage III KIR associated with the emergence of MHC-C1. J Immunol. 2007;179:491–504. doi: 10.4049/jimmunol.179.1.491. [DOI] [PubMed] [Google Scholar]
- 16.Robinson J, Waller MJ, Stoehr P, Marsh SG. IPD-the immuno polymorphism database nucleic acids res. 2005;33:523–526. doi: 10.1093/nar/gki032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook JG, Bashirova A, Andersen H, Piatak M, Vernikos GS, Coggill P, et al. Identification of the ancestral killer immunoglobulin-like receptor gene in primates. BMC Genomics. 2006;7:209. doi: 10.1186/1471-2164-7-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piatak M, Vernikos GS, Coggill P, Lifson JD, Carrington M, Beck S. Identification of the ancestral killer immunoglobulin-like receptor gene in primates. BMC Genomics. 2006;15:209. doi: 10.1186/1471-2164-7-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Springer MS, Murphy WJ, Eizirik E, O'Brien SJ. Placental mammal diversification and the Cretaceous-Tertiary boundary. Proc Natl Acad Sci USA. 2003;100:1056–1061. doi: 10.1073/pnas.0334222100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith HR, Karlhofer FM, Yokoyama WM. Ly-49 multigene family expressed by IL2-activated NK cells. J Immunol. 1994;153:1068–1079. [PubMed] [Google Scholar]
- 21.Kelley J, Walter L, Trowsdale J. Comparative genomics of natural killer cell receptor gene clusters. PLoS Genet. 2005;1:129–139. doi: 10.1371/journal.pgen.0010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raulet DH, Vance RE. Self-tolerance of natural killer cells. Nat Rev Immunol. 2006;6:520–531. doi: 10.1038/nri1863. [DOI] [PubMed] [Google Scholar]
- 23.Mager DL, McQueen KL, Wee V, Freeman JD. Evolution of natural killer cell receptors, coexistence of functional Ly49 and KIR genes in baboons. Curr Biol. 2001;11:626. doi: 10.1016/s0960-9822(01)00148-8. [DOI] [PubMed] [Google Scholar]
- 24.Westgaard IH, Berg SF, Orstavik S, Fossum S, Dissen E. Identification of a human member of the Ly-49 multigene family. Eur J Immunol. 1998;28:1839–1846. doi: 10.1002/(SICI)1521-4141(199806)28:06<1839::AID-IMMU1839>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 25.Hoelsbrekken SE, Nylenna Ø, Saether PC, Slettedal IO, Ryan JC, Fossum S, et al. Cutting edge: molecular cloning of a killer cell Ig-like receptor in the mouse and rat. J Immunol. 2003;170:2259–2263. doi: 10.4049/jimmunol.170.5.2259. [DOI] [PubMed] [Google Scholar]
- 26.Welch AY, Kasahara M, Spain LM. Identification of the mouse killer immunoglobulin-like receptor-like (Kirl) gene family mapping to chromosome X. Immunogenetics. 2003;54:782–790. doi: 10.1007/s00251-002-0529-6. [DOI] [PubMed] [Google Scholar]
- 27.Roos C, Schmitz J, Zischler H. Primate jumping genes elucidate strepsirrhine phylogeny. Proc Natl Acad Sci USA. 2004;101:10651–10654. doi: 10.1073/pnas.0403852101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borrego F, Ulbrecht M, Weiss EH, Coligan JE, Brooks AG. Recognition of human histocompatibility leukocyte antigen (HLA)-E complexed with HLA class I signal sequence-derived peptides by CD94/NKG2 confers protection from natural killer cell-mediated lysis. J Exp Med. 1998;187:813–818. doi: 10.1084/jem.187.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee N, Llano M, Carretero M, Ishitani A, Navarro F, Lòpez-Botet M, et al. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc Natl Acad Sci USA. 1998;95:5199–5204. doi: 10.1073/pnas.95.9.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carretero M, Cantoni C, Bellón T, Bottino C, Biassoni R, Rodríguez A, et al. The CD94 and NKG2-A C-type lectins covalently assamble to form a a NK cell inhibitory receptor for HLA class I molecules. Eur J Immunol. 1997;27:563–567. doi: 10.1002/eji.1830270230. [DOI] [PubMed] [Google Scholar]
- 31.Braud VM, Allan DSJ, O'Callaghan CA, Soderstrom K, D'Andrea A, Ogg GS, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 32.Vance RE, Kraft JR, Altman JD, Jensen PE, Raulet DH. Mouse CD94/NKG2A is a natural killer cell receptor for the nonclassical major histocompatibility complex (MHC) class I molecule Qa-1(b) J Exp Med. 1998;188:1841–1848. doi: 10.1084/jem.188.10.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rajalingam R, Parham P, Abi-Rached L. Domain shuffling has been the main mechanism forming new hominoid killer cell Ig-like receptors. J Immunol. 2004;172:356–369. doi: 10.4049/jimmunol.172.1.356. [DOI] [PubMed] [Google Scholar]
- 34.Schneider H, Schneider MP, Sampaio I, Harada ML, Stanhope M, Czelusniak J, et al. Molecular phylogeny of the new world monkeys (Platyrrhini primates) Mol Phylogenet Evol. 1993;2:225–242. doi: 10.1006/mpev.1993.1022. [DOI] [PubMed] [Google Scholar]
- 35.Cadavid LF, Lun CM. Lineage-specific diversification of killer cell Ig-like receptors in the owl monkey, a new world primate. Immunogenetics. 2009;61:27–41. doi: 10.1007/s00251-008-0342-y. [DOI] [PubMed] [Google Scholar]
- 36.Ciccone E, Pende D, Viale O, Di Donato C, Orengo AM, Biassoni R, et al. Involvement of HLA class I alleles in NK cell specific functions: expression of HLA-Cw3 confers selective protection from lysis by alloreactive NK clones displaying a defined specificity (specificity 2) J Exp Med. 1992;176:963–971. doi: 10.1084/jem.176.4.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagtmann N, Biassoni R, Cantoni C, Verdiani S, Malnati M, Vitale M, et al. Molecular clones of the p58 natural killer cell receptor reveal immunoglobulin-related molecules with diversity in both the extra- and intracellular domains. Immunity. 1995;2:439–449. doi: 10.1016/1074-7613(95)90025-x. [DOI] [PubMed] [Google Scholar]
- 38.Colonna M, Samaridis J. Cloning of immunoglobulin-superfamily members associated with HLA-C and HLA-B recognition by human natural killer cells. Science. 1995;268:405–408. doi: 10.1126/science.7716543. [DOI] [PubMed] [Google Scholar]
- 39.Biassoni R, Falco M, Cambiaggi A, Costa P, Verdiani S, Pende D, et al. Single aminoacidic substitutions can influence the NK-mediated recognition of HLA-C molecules. Role of serine-77 and lysine-80 in the target cell protection from lysis mediated by “group 2” or “group 1” NK clones. J Exp Med. 1995;182:605–609. doi: 10.1084/jem.182.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biassoni R, Cantoni C, Falco M, Verdiani S, Bottino C, Vitale M, et al. The human leukocyte antigen (HLA)-C-specific “activatory” or “inhibitory” natural killer cell receptors display highly homologous extracellular domains but differ in their transmembrane and intracytoplasmic portions. J Exp Med. 1996;183:645–650. doi: 10.1084/jem.183.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moretta A, Bottino C, Vitale M, Pende D, Biassoni R, Mingari MC, Moretta L. Receptors for HLA class I molecules in human natural killer cells. Annu Rev Immunol. 1996;14:619–648. doi: 10.1146/annurev.immunol.14.1.619. [DOI] [PubMed] [Google Scholar]
- 42.Pende D, Biassoni R, Cantoni C, Verdiani S, Falco M, Di Donato C, et al. The natural killer cell receptor specific for HLA-A allotypes: a novel member of the p58/p70 family of inhibitory receptors which is characterized by three ig-like domains and is expressed as a 140 kd disulphide-linked dimer. J Exp Med. 1996;184:505–518. doi: 10.1084/jem.184.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bottino C, Sivori S, Vitale M, Cantoni C, Falco M, Pende D, et al. A novel surface molecule homologous to the p58/p50 family of receptors is selectively expressed on a subset of human natural killer cells and induces both triggering of cell functions and proliferation. Eur J Immunol. 1996;26:1816–1824. doi: 10.1002/eji.1830260823. [DOI] [PubMed] [Google Scholar]
- 44.Biassoni R, Pessino A, Malaspina A, Cantoni C, Bottino C, Sivori S, et al. Role of amino acid position 70 in the binding affinity of p50.1 and p58.1 receptors for HLA-Cw4 molecules. Eur J Immunol. 1997;27:3095–3099. doi: 10.1002/eji.1830271203. [DOI] [PubMed] [Google Scholar]
- 45.Cantoni C, Verdiani S, Falco M, Pessino A, Cilli M, Conte R, et al. p49, a novel putative HLA-class I-specific inhibitory NK receptor belonging to the immunoglobulin superfamily. Eur J Immunol. 1998;28:1980–1990. doi: 10.1002/(SICI)1521-4141(199806)28:06<1980::AID-IMMU1980>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 46.Deng L, Mariuzza RA. Structural basis for recognition of MHC and MHC-like ligands by natural killer cell receptors. Semin Immunol. 2006;18:159–166. doi: 10.1016/j.smim.2006.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boyington JC, Motyka SA, Schuck P, Brooks AG, Sun PD. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature. 2000;405:537–543. doi: 10.1038/35014520. [DOI] [PubMed] [Google Scholar]
- 48.Cosman D, Fanger N, Borges L, Kubin M, Chin W, Peterson L, Hsu ML. A novel immunoglobulin superfamily receptor for cellular and viral MHC class I molecules. Immunity. 1997;7:273–282. doi: 10.1016/s1074-7613(00)80529-4. [DOI] [PubMed] [Google Scholar]
- 49.Borges L, Hsu M-L, Fanger N, Kubin M, Cosman D. A family of human lymphoid and myeloid Ig-like receptors, some of which bind to MHC class I molecules. J Immunol. 1997;159:5192–5196. [PubMed] [Google Scholar]
- 50.Colonna M, Navarro F, Bellón T, Llano M, Pilar G, Samaridis J, et al. A common inhibitory receptor for major histocompatibility complex class I molecules on human lymphoid and myelomonocytic cells. J Exp Med. 1997;186:1809–1818. doi: 10.1084/jem.186.11.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vitale M, Castriconi R, Parolini S, Pende D, Hsu ML, Moretta L, et al. The leukocyte Ig-like receptor (LIR)-1 for the cytomegalovirus UL18 protein displays a broad specificity for different HLA class I alleles: Analysis of LIR-1+ NK cell clones. Int Immunol. 1999;11:29–35. doi: 10.1093/intimm/11.1.29. [DOI] [PubMed] [Google Scholar]
- 52.Chapman TL, Heikeman AP, Bjorkman PJ. The inhibitory receptor LIR-1 uses a common binding interaction to recognize class I MHC molecules and the viral homolog UL18. Immunity. 1999;11:603–613. doi: 10.1016/s1074-7613(00)80135-1. [DOI] [PubMed] [Google Scholar]
- 53.Shiroishi M, Kuroki K, Rasubala L, Tsumoto K, Kumagai I, Kurimoto E, et al. Structural basis for recognition of the nonclassical MHC molecule HLA-G by the leukocyte Ig-like receptor B2 (LILRB2/LIR2/ILT4/CD85d) Proc Natl Acad Sci USA. 2006;103:16412–16417. doi: 10.1073/pnas.0605228103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willcox BE, Thomas LM, Bjorkman PJ. Crystal structure of HLA-A2 bound to LIR-1, a host and viral major histocompatibility complex receptor. Nat Immunol. 2003;4:913–919. doi: 10.1038/ni961. [DOI] [PubMed] [Google Scholar]
- 55.Chen Y, Shi Y, Cheng H, An YQ, Gao GF. Structural immunology and crystallography help immunologists see the immune system in action: how T and NK cells touch their ligands. IUBMB Life. 2009;61:579–590. doi: 10.1002/iub.208. [DOI] [PubMed] [Google Scholar]
- 56.Llano M, Lee N, Navarro F, García P, Albar JP, Geraghty DE, López-Botet M. HLA-E-bound peptides influence recognition by inhibitory and triggering CD94/NKG2 receptors: preferential response to an HLA-G-derived nonamer. Eur J Immunol. 1998;28:2854–2863. doi: 10.1002/(SICI)1521-4141(199809)28:09<2854::AID-IMMU2854>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 57.Petrie EJ, Clements CS, Lin J, Sullivan LC, Johnson D, Huyton T, et al. CD94-NKG2A recognition of human leukocyte antigen (HLA)-E bound to an HLA class I leader sequence. J Exp Med. 2008;205:725–735. doi: 10.1084/jem.20072525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Birch J, Ellis SA. Complexity in the cattle CD94/NKG2 gene families. Immunogenetics. 2007;59:273–280. doi: 10.1007/s00251-006-0189-z. [DOI] [PubMed] [Google Scholar]
- 59.Cantoni C, Biassoni R, Pende D, Sivori S, Accame L, Pareti L, et al. The activating form of CD94 receptor complex: CD94 covalently associates with the Kp39 protein that represents the product of the NKG2-C gene. Eur J Immunol. 1998;28:327–338. doi: 10.1002/(SICI)1521-4141(199801)28:01<327::AID-IMMU327>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 60.Biassoni R. Natural killer cell receptors. Adv Exp Med Biol. 2008;640:35–52. doi: 10.1007/978-0-387-09789-3_4. [DOI] [PubMed] [Google Scholar]
- 61.Abi-Rached L, Parham P. Natural selection drives recurrent formation of activating killer cell immunoglobulin-like receptor and Ly49 from inhibitory homologues. J Exp Med. 2005;201:1319–1332. doi: 10.1084/jem.20042558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sambrook JG, Bashirova A, Palmer S, Sims S, Trowsdale J, Abi-Rached L, et al. Single haplotype analysis demonstrates rapid evolution of the killer immunoglobulin-like receptor (KIR) loci in primates. Genome Res. 2005;15:25–35. doi: 10.1101/gr.2381205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berg SF, Dissen E, Westgaard IH, Fossum S. Two genes in the rat homologous to human NKG2. Eur J Immunol. 1998;28:444–450. doi: 10.1002/(SICI)1521-4141(199802)28:02<444::AID-IMMU444>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 64.Silver ET, Lau JC, Kane KP. Molecular cloning of mouse NKG2A and C. Immunogenetics. 1999;49:727–730. doi: 10.1007/s002510050674. [DOI] [PubMed] [Google Scholar]
- 65.Biassoni R, Fogli M, Cantoni C, Costa P, Conte R, Koopman G, et al. Molecular and functional characterization of NKG2D, NKp80 and NKG2C triggering NK cell receptors in Rhesus and Cynomolgus macaques: monitoring of NK cell function during SHIV infection. J Immunol. 2005;174:5695–5705. doi: 10.4049/jimmunol.174.9.5695. [DOI] [PubMed] [Google Scholar]
- 66.Shum BP, Flodin LR, Muir DG, Rajalingam R, Khakoo SI, Cleland S, et al. Conservation and variation in human and common chimpanzee CD94 and NKG2 genes. J Immunol. 2002;168:240–252. doi: 10.4049/jimmunol.168.1.240. [DOI] [PubMed] [Google Scholar]
- 67.Kosiol C, Vinar T, da Fonseca RR, Hubisz MJ, Bustamante CD, Nielsen R, Siepel A. Patterns of positive selection in six mammalian genomes. PLoS Genet. 2008;4:1000144. doi: 10.1371/journal.pgen.1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pessino A, Sivori S, Bottino C, Malaspina A, Morelli L, Moretta L, et al. Molecular cloning of NKp46: a novel member of the immunoglobulin superfamily involved in triggering of natural cytotoxicity. J Exp Med. 1998;188:953–960. doi: 10.1084/jem.188.5.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moretta A, Bottino C, Vitale M, Pende D, Cantoni C, Biassoni R, et al. Activating receptors and co-receptors involved in the natural cytotoxicity. Annu Rev Immunol. 2001;19:197–223. doi: 10.1146/annurev.immunol.19.1.197. [DOI] [PubMed] [Google Scholar]
- 70.Biassoni R, Pessino A, Bottino C, Pende D, Moretta L, Moretta A. The murine homologue of the human NKp46, a triggering receptor involved in the induction of natural cytotoxicity. Eur J Immunol. 1999;29:1014–1020. doi: 10.1002/(SICI)1521-4141(199903)29:03<1014::AID-IMMU1014>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 71.Falco M, Cantoni C, Bottino C, Moretta A, Biassoni R. Identification of the rat homologue of the human NKp46 triggering receptor. Immunol Lett. 1999;68:411–414. doi: 10.1016/s0165-2478(99)00052-8. [DOI] [PubMed] [Google Scholar]
- 72.De Maria A, Biassoni R, Fogli M, Rizzi M, Cantoni C, Costa P, et al. Identification, molecular cloning and functional characterization of NKp46 and NKp30 natural cytotoxicity receptors in Macaca fascicularis (Macaca Rhesus) NK cells. Eur J Immunol. 2001;31:3546–3556. doi: 10.1002/1521-4141(200112)31:12<3546::aid-immu3546>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 73.Storset AK, Slettedal IO, Williams JL, Law A, Dissen E. Natural killer cell receptors in cattle: A bovine killer cell immunoglobulin-like receptor multigene family contains members with divergent signaling motifs. Eur J Immunol. 2003;33:980–990. doi: 10.1002/eji.200323710. [DOI] [PubMed] [Google Scholar]
- 74.Westgaard IH, Berg SF, Vaage JT, Wang LL, Yokoyama WM, Dissen E, Fossum S. Rat NKp46 activates natural killer cell cytotoxicity and is associated with FcepsilonRIgamma and CD3zeta. J Leukoc Biol. 2004;76:1200–1206. doi: 10.1189/jlb.0903428. [DOI] [PubMed] [Google Scholar]
- 75.Walzer T, Bléry M, Chaix J, Fuseri N, Chasson L, Robbins SH, et al. Identification, activation and selective in vivo ablation of mouse NK cells via NKp46. Proc Natl Acad Sci USA. 2007;104:3384–3389. doi: 10.1073/pnas.0609692104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rutjens E, Mazza S, Biassoni R, Koopman G, Radic' L, Fogli M, et al. Differential NKp30 inducibility is associated with conserved NK cell phenotype and function in AIDS resistant chimpanzees. J Immunol. 2007;178:1702–1712. doi: 10.4049/jimmunol.178.3.1702. [DOI] [PubMed] [Google Scholar]
- 77.Boysen P, Olsen I, Berg I, Kulberg S, Johansen GM, Storset AK. Bovine CD2−/NKp46+ cells are fully functional natural killer cells with a high activation status. BMC Immunol. 2006;7:10. doi: 10.1186/1471-2172-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ponassi M, Cantoni C, Biassoni R, Conte R, Spallarossa A, Pesce A, et al. Structure of the human NK cell triggering receptor NKp46 ectodomain. Biochem Biophys Res Commun. 2003;309:317–323. doi: 10.1016/j.bbrc.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 79.Sivori S, Pende D, Bottino C, Marcenaro E, Pessino A, Biassoni R, et al. NKp46 is the major triggering receptor involved in the natural cytotoxicity of fresh or cultured human NK cells. Correlation between surface density of NKp46 and natural cytotoxicity against autologous, allogeneic or xenogeneic target cells. Eur J Immunol. 1999;29:1656–1666. doi: 10.1002/(SICI)1521-4141(199905)29:05<1656::AID-IMMU1656>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 80.Cantoni C, Bottino C, Vitale M, Pessino A, Augugliaro R, Malaspina A, et al. NKp44, a triggering receptor involved in tumor cell lysis by activated human natural killer cells, is a novel member of the immunoglobulin superfamily. J Exp Med. 1999;189:787–796. doi: 10.1084/jem.189.5.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Allcock RJ, Barrow AD, Forbes S, Beck S, Trowsdale J. The human TREM gene cluster at 6p21.1 encodes both activating and inhibitory single IgV domain receptors and includes NKp44. Eur J Immunol. 2003;33:567–577. doi: 10.1002/immu.200310033. [DOI] [PubMed] [Google Scholar]
- 82.De Maria A, Ugolotti E, Rutjens E, Mazza S, Radic' L, Faravelli A, et al. NKp44 expression, phylogenesis and function in non-human primate NK cells. Int J Immunol. 2009;21:245–255. doi: 10.1093/intimm/dxn144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cantoni C, Ponassi M, Biassoni R, Conte R, Spallarossa A, Moretta A, et al. The three-dimensional structure of the human NK cell receptor NKp44, a triggering partner in natural cytotoxicity. Structure. 2003;11:725–734. doi: 10.1016/s0969-2126(03)00095-9. [DOI] [PubMed] [Google Scholar]
- 84.Grimm EA, Robb RJ, Roth JA, Neckers LM, Lachman LB, Wilson DJ, et al. Lymphokine-activated killer cell phenomenon III. Evidence that IL-2 is sufficient for direct activation of peripheral blood lymphocytes into lymphokine-activated killer cells. J Exp Med. 1983;158:1356–1361. doi: 10.1084/jem.158.4.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Byrd A, Hoffmann SC, Jarahian M, Momburg F, Watzl C. Expression analysis of the ligands for the natural killer cell receptors NKp30 and NKp44. PLoS ONE. 2007;2:1339. doi: 10.1371/journal.pone.0001339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Esin S, Batoni G, Counoupas C, Stringaro A, Brancatisano FL, Colone M, et al. Direct binding of human NK cell natural cytotoxicity receptor NKp44 to the surfaces of mycobacteria and other bacteria. Infect Immun. 2008;76:1719–1727. doi: 10.1128/IAI.00870-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hershkovitz O, Jivov S, Bloushtain N, Zilka A, Landau G, Bar-Ilan A, et al. Characterization of the recognition of tumor cells by the natural cytotoxicity receptor, NKp44. Biochemistry. 2007;46:7426–7436. doi: 10.1021/bi7000455. [DOI] [PubMed] [Google Scholar]
- 88.Arnon TI, Lev M, Katz G, Chernobrov Y, Porgador A, Mandelboim O. Recognition of viral hemagglutinins by NKp44 but not by NKp30. Eur J Immunol. 2001;31:2680–2689. doi: 10.1002/1521-4141(200109)31:9<2680::aid-immu2680>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 89.Arnon TI, Achdout H, Lieberman N, Gazit R, Gonen-Gross T, Katz G, et al. The mechanisms controlling the recognition of tumor- and virus-infected cells by NKp46. Blood. 2004;103:664–672. doi: 10.1182/blood-2003-05-1716. [DOI] [PubMed] [Google Scholar]
- 90.Mandelboim O, Lieberman N, Lev M, Paul L, Arnon TI, Bushkin Y, et al. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature. 2001;409:1055–1060. doi: 10.1038/35059110. [DOI] [PubMed] [Google Scholar]
- 91.Gazit R, Gruda R, Elboim M, Arnon TI, Katz G, Achdout H, et al. Lethal influenza infection in the absence of the natural killer cell receptor gene Ncr1. Nat Immunol. 2006;7:517–523. doi: 10.1038/ni1322. [DOI] [PubMed] [Google Scholar]
- 92.Ho JW, Hershkovitz O, Peiris M, Zilka A, Bar-Ilan A, Nal B, et al. H5-type influenza virus hemagglutinin is functionally recognized by the natural killer-activating receptor NKp44. J Virol. 2008;82:2028–2032. doi: 10.1128/JVI.02065-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pende D, Parolini S, Pessino A, Sivori S, Augugliaro R, Morelli L, et al. Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. J Exp Med. 1999;190:1505–1516. doi: 10.1084/jem.190.10.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Backman-Petersson E, Miller JR, Hollyoake M, Aguado B, Butcher GW. Molecular characterization of the novel rat NK receptor 1C7. Eur J Immunol. 2003;33:342–351. doi: 10.1002/immu.200310008. [DOI] [PubMed] [Google Scholar]
- 95.Hollyoake M, Campbell RD, Aguado B. NKp30 (NCR3) is a pseudogene in 12 inbred and wild mouse strains, but an expressed gene in Mus caroli. Mol Biol Evol. 2005;22:1661–1672. doi: 10.1093/molbev/msi162. [DOI] [PubMed] [Google Scholar]
- 96.Trowsdale J, Barten R, Haude A, Stewart CA, Beck S, Wilson MJ. The genomic context of natural killer receptor extended gene families. Immunol Rev. 2001;181:20–38. doi: 10.1034/j.1600-065x.2001.1810102.x. [DOI] [PubMed] [Google Scholar]
- 97.Call ME, Wucherpfennig KW. Common themes in the assembly and architecture of activating immune receptors. Nat Rev Immunol. 2007;7:841–850. doi: 10.1038/nri2186. [DOI] [PubMed] [Google Scholar]
- 98.Spaggiari GM, Carosio R, Pende D, Marcenaro S, Rivera P, Zocchi MR, et al. NK cell-mediated lysis of autologous antigen-presenting cells is triggered by the engagement of the phosphatidylinositol 3-kinase upon ligation of the natural cytotoxicity receptors NKp30 and NKp46. Eur J Immunol. 2001;31:1656–1665. doi: 10.1002/1521-4141(200106)31:6<1656::aid-immu1656>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 99.Brandt CS, Baratin M, Yi EC, Kennedy J, Gao Z, Fox B, et al. The B7 family member B7-H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. J Exp Med. 2009;206:1495–1503. doi: 10.1084/jem.20090681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pogge von Strandmann E, Simhadri VR, von Tresckow B, Sasse S, Reiners KS, Hansen HP, et al. Human leukocyte antigen-B-associated transcript 3 is released from tumor cells and engages the NKp30 receptor on natural killer cells. Immunity. 2007;27:965–974. doi: 10.1016/j.immuni.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 101.Arnon TI, Achdout H, Levi O, Markel G, Saleh N, Katz G, et al. Inhibition of the NKp30 activating receptor by pp65 of human cytomegalovirus. Nat Immunol. 2005;6:515–523. doi: 10.1038/ni1190. [DOI] [PubMed] [Google Scholar]
- 102.Moretta A, Marcenaro E, Sivori S, Della Chiesa M, Vitale M, Moretta L. Early liaisons between cells of the innate immune system in inflamed peripheral tissues. Trends Immunol. 2005;26:668–675. doi: 10.1016/j.it.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 103.Ferlazzo G, Semino C, Melioli G. HLA class I molecule expression is upregulated during maturation of dendritic cells, protecting them from natural killer cell-mediated lysis. Immunol Lett. 2001;76:37–41. doi: 10.1016/s0165-2478(00)00323-0. [DOI] [PubMed] [Google Scholar]
- 104.Ferlazzo G, Tsang ML, Moretta L, Melioli G, Steinman RM, Münz C. Human dendritic cells activate resting natural killer (NK) cells and are recognized via the NKp30 receptor by activated NK cells. J Exp Med. 2002;195:343–351. doi: 10.1084/jem.20011149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ferlazzo G. Natural killer and dendritic cell liaison: Recent insights and open questions. Immunol Lett. 2005;101:12–17. doi: 10.1016/j.imlet.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 106.Vitale M, Della Chiesa M, Carlomagno S, Pende D, Aricò M, Moretta L, et al. NK-dependent DC maturation is mediated by TNFalpha and IFNgamma released upon engagement of the NKp30 triggering receptor. Blood. 2005;106:566–571. doi: 10.1182/blood-2004-10-4035. [DOI] [PubMed] [Google Scholar]
- 107.Houchins JP, Yabe T, McSherry C, Bach FH. DNA sequence analysis of NKG2, a family of related cDNA clones encoding type II integral membrane proteins on human natural killer cells. J Exp Med. 1991;173:1017–1020. doi: 10.1084/jem.173.4.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vance RE, Tanamachi DM, Hanke T, Raulet DH. Cloning of a mouse homolog of CD94 extends the family of C-type lectins on murine natural killer cells. Eur J Immunol. 1997;27:3236–3241. doi: 10.1002/eji.1830271222. [DOI] [PubMed] [Google Scholar]
- 109.Berg SF, Dissen E, Westgaard IH, Fossum S. Molecular characterization of rat NKR-P2, a lectin-like receptor expressed by NK cells and resting T cells. Int Immunol. 1998;10:379–385. doi: 10.1093/intimm/10.4.379. [DOI] [PubMed] [Google Scholar]
- 110.Govaerts MM, Goddeeris BM. Homologues of natural killer cell receptors NKG2-D and NKR-P1 expressed in cattle. Vet Immunol Immunopathol. 2001;80:339–344. doi: 10.1016/s0165-2427(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 111.Yim D, Jie HB, Sotiriadis J, Kim YS, Kim KS, Rothschild MF, et al. Molecular cloning and characterization of pig immunoreceptor DAP10 and NKG2D. Immunogenetics. 2001;53:243–249. doi: 10.1007/s002510100321. [DOI] [PubMed] [Google Scholar]
- 112.Watzl C. The NKG2D receptor and its ligands recognition beyond the “missing self”? Microbes Infect. 2003;5:31–37. doi: 10.1016/s1286-4579(02)00057-6. [DOI] [PubMed] [Google Scholar]
- 113.Garrity D, Call ME, Feng J, Wucherpfennig KW. The activating NKG2D receptor assembles in the membrane with two signaling dimers into a hexameric structure. Proc Natl Acad Sci USA. 2005;102:7641–7646. doi: 10.1073/pnas.0502439102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chang C, Dietrich J, Harpur AG, Lindquist JA, Haude A, Loke YW, et al. Cutting edge: KAP10, a novel transmembrane adapter protein genetically linked to DAP12 but with unique signaling properties. J Immunol. 1999;163:4651. [PubMed] [Google Scholar]
- 115.Wu J, Song Y, Bakker AB, Bauer S, Spies T, Lanier LL, et al. An activating receptor complex on natural killer and T cells formed by NKG2D and DAP10. Science. 1999;285:730–732. doi: 10.1126/science.285.5428.730. [DOI] [PubMed] [Google Scholar]
- 116.Wu J, Cherwinski H, Spies T, Phillips JH, Lanier LL. DAP10 and DAP12 form distinct, but functionally cooperative, receptor complexes in natural killer cells. J Exp Med. 2000;192:1059–1068. doi: 10.1084/jem.192.7.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Groh V, Bahram S, Bauer S, Herman A, Beauchamp M, Spies T. Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc Natl Acad Sci USA. 1996;93:12445–12450. doi: 10.1073/pnas.93.22.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Groh V, Rhinehart R, Secrist H, Bauer S, Grabstein KH, Spies T. Broad tumor-associated expression and recognition by tumor-derived gamma delta T cells of MICA and MICB. Proc Natl Acad Sci USA. 1999;96:6879–6884. doi: 10.1073/pnas.96.12.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bauer S, Groh V, Wu J, Steinle A, Phillips JH, Lanier LL, et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285:727–729. doi: 10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- 120.Bahram S, Bresnahan M, Geraghty DE, Spies T. A second lineage of mammalian major histocompatibility complex class I genes. Proc Natl Acad Sci USA. 1994;91:6259–6263. doi: 10.1073/pnas.91.14.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bahram S. MIC genes: From genetics to biology. Adv Immunol. 2000;76:1–60. doi: 10.1016/s0065-2776(01)76018-x. [DOI] [PubMed] [Google Scholar]
- 122.Gleimer M, Parham P. Stress management: MHC class I and class I-like molecules as reporters of cellular stress. Immunity. 2003;19:469–477. doi: 10.1016/s1074-7613(03)00272-3. [DOI] [PubMed] [Google Scholar]
- 123.Stern-Ginossar N, Mandelboim O. An integrated view of the regulation of NKG2D ligands. Immunology. 2009;128:1–6. doi: 10.1111/j.1365-2567.2009.03147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Seo JW, Bontrop R, Walter L, Günther E. Major histocompatibility complex-linked MIC genes in rhesus macaques and other primates. Immunogenetics. 1999;50:358–362. doi: 10.1007/s002510050614. [DOI] [PubMed] [Google Scholar]
- 125.Cosman D, Mullberg J, Sutherland CL, Chin W, Armitage R, Fanslow W, et al. ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity. 2001;14:123–133. doi: 10.1016/s1074-7613(01)00095-4. [DOI] [PubMed] [Google Scholar]
- 126.Radosavljevic M, Cuillerier B, Wilson MJ, Clement O, Wicker S, Gilfillan S, et al. A cluster of ten novel MHC class I related genes on human chromosome 6q24.2-q25.3. Genomics. 2002;79:114–123. doi: 10.1006/geno.2001.6673. [DOI] [PubMed] [Google Scholar]
- 127.Chalupny NJ, Sutherland CL, Lawrence WA, Rein-Weston A, Cosman D. ULBP4 is a novel ligand for human NKG2D. Biochem Biophys Res Commun. 2003;305:129–135. doi: 10.1016/s0006-291x(03)00714-9. [DOI] [PubMed] [Google Scholar]
- 128.Bacon L, Eagle RA, Meyer M, Easom N, Young NT, Trowsdale J. Two human ULBP/RAET1 molecules with transmembrane regions are ligands for NKG2D. J Immunol. 2004;173:1078–1084. doi: 10.4049/jimmunol.173.2.1078. [DOI] [PubMed] [Google Scholar]
- 129.López-Larrea C, Suárez-Alvarez B, López-Soto A, López-Vázquez A, Gonzalez S. The NKG2D receptor: sensing stressed cells. Trends Mol Med. 2008;14:179–189. doi: 10.1016/j.molmed.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 130.Nomura M, Zou Z, Joh T, Takihara Y, Matsuda Y, Shimada K. Genomic structures and characterization of Rae1 family members encoding GPI-anchored cell surface proteins and expressed predominantly in embryonic mouse brain. J Biochem. 1996;120:987–995. doi: 10.1093/oxfordjournals.jbchem.a021517. [DOI] [PubMed] [Google Scholar]
- 131.Zou Z, Nomura M, Takihara Y, Yasunaga T, Shimada K. Isolation and characterization of retinoic acid-inducible cDNA clones in F9 cells: a novel cDNA family encodes cell surface proteins sharing partial homology with MHC class I molecules. J Biochem. 1996;119:319–328. doi: 10.1093/oxfordjournals.jbchem.a021242. [DOI] [PubMed] [Google Scholar]
- 132.Malarkannan S, Shih PP, Eden PA, Horng T, Zuberi AR, Christianson G, et al. The molecular and functional characterization of a dominant minor H antigen, H60. J Immunol. 1998;161:3501–3509. [PubMed] [Google Scholar]
- 133.Carayannopoulos LN, Naidenko OV, Fremont DH, Yokoyama WM. Cutting edge: Murine UL16-binding protein-like transcript 1: A newly described transcript encoding a high-affinity ligand for murine NKG2D. J Immunol. 2002;169:4079–4083. doi: 10.4049/jimmunol.169.8.4079. [DOI] [PubMed] [Google Scholar]
- 134.Cerwenka A, Bakker AB, McClanahan T, Wagner J, Wu J, Phillips JH, Lanier LL. Retinoic acid early inducible genes define a ligand family for the activating NKG2D receptor in mice. Immunity. 2000;12:721–727. doi: 10.1016/s1074-7613(00)80222-8. [DOI] [PubMed] [Google Scholar]
- 135.Diefenbach A, Jensen ER, Jamieson AM, Raulet DH. Rae1 and H60 ligands of the NKG2D receptor stimulate tumour immunity. Nature. 2001;413:165–171. doi: 10.1038/35093109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Diefenbach A, Hsia JK, Hsiung MY, Raulet DH. A novel ligand for the NKG2D receptor activates NK cells and macrophages and induces tumor immunity. Eur J Immunol. 2003;33:381–391. doi: 10.1002/immu.200310012. [DOI] [PubMed] [Google Scholar]
- 137.Li P, Morris DL, Willcox BE, Steinle A, Spies T, Strong RK. Complex structure of the activating immunoreceptor NKG2D and its MHC class I-like ligand MICA. Nat Immunol. 2001;2:443–451. doi: 10.1038/87757. [DOI] [PubMed] [Google Scholar]
- 138.Radaev S, Kattah M, Zou Z, Colonna M, Sun PD. Making sense of the diverse ligand recognition by NKG2D. J Immunol. 2002;169:6279–6285. doi: 10.4049/jimmunol.169.11.6279. [DOI] [PubMed] [Google Scholar]
- 139.McFarland BJ, Strong RK. Thermodynamic analysis of degenerate recognition by the NKG2D immunoreceptor: Not induced fit but rigid adaptation. Immunity. 2003;19:803–812. doi: 10.1016/s1074-7613(03)00320-0. [DOI] [PubMed] [Google Scholar]
- 140.Stephens HA. MICA and MICB genes: can the enigma of their polymorphism be resolved? Trends Immunol. 2001;22:378–385. doi: 10.1016/s1471-4906(01)01960-3. [DOI] [PubMed] [Google Scholar]
- 141.Bahram S, Inoko H, Shiina T, Shiina T, Radosavljevic M. MIC and other NKG2D ligands: from none to too many. Curr Opin Immunol. 2005;17:505–509. doi: 10.1016/j.coi.2005.07.016. [DOI] [PubMed] [Google Scholar]
- 142.Burgess SJ, Maasho K, Masilamani M, Narayanan S, Borrego F, Coligan JE. The NKG2D receptor: immunobiology and clinical implications. Immunol Res. 2008;40:18–34. doi: 10.1007/s12026-007-0060-9. [DOI] [PubMed] [Google Scholar]
- 143.Groh V, Steinle A, Bauer S, Spies T. Recognition of stress-induced MHC molecules by intestinal epithelial gammadelta T cells. Science. 1998;279:1737–1740. doi: 10.1126/science.279.5357.1737. [DOI] [PubMed] [Google Scholar]
- 144.Groh V, Rhinehart R, Randolph-Habecker J, Topp MS, Riddell SR, Spies T. Costimulation of CD8alphabeta T cells by NKG2D via engagement by MIC induced on virus-infected cells. Nat Immunol. 2001;2:255–260. doi: 10.1038/85321. [DOI] [PubMed] [Google Scholar]
- 145.Gasser S, Orsulic S, Brown EJ, Raulet DH. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 2005;436:1186–1190. doi: 10.1038/nature03884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Venkataraman GM, Suciu D, Groh V, Boss JM, Spies T. Promoter region architecture and transcriptional regulation of the genes for the MHC class I-related chain A and B ligands of NKG2D. J Immunol. 2007;178:961–969. doi: 10.4049/jimmunol.178.2.961. [DOI] [PubMed] [Google Scholar]
- 147.Seo JW, Walter L, Gunther E. Genomic analysis of MIC genes in rhesus macaques. Tissue Antigens. 2001;58:159–165. doi: 10.1034/j.1399-0039.2001.580303.x. [DOI] [PubMed] [Google Scholar]
- 148.Heeney JL, Dalgleish AG, Weiss RA. Origins of HIV and the evolution of resistance to AIDS. Science. 2006;313:462–466. doi: 10.1126/science.1123016. [DOI] [PubMed] [Google Scholar]
- 149.Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 150.Knapp S, Warshow U, Hegazy D, Brackenbury L, Guha IN, Fowell A, et al. Consistent beneficial effects of killer cell immunoglobulin-like receptor 2DL3 and group 1 human leukocyte antigen-C following exposure to hepatitis C virus. Hepatology. 2010;51:1168–1175. doi: 10.1002/hep.23477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Askar M, Avery R, Corey R, Lopez R, Thomas D, Pidwell D, et al. Lack of killer immunoglobulin-like receptor 2DS2 (KIR2DS2) and KIR2DL2 is associated with poor responses to therapy of recurrent hepatitis C virus in liver transplant recipients. Liver Transplan. 2009;15:1557–1563. doi: 10.1002/lt.21878. [DOI] [PubMed] [Google Scholar]
- 152.de Arias AE, Haworth SE, Belli LS, Burra P, Pinzello G, Vangeli M, et al. Killer cell immunoglobulin-like receptor genotype and killer cell immunoglobulin-like receptor-human leukocyte antigen C ligand compatibility affect the severity of hepatitis C virus recurrence after liver transplantation. Liver Transplan. 2009;15:390–399. doi: 10.1002/lt.21673. [DOI] [PubMed] [Google Scholar]
- 153.Boonstra A, van der Laan LJ, Vanwolleghem T, Janssen HL. Experimental models for hepatitis C viral infection. Hepatology. 2009;50:1646–1655. doi: 10.1002/hep.23138. [DOI] [PubMed] [Google Scholar]
- 154.Youn JW, Hu YW, Tricoche N, Pfahler W, Shata MT, Dreux M, et al. Evidence for protection against chronic hepatitis C virus infection in chimpanzees by immunization with replicating recombinant vaccinia virus. J Virol. 2008;82:10896–10905. doi: 10.1128/JVI.01179-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Moesta AK, Abi-Rached L, Norman PJ, Parham P. Chimpanzees use more varied receptors and ligands than humans for inhibitory killer cell Ig-like receptor recognition of the MHC-C1 and MHC-C2 epitopes. J Immunol. 2009;182:3628–3637. doi: 10.4049/jimmunol.0803401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ploss A, Rice CM. Towards a small animal model for hepatitis C. EMBO Rep. 2009;10:1220–1227. doi: 10.1038/embor.2009.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Vidal-Castiñeira JR, López-Vázquez A, Díaz-Peña R, Alonso-Arias R, Martínez-Borra J, Pérez R, et al. Effect of killer immunoglobulin-like receptors in the response to combined treatment in patients with chronic hepatitis C virus infection. J Virol. 84:475–481. doi: 10.1128/JVI.01285-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bonagura VR, Du Z, Ashouri E, Luo L, Hatam LJ, Devoti JA, et al. Activating killer cell immunoglobulin-like receptors 3DS1 and 2DS1 protect against developing the severe form of recurrent respiratory papillomatosis. Human Immunol. 2010;71:212–217. doi: 10.1016/j.humimm.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Paladino N, Flores AC, Marcos CY, Fainboim H, Theiler G, Arruvito L, et al. Increased frequencies of activating natural killer receptors are associated with liver injury in individuals who do not eliminate hepatitis C virus. Tissue Antigens. 2007;69:109–111. doi: 10.1111/j.1399-0039.2006.762_7.x. [DOI] [PubMed] [Google Scholar]
- 160.Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 161.Alter G, Martin MP, Teigen N, Carr WH, Suscovich TJ, Schneidewind A, et al. Differential natural killer cell mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med. 2007;204:3027–3036. doi: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Migueles SA, Sabbaghian MS, Shupert WL, Bettinotti MP, Marincola FM, Martino L, et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci USA. 2000;97:2709–2714. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.O'Connell KA, Han Y, Williams TM, Siliciano RF, Blankson JN. The role of natural killer cells in a cohort of elite suppressors: low frequency of the protective KIR3DS1 allele and limited inhibition of HIV-1 replication in vitro. J Virol. 2009;83:5028–5034. doi: 10.1128/JVI.02551-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.O'Connor GM, Holmes A, Mulcahy F, Gardiner CM. Natural Killer cells from long-term non-progressor HIV patients are characterized by altered phenotype and function. Clin Immunol. 2007;124:277–283. doi: 10.1016/j.clim.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 165.Pandrea I, Silvestri G, Apetrei C. AIDS in african non-human primate hosts of SIVs: a new paradigm of SIV infection. Curr HIV Res. 2009;7:57–72. doi: 10.2174/157016209787048456. [DOI] [PubMed] [Google Scholar]
- 166.Boberg A, Bråve A, Johansson S, Wahren B, Hinkula J. Rollman. Murine models for HIV vaccination and challenge. Expert Rev Vaccines. 2008;7:117–130. doi: 10.1586/14760584.7.1.117. [DOI] [PubMed] [Google Scholar]
- 167.Denton P, Garcia J. Novel humanized murine models for HIV research. Curr HIV/AIDS Rep. 2009;6:13–19. doi: 10.1007/s11904-009-0003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Baroncelli S, Negri DR, Michelini Z, Cara A. Macaca mulatta, fascicularis and nemestrina in AIDS vaccine development. Expert Rev Vaccines. 2008;7:1419–1434. doi: 10.1586/14760584.7.9.1419. [DOI] [PubMed] [Google Scholar]
- 169.Shedlock D, Silvestri G, Weiner D. Monkeying around with HIV vaccines: using rhesus macaques to define ‘gatekeepers’ for clinical trials. Nat Rev. 2009;9:717–728. doi: 10.1038/nri2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Vieillard V, Strominger JL, Debré P. NK cytotoxicity against CD4+ T cells during HIV-1 infection: a gp41 peptide induces the expression of an NKp44 ligand. Proc Natl Acad Sci USA. 2005;102:10981–10986. doi: 10.1073/pnas.0504315102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Fausther-Bovendo H, Sol-Foulon N, Candotti D, Agut H, Schwartz O, Debré P, Vieillard V. HIV escape from natural killer cytotoxicity: nef inhibits NKp44L expression on CD4+ T cells. AIDS. 2009;23:1077–1087. doi: 10.1097/QAD.0b013e32832cb26b. [DOI] [PubMed] [Google Scholar]
- 172.De Maria A, Fogli M, Costa P, Murdaca G, Puppo F, Mavilio D, et al. The impaired NK cell cytolytic function in viremic HIV-1 infection is associated with a reduced surface expression of natural cytotoxicity receptors (NKp46, NKp30 and NKp44) Eur J Immunol. 2003;33:2410–2418. doi: 10.1002/eji.200324141. [DOI] [PubMed] [Google Scholar]