Abstract
Motivation: Functional genomics research has expanded enormously in the last decade thanks to the cost reduction in high-throughput technologies and the development of computational tools that generate, standardize and share information on gene and protein function such as the Gene Ontology (GO). Nevertheless, many biologists, especially working with non-model organisms, still suffer from non-existing or low-coverage functional annotation, or simply struggle retrieving, summarizing and querying these data.
Results: The Blast2GO Functional Annotation Repository (B2G-FAR) is a bioinformatics resource envisaged to provide functional information for otherwise uncharacterized sequence data and offers data mining tools to analyze a larger repertoire of species than currently available. This new annotation resource has been created by applying the Blast2GO functional annotation engine in a strongly high-throughput manner to the entire space of public available sequences. The resulting repository contains GO term predictions for over 13.2 million non-redundant protein sequences based on BLAST search alignments from the SIMAP database. We generated GO annotation for approximately 150 000 different taxa making available 2000 species with the highest coverage through B2G-FAR. A second section within B2G-FAR holds functional annotations for 17 non-model organism Affymetrix GeneChips.
Conclusions: B2G-FAR provides easy access to exhaustive functional annotation for 2000 species offering a good balance between quality and quantity, thereby supporting functional genomics research especially in the case of non-model organisms.
Availability: The annotation resource is available at http://www.b2gfar.org.
Contact: aconesa@cipf.es; sgoetz@cipf.es
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Functional genomics research has gained importance in the last decade thanks to the fast improvement and cost reduction in high-throughput technologies. Beyond traditional model species, also non-model organisms are in the genomics race and today it is hard to find a biological domain without a functional genomics initiative. This expansion would not have been that successful without the accompanying development of computational tools that generate, standardize and share information on gene and protein function. The Gene Ontology (GO) project is one such standard. GO is a collaborative effort aiming at the establishment of a controlled vocabulary that provides biologically meaningful annotations for gene (products) across species (Ashburner et al., 2000). There are two main aspects of this project: (a) the definition of a comprehensive ontology of functional terms and (b) the generation of an annotation database containing the assignment of GO terms to genes and proteins (The Gene Ontology Consortium, 2008). Annotation for each organism in the GO database is supplied and maintained by the respective consortium member. Evidence codes (ECs) are added to each individual annotation to reflect the information source used in a GO term assignment. ECs indicate if the annotation is supported by some (and which) experimental evidence, whether it was transferred (and how) from related genes or if it was generated by other prediction methods. The large majority of GO annotations is centralized in the Gene Ontology Annotation (GOA) Database (Camon et al., 2004). This resource contains high-quality functional annotations for proteins mostly obtained from the UniProt Knowledgebase (The Uniprot Consortium, 2007). However, the great majority (∼95%) of GO corresponds to automatically transferred annotations based on InterProScan (Quevillon et al., 2005) results. Currently, only a small number of assignments have experimental evidence or are revised by expert curators. While the GO project is improving the ontology definition and quality of the GOA database (Barrell et al., 2009), it is still far from providing extensive functional annotation for the wealth of sequence data that populate public databases. However, thanks to the structured and universal nature of GO, large-scale annotation using an automated process is conceivable and could be feasible given that adequate computational resources are available. Such an annotation effort complement current established annotation initiatives by generating preliminary functional labels for sequences not yet covered by any of the reference projects of the GO consortium. Examples of functional data-intensive resources in the field of functional genomics such as the Integr8 (Kersey et al., 2005) and PEDANT (Riley, 1993) projects, which offer genome-oriented views on different types of annotation data, can be found. An example of a specialized resource in the area of functional plant genomics is the PlexDB (Wise et al., 2007), a project to support transcriptomics studies on plants and plant pathogens. Although these resources provide comprehensive information on many genomic aspects, they do not represent a universal resource where up-to-date large-scale functional data on species can be readily accessed and analyzed. The Blast2GO project started in 2005 to build an universal bioinformatics platform for the functional annotation and analysis of novel sequence data (Conesa et al., 2005). The tool was designed to provide high-throughput and quality assignments with a user interface strongly based on visualization and intuitive use (Götz et al., 2008) (Conesa and Götz, 2008). So far, Blast2GO has been used in nearly 300 functional profiling projects involving a wide range of model and non-model species, from bacteria and fungi, through arthropods and mollusks, to plants, avian, fish and mammals, which evidences the strong need for functional annotations beyond that provided by public sequence databases. During the last 5 years of Blast2GO, we have witnessed many cases of redundant annotation of public data by different labs and limitations for keeping results updated. These facts, in combination with the annotation gaps mentioned above, have encouraged us to consider the generation of an extensive repository of automatic functional annotation making use of the widely accepted functional annotation engine Blast2GO. Lastly advances in supercomputing and distributed computing have paved the way for undertaking highly intensive genome analysis tasks as demonstrated by many examples in different research fields such as proteomics (Marti-Renom et al., 2007), phylogenomics (Huerta-Cepas et al., 2007; Sjölander, 2004) and sequence analysis (Rattei et al., 2008). In this work, we have made use of these technologies to create the Functional Annotation Repository (B2G-FAR), a web-based large-scale resource of precomputed functional annotations. Generated by pipelines of intensive computing, B2G-FAR is a centralized repository of automatically annotated sequence data, which can directly be downloaded and used, saving processing time and avoiding redundant processing. The resource covers all biological kingdoms and is structured in such a way that easy download and processing of data for a species of interest is possible. The B2G-FAR follows the philosophy of Blast2GO development: (i) high throughput: over 25 million sequences have been processed and 2000 species are now present in the B2G-FAR database; (ii) the application to functional genomics projects: the repository contains GO term predictions for both, gene products and probe set collections of Affymetrix GeneChips; and (iii) user friendly: annotations are provided in the form of plain text files for direct download. The main benefit is obtained in the case of non-model species where functional information and bioinformatics resources are limited.
2 METHODS
2.1 Construction
The data contained in the B2G-FAR repository consists of two parts: species-centered functional annotations obtained through the Simap2GO project and a collection of microarray probe set annotations. Although generated by different pipelines, both data sources implement the same basic annotation scheme as established by Blast2GO (Götz et al., 2008): (i) sequence similarity search (BLAST) (Altschul et al., 1990) to obtain a list of potential homologues;, (ii) mapping of the corresponding GO terms and their evidence codes for all BLAST hits; and (iii) the annotation step applying the Blast2GO annotation rule taking into account GO evidence codes, the GO hierarchy and the degree of sequence similarity (an extensive explanation of the Blast2GO annotation rule can be found in the Supplementary Material). In a final step, protein domains, families and motifs are identified through InterProScan and the corresponding GO terms are merged with the already transferred annotations taking care of redundancies and GO hierarchy.
Simap2GO data and processing: Simap2GO is the result of the collaboration between the SIMAP (Rattei et al., 2008) and Blast2GO projects. SIMAP is a database first published in 2005 (Arnold et al., 2005), which provides an all-against-all sequence similarity matrix containing precalculated pairwise comparisons of almost all publicly available proteins. Computations are accomplished by volunteer grid computing based on the Berkeley Open Infrastructure for Network Computing (BOINC). By May 2010, SIMAP contained over 29 million non-redundant sequences covering the content of all major public sequence resources. The Simap2GO project, with the ambitious goal of functionally annotating this comprehensive sequence space, adopted and applied successfully the Blast2GO annotation methodology to the SIMAP resource. Blast2GO is primarily based on sequence similarity and therefore perfectly suited to be run on the SIMAP database.
The computation of the annotations is run in parallel for the entire sequence space. For each uncharacterized sequence, GO annotated (only non-electronic annotations) homologues are retrieved from the SIMAP sequence similarity matrix. The applied Blast2GO method selects the GO terms above a given confidence threshold according to the established annotation rule. Here, the algorithm takes into account the actual sequence similarity, the original GO evidence code and the location of the potentially transferred GO term within the hierarchy (see the Supplementary Material for a more detailed description of the Blast2GO annotation strategy and annotation parameters). Apart from the annotation transfer through potential homologues, Simap2GO makes use of the InterPro domain information also provided in a precalculated manner by SIMAP. This information is merged with the already predicted assignments to improve and increase the GO term assignment. The sequence annotations generated in this way are stored in the B2G-FAR database and the routine continues with the next unprocessed sequence until completion (see Fig. 1). One entire annotation process takes approximately 3 days with 10 CPUs running in parallel and with direct access to the SIMAP database. The annotation of the same amount of sequence data without precalculated alignments would have taken over half a year on a 150 CPU cluster. All annotations are further processed to summarize and present species-centered information online. Available charts and data files are given in the Supplementary Table S1.
Fig. 1.
The B2G-FAR annotation pipeline. The scheme shows how the different data-sources are related and contribute to the generation of the B2G-FAR annotations, all passing through the Blast2GO annotation algorithm.
Micorarray probe set data: the probe-set collection of 17 Affymetrix GeneChip designs corresponding to non-model species was annotated with Blast2GO. As GeneChip probe sets do not necessarily target protein sequences available in public databases, their functional annotation cannot be recovered by Simap2GO and therefore has been computed using local resources. FASTA files containing the target sequences of the probe sets were downloaded from the official Affymetrix web site. The annotation pipeline started by splitting source FASTA files into smaller chunks and launching them against a distributed BLAST setup. A 150 CPU cluster at the CIPF Bioinformatics and Genomics Department was used to run BLAST searches against the NCBI non-redundant (NR) database. Simultaneously, protein domain information was obtained through a local installation of InterProScan (Quevillon et al., 2005). Once BLAST and InterProScan searches were completed, results for every species were gathered and processed within Blast2GO for automatic function prediction. Charts were generated during the annotation process and are provided online in Supplementary Table S3. Finally, to assess the coverage of annotation results, each GeneChip was compared with the GO information provided by Affymetrix. All currently annotated and available datasets are listed in the Supplementary Table S2.
2.2 Contents
B2G-FAR presents contents in a user-friendly data sheet concept based on Wiki technology. All the given information (annotations, data files, images) is generated beforehand by the B2G-FAR annotation pipeline and is summarized on automatically generated web pages. This facilitates fast access to data files, images and charts describing genome-wide information. Annotation data can be further visualized and analyzed through its upload into the Blast2GO application (see below: Download and query options). B2G-FAR is periodically updated every 6 months.
Species annotations: by applying the above-described steps, we could assign GO terms to 14 million sequences that represents ∼56.4% of the entire SIMAP database (excluding metagenomic data). The remaining sequences are entries without significant alignments (35.7%) or that did not surpass the annotations quality threshold (7.7%). Sequences from ∼150 000 taxa were functionally annotated and the 2000 most represented species are now available through B2G-FAR. Table 1 contains the numbers of annotated sequences compared with the whole SIMAP dataset and the available source annotations by the GO. Species can be accessed directly by their scientific name or NCBI taxa ID through a search function. For every species, several precalculated files and statistical charts are available. These include a GO annotation flat file and its corresponding GO-Slim version. Statistical charts provide information about GO annotation distributions, GO level distributions or about the most abundant functional terms within one of the three GO categories.
Table 1.
Simap2GO annotation coverage: the table shows the number of Blast2GO-annotated sequences in relation to the whole SIMAP dataset (May 2010) and the number of GO sequences which has been used as annotation source/reference dataset
| Data source | Unique sequences |
|---|---|
| Whole Simap | 29 906 548 |
| Simap without metagenomes | 25 099 929 |
| Simap protein sequences annotated by Blast2GO | 14 175 984 |
| Sequences which do not surpass the annotation threshold | 1 938 862 |
| Sequences without sequence alignment | 8 985 083 |
| GO annotation source sequences (only sequences with non-electronic annotations) | 465 677 |
Only sequences with at least one non-electronic annotations (non-IEA) were used (GO-Lite data-set). Additionally, the number of sequences which could not be annotated is given, i.e. sequences without sequence alignments and sequences whose annotations did not surpass the annotation threshold.
Microarray annotations: This section is organized as annotation sheets for each probe-set collection corresponding to the 17 non-model Affymetrix GeneChips. Model species Affymetrix chips were purposely not included in the repository as there already exist extensive functional annotation projects. The annotation sheet contains detailed information on the Blast2GO annotation process from the BLAST step up to the augmentation by ANNEX [a data mining procedure to annotate from links between molecular function and biological process/cellular component GO terms (Myhre et al., 2006)] and InterProScan. In contrast to the previous section, which provides only final annotation records, the microarray probe-set annotation sheets include a great variety of descriptive charts that offer a comprehensive view of the functional information contents gathered throughout the annotation pipeline. Likewise, Blast2GO project files are provided. The charts and files included in the annotation sheets are listed in the Supplementary Table S3.
Download and query options: in both Species and Microarray sections, final annotation files are provided in plaintext format as GO and GO-Slim data. The text file format allows direct upload into the Blast2GO application for further analysis of annotation results as well as integration in other applications accepting GO annotation data. Additionally, all species annotations are available in the standard GO annotation format. Some descriptive charts are included in B2G-FAR for a quick overview of the results. Dynamic access to the data is provided by the Blast2GO Java application. This guarantees optimal reutilization and synchrony within Blast2GO developments. For example, new query options have been incorporated into Blast2GO to support diverse access to B2G-FAR data (see online tutorial available as Supplementary Material). Annotated sequences can be queried and filtered by their name/id, description, GO code and GO name, either as exact or ‘contains’ matches. Existing Blast2GO functions such as the generation of summary charts, single or combined graphs, annotation pies and enrichment analysis can be performed for the sequences selected by the user. Moreover, the .annot files from B2G-FAR are fully compatible with the Babelomics suite (Al-Shahrour et al., 2008; http://www.babelomics.org) for functional profiling analysis, where additional statistical methods for pathway analysis [FatiGO (Al-Shahrour et al., 2004) and FatiScan (Al-Shahrour et al., 2007)] are available. This is especially interesting in the case of microarray probe files or when a functional enrichment needs to be assessed with experimental data involving any of the non-model species included in the repository.
Comparison of B2G-FAR annotations with GO annotations: the quality of the Blast2GO annotation method has been extensively assessed and proved in previous works (Conesa and Götz, 2008; Conesa et al., 2005; Götz et al., 2008). However, we performed an additional evaluation of the annotation process to provide B2G-FAR users with a general feeling of the performance and nature of the annotations contained in the repository. We selected 10 000 random sequences from B2G-FAR which were also present in the GO database and compared their annotations. We recorded the number of exact GO term matches, more specific or more general terms (different specificity levels of the annotation), other branch or other GO category (true novel annotations) as described previously (Götz et al., 2008). Results are given in Table 2. The comparison study revealed that most of the original GO annotations (93.5%) were contained in the B2G-FAR repository as exact matches and more specific/general terms and only a small fraction (6.5%) were lost (other GO branch and category annotations) during the annotation process, presumably due to GO version differences or the removal of root category terms in the B2G-FAR repository. When comparing in the opposite direction, we observed that 49% of the B2F-FAR annotations were represented as exact matches in the GOA, and an additional 13% of terms are provided as more specific concepts. The remaining 38% are terms in other branches and in other main GO categories. To have an impression on the nature of these novel B2G-FAR annotations, we checked manually 20 randomly selected sequences for which differences between the two databases were found (see manual_evaluation.xls in Supplementary Material). Curation of the novel GO terms implied contrasting against scientific papers and established functional databases, such as UniProt, Tair, Saccharomyces Genome Database, Entrez, etc. From these 20 sequences one (AT5G35370.1) resulted to have doubtful sequence identity and was not considered in further computations. The remaining 19 sequences accounted for 109 novel GO terms, 9 of which could not be verified from the available literature. One sequence (Cyclin CLB2 of S.cerevisiae) obtained 4 presumably false GO functions due to sequence similarity to a paralogue with different functional specification. The remaining 96 GO terms (88%) were confirmed from literature data and assessed as valid annotations. These results evidence the quality of the GO term assignments contained in B2G-FAR.
Table 2.
Functional annotation of 10 000 random sequences from the GO and B2G-FAR compared against each other (annotation score ≥ 70, evalue ≤1 × E−10, GOw = 5, 5 BLAST hits)
| Compared | GO versus FAR* | FAR versus GO |
|---|---|---|
| Compared terms | 46 414 (GO) | 61 176 (B2G-FAR) |
| Exact GO term match | 29 446 | 29 446 |
| More specific GO terms | 510 | 7960 |
| More general GO terms | 13 457 | 156 |
| Other GO branch | 1126 | 16 193 |
| Other GO category | 1875 | 7421 |
*Comparisons are given as reference database versus comparing database, and numbers refer to the reference database.
3 UTILITY
We illustrate the utility of the B2G-FAR on two examples of functional genomics studies taken from the literature and show how B2G-FAR can speed up or facilitate new data analyses.
The first example is in the field of next-generation sequencing (NGS). These methods are rapidly extending within the genomics community as they greatly outperform both in sensitivity and accuracy hybridization-based approaches. B2G-FAR can support functional assessment in NGS research. In a pioneering study, Holt and colleagues analyzed genome variation and evolution in Salmonella typhi using NGS (Holt et al., 2008). The authors applied 454 and Solexa technologies to resequence 19 different S.typhi strains and isolates. The authors carried out a phylogenetic analysis of SNPs variance and identified genome insertions, deletions and modified genes across strains. However, although the impact of genomic changes on certain coding regions was discussed, no genome-wide functional analysis of strain variations was attempted. By typing S.typhi on the B2G-FAR species search box, we can readily locate the annotation file for this species, which contains GO assignments for 3917 genes (Supplementary Fig. S1). GeneBank IDs included in the annotation file provide the means for matching functional and genomic variation data. This annotation file can be opened with the Blast2GO software and by uploading each list of strain-specific varying genes, Blast2GO functions can be used to interrogate data for significant functional differences between isolates at the genome level, and to obtain the functional profiling of the genomic alterations or to locate mutated genes in metabolic pathways. This example illustrates how readily available functional data can complement the analysis of experimental results with little additional effort.
The second example relates to the use of Affymetrix probe-set annotation data available at the repository. B2G-FAR offers an annotation coverage which is substantially higher than the NetAffx GO annotations provided by the manufacturer and also has fast and reliable access to functional data for these GeneChips. The study by Espinoza et al. (2007) can serve as an illustrative example for this section of the repository. The article presents a transcriptomics analysis of viral infection in wine grape cultivars using the Affymetrix Grape GeneChip. In this study, authors generated functional annotations for up- and downregulated gene groups through similarity-based function transfer from Arabidopsis thaliana by WU-BLAST, GO terms being directly transferred for all retrieved alignments. The obtained annotation was summarized to reflect the abundance of distinct functional classes within regulated genes. Although valid, this basic functional description does not allow the identification of those functional categories which are specifically activated at viral infection. For this, a functional comparison to the whole genome represented in the array would be required, which implies that functional data for all probes would be needed. This information, absent in the article and presumably costly for the authors to obtain, is readily available from the B2G-FAR site. The B2G-FAR annotation file for the Grape GeneChip contains 54 841 GO terms and covers 11 971 probe sets. The list of differentially expressed genes provided in the article as Supplementary Material was used in Blast2GO to perform a GO term enrichment analysis based on the B2G-FAR annotation file. The analysis indicated a significant overrepresentation of chloroplast genes in the Camnre downregulated gene set (adjusted P-value: 1.2 × 10−5) (see Supplementary Fig. S2a) and only a slight enrichment of membrane, l-arginine and l-glutamate import and other membrane transport activities (P-values: 6 × 10−3) for the upregulated gene set (see Supplementary Fig. S2b).
4 DISCUSSION
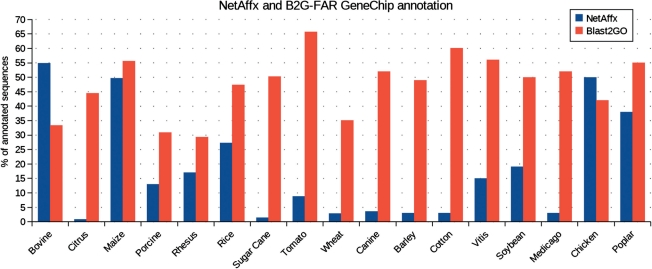
The major purpose of B2G-FAR is to offer biologists easy access to functional information. B2G-FAR has been conceived as a repository of automatic annotations generated by Blast2GO using high-throughput computing technologies to save annotation time to the functional genomics community. The B2G-FAR is species centered, which means that data can readily be obtained for any of the 2000 organisms present in the database. The Blast2GO annotation strategy has shown to render good recall values for sequence similarity function transfer methods and to match functional assignments by curated computational analysis (Götz et al., 2008). The B2G-FAR retains these quality levels: we showed that the majority of B2G-FAR assignments are identical or functionally related to GO Database annotations for sequences present in this database and, additionally, novel predictions are generally supported by the available literature. It should be stressed, however, that the quality of B2G-FAR is closely linked to the completeness and accuracy of the GO and InterPro databases. B2G-FAR complements the GO effort by offering high-throughput automatic annotations on a species basis. Compared with GOA, where automatically generated annotations are to a big extent based on protein domains, B2G-FAR combines both sequence similarity-based annotations through Blast2GO together with domain-based information through InterProScan. In this way, GO term assignments could be increased in number and the amount of available annotated sequences could be nearly doubled. Comparing the generated Affymetrix GeneChip annotations to the current GO annotation available at the NetAffx site, the B2G-FAR resource increased the coverage of functional annotations from an average of 7.89% (NetAffx) to 40.89% (Blast2GO) (Fig. 2). Only the Bovine and Chicken NetAffx annotations were richer than the ones generated by Blast2GO due to intensive proteome annotation efforts of GOA in collaboration with the International Protein Index (Barrell et al., 2009). Currently, most of the GeneChips of non-model species processed in B2G-FAR contain sufficient annotation coverage for a successful evaluation of microarray results in terms of pathways and biological functions. Moreover, the compatibility of B2G-FAR file formats with functional profiling tools make functional assessment methods readily accessible for a much larger diversity of organisms. Finally, B2G-FAR should not be understood as a competitive annotation source to annotation projects as carried out within the GO consortium, nor as a replacement to high-quality manual annotation of single-gene products, but as a complementing resource. Although automated annotation is by nature more error prone than manually curated one, B2G-FAR offers novel valuable information, making functional data accessible to a large users community working on different species.
Fig. 2.
Comparison between NetAffx and Blast2GO generated annotations for GeneChips contained in B2G-FAR.
5 CONCLUSIONS
B2G-FAR provides easy access to exhaustive functional information for a broad range of species encompassing most organisms under genome investigation. The repository is simple in architecture and still offers many analysis possibilities through the proximity to the Blast2GO software. In its current form, the resource is species centric. Future developments will consider multispecies scenarios such as metagenomics data or comparisons across taxa.
6 AVAILABILITY AND REQUIREMENTS
The annotation resource is freely available at http://b2gfar.bioinfo.cipf.es, is based on the DokuWiki framework and works with any common web browser. There are no other requirements or plugins needed to use the repository. Data files can be downloaded and unzipped or directly uploaded into the Blast2GO application through Java WebStart technology. Therefore, Java has to be installed. For both, B2G-FAR and Blast2GO, tutorials and quick-start sections are provided online.
Funding: Spanish Ministry of Science and Innovation (MICINN) (grants BIO2008-04638-E, BIO2008-05266-E, BIO2008-04212, BIO2009-10799 and CEN-20081002) and the PlanE Program; GVA-FEDER (PROMETEO/2010/001); Red Tematica de Investigacion Cooperativa en Cancer (RTICC), ISCIII, MICINN (grant RD06/0020/1019, in part). Further financial support was granted by the European Science Foundation (ESF) with the activity entitled ‘Frontiers of Functional Genomics’.
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Al-Shahrour F., et al. Fatigo: a web tool for finding significant associations of gene ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- Al-Shahrour F., et al. From genes to functional classes in the study of biological systems. BMC Bioinformatics. 2007;8:114–131. doi: 10.1186/1471-2105-8-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shahrour F., et al. Babelomics: advanced functional profiling of transcriptomics, proteomics and genomics experiments. Nucleic Acids Res. 2008;36(Suppl. 2):W341–W346. doi: 10.1093/nar/gkn318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., et al. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arnold R., et al. Simap–the similarity matrix of proteins. Bioinformatics. 2005;21(Suppl. 2):ii42–ii46. doi: 10.1093/bioinformatics/bti1107. [DOI] [PubMed] [Google Scholar]
- Ashburner M., et al. Gene ontology: tool for the unification of biology. the gene ontology consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrell D., et al. The goa database in 2009–an integrated gene ontology annotation resource. Nucleic Acids Res. 2009;37:D396–D403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camon E., et al. The gene ontology annotation (goa) database: sharing knowledge in uniprot with gene ontology. Nucleic Acids Res. 2004;32(Suppl. 1):D262–D266. doi: 10.1093/nar/gkh021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A., Götz S. Blast2go: a comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics. 2008;2008:1–13. doi: 10.1155/2008/619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A., et al. Blast2go: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Espinoza C., et al. Gene expression associated with compatible viral diseases in grapevine cultivars. Funct. Integr. Genomics. 2007;7:95–110. doi: 10.1007/s10142-006-0031-6. [DOI] [PubMed] [Google Scholar]
- Götz S., et al. High-throughput functional annotation and data mining with the blast2go suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt K.E., et al. High-throughput sequencing provides insights into genome variation and evolution in salmonella typhi. Nat. Genet. 2008;40:987–993. doi: 10.1038/ng.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J., et al. The human phylome. Genome Biol. 2007;8:R109–R125. doi: 10.1186/gb-2007-8-6-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey P., et al. Integr8 and genome reviews: integrated views of complete genomes and proteomes. Nucleic Acids Res. 2005;33:D297–D302. doi: 10.1093/nar/gki039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti-Renom M.A., et al. The annolite and annolyze programs for comparative annotation of protein structures. BMC Bioinformatics. 2007;8(Suppl. 4):1–12. doi: 10.1186/1471-2105-8-S4-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myhre S., et al. Additional gene ontology structure for improved biological reasoning. Bioinformatics. 2006;22:2020–2027. doi: 10.1093/bioinformatics/btl334. [DOI] [PubMed] [Google Scholar]
- Quevillon E., et al. Interproscan: protein domains identifier. Nucleic Acids Res. 2005;33(Suppl. 2):W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattei T., et al. Simap structuring the network of protein similarities. Nucleic Acids Res. 2008;36(Suppl. 1):D289–D292. doi: 10.1093/nar/gkm963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M. Functions of the gene products of escherichia coli. Microbiol. Mol. Biol. Rev. 1993;57:862–952. doi: 10.1128/mr.57.4.862-952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjölander K. Phylogenomic inference of protein molecular function: advances and challenges. Bioinformatics. 2004;20:170–179. doi: 10.1093/bioinformatics/bth021. [DOI] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. The gene ontology project in 2008. Nucleic Acids Res. 2008;36:D440–D444. doi: 10.1093/nar/gkm883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Uniprot Consortium. The universal protein resource (uniprot) Nucleic Acids Res. 2007;35:D193–D197. doi: 10.1093/nar/gkl929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise R.P., et al. Barleybase/plexdb. Methods Mol. Biol. 2007;406:347–363. doi: 10.1007/978-1-59745-535-0_17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.