Abstract
Summary: The SFF file format produced by Roche's 454 sequencing technology is a compact, binary format that contains the flow values that are used for base and quality calling of the reads. Applications, e.g. in metagenomics, often depend on accurate sequence information, and access to flow values is important to estimate the probability of errors. Unfortunately, the programs supplied by Roche for accessing this information are not publicly available. Flower is a program that can extract the information contained in SFF files, and convert it to various textual output formats.
Availability: Flower is freely available under the General Public License.
Contact: ketil.malde@imr.no
1 INTRODUCTION
After pyrosequencing was first introduced (Margulies et al., 2005), the use of this technology has grown rapidly. Data from pyrosequencing can be analyzed with traditional tools for sequence analysis (typically by first converting data into the Fasta format), but native formats usually contain more information and for many analyses this information can be used to produce more accurate results.
Pyrosequencing data from the Roche 454 sequencing technology comes in the form of SFF files, a relatively compact, binary format. This format contains the sequence of flow values for each read, corresponding to the light intensities, resulting from the ligation process during sequencing. In addition, the SFF file contains the base-called sequence with associated quality information, and an index linking each called base to the corresponding flow value. The SFF file also contains clipping information for each read, and various metadata.
Roche provides some utilities for working with SFF files, and these have so far been the most convenient option for pipelines like Pyronoise (Quince et al., 2009) and QIIME (Caporaso et al., 2010) that need to access the raw flow values and other information from SFF files. But as Roche's tools are not publicly available and may not be modified or redistributed, this complicates distribution of integrated pipelines. As a consequence, usage and development of more effective analysis tools is hampered. Other tools like sff_extract (Jose Blanca, unpublished), a utility supplied with the Mira assembler (Chevreux et al., 1999), is limited to extracting sequence information.
Flower is an efficient, portable and freely distributable software utility that reads SFF files, and outputs information in textual form. It uses an independent reimplementation of the SFF format based only on published documentation (Roche, 2009), and is freely redistributable and modifiable under the General Public License.
2 FEATURES
Flower is implemented as a command line tool that reads one or more SFF files, and produces output in one or more formats, either to files or to standard output.
The default output format is a textual representation of each read in the SFF file. The information is presented as one field per line, a tab character separates the label from the contents. Some fields are optional, so if for instance the read name does not follow 454 encoding conventions, no Info field will be present. An example of this output (truncated in length) is shown in Figure 1.
Fig. 1.
Example textual output for one read. The Info field displays the information embedded in the read name, and includes date, time and picotiter plate region and coordinates.
An alternative output is the tabular flowgram output (‘-F’ option), shown in Figure 2, that generates one line per flow value, with read name, flowgram position, nucleotide and quality calls. Although verbose, this makes it easy to link quality calling with the corresponding flow value.
Fig. 2.
Example tabular output. The columns are read name, flow number, nucleotide (lower case if masked), flow value and list of associated quality values.
Flower can also extract the sequence data to the Fasta format (using the -f option), associated qualities (-q) and the Fastq (Cock et al., 2010) format using either Phred (-Q) or Illumina (-I) type qualities. Summary information about the contents of SFF files is available as either the header information (with the -i option) or one line per read (with -s).
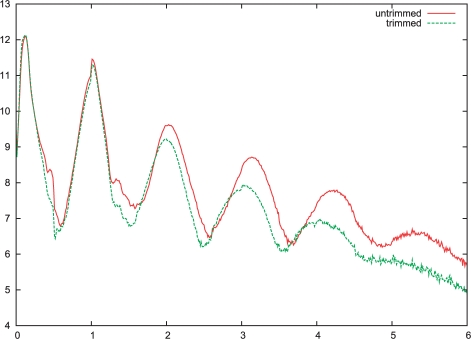
One common graphical representation of a sequencing run is a histogram of flow values in the data (an example is shown in Fig. 3). Although this is straightforward to calculate from the textual output, flower has specific functionality for accumulating these. Since the output size is greatly reduced, this is faster.
Fig. 3.
Plot showing the flow values before and after trimming, using the -h option.
SFF files contain trimming information, indicating parts of each read that are either low quality or part of synthetic sequences like adapter sequences or the four-letter key that prefixes each read. Flower will by default output the trimmed sequence parts as lower case letters and the untrimmed as upper case, but there are also options for removing the unwanted sequence parts, with the corresponding quality and flowgram information. The –trim option removes all trimmed sequence, and the –trimkey option removes only the key. This affects all output options.
3 DISCUSSION
Running time depends to a large degree on the output formats chosen, as some formats like textual or tabular generate a large amount of output, and this dominates running time. For example, converting an 2.1 GB SFF file (containing ∼630 000 reads) to Fasta or FastQ format takes about 20 s on a 3.4 GHz Xeon, processing the SFF file at about 100 MB/s, which is faster than a typical hard disk can deliver it. Generating text output for the same file is substantially slower at 476 s.1
Although it is likely that these numbers can be improved somewhat, in most cases any subsequent analysis will be much more computationally expensive. The memory footprint is generally low (less than 5 MB resident size as measured by top in the experiments described above) and, as flower works incrementally (streaming), memory consumption is independent on total input or output size.
4 AVAILABILITY
Flower is available under the General Public License, and is freely available for any purpose. It may be modified, and the modified version is redistributed according to the conditions in the license.
Flower is implemented in Haskell, and the source code is available from the Hackage repository at http://hackage.haskell.org/package/flower. Further documentation can be found at http://biohaskell.org/Applications/flower
ACKNOWLEDGEMENTS
I would like to thank Susanne Balzer, Christopher Quince and Erick Matsen for valuable suggestions and discussion.
Funding: This work is supported by The National Program for Research in Functional Genomics in Norway (FUGE) and the Research Council of Norway.
Conflict of Interest: none declared.
Footnotes
1See the flower web page for a performance comparison of alternative programs.
REFERENCES
- Caporaso J.G., et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevreux B., et al. Genome sequence assembly using trace signals and additional sequence information. German Conference on Bioinformatics. 1999 [Google Scholar]
- Cock P.J.A., et al. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010;38:1767–1771. doi: 10.1093/nar/gkp1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C., et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat. Methods. 2009;6:639–641. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- Roche Genome Sequencer FLX System Software Manual, version 2.3, General Overview and File Formats. 2009 Roche/454 Life Sciences. [Google Scholar]