Abstract
Summary: A motif is a short DNA or protein sequence that contributes to the biological function of the sequence in which it resides. Over the past several decades, many computational methods have been described for identifying, characterizing and searching with sequence motifs. Critical to nearly any motif-based sequence analysis pipeline is the ability to scan a sequence database for occurrences of a given motif described by a position-specific frequency matrix.
Results: We describe Find Individual Motif Occurrences (FIMO), a software tool for scanning DNA or protein sequences with motifs described as position-specific scoring matrices. The program computes a log-likelihood ratio score for each position in a given sequence database, uses established dynamic programming methods to convert this score to a P-value and then applies false discovery rate analysis to estimate a q-value for each position in the given sequence. FIMO provides output in a variety of formats, including HTML, XML and several Santa Cruz Genome Browser formats. The program is efficient, allowing for the scanning of DNA sequences at a rate of 3.5 Mb/s on a single CPU.
Availability and Implementation: FIMO is part of the MEME Suite software toolkit. A web server and source code are available at http://meme.sdsc.edu.
Contact: t.bailey@imb.uq.edu.au; t.bailey@imb.uq.edu.au
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
A DNA or protein sequence motif is a short pattern that is conserved by purifying selection. In DNA, a motif may correspond to a protein binding site; in proteins, a motif may correspond to the active site of an enzyme or a structural unit necessary for proper folding of the protein. Thus, sequence motifs are one of the basic functional units of molecular evolution. Consequently, identifying and understanding these motifs is fundamental to building models of cellular processes at the molecular scale and to understanding the mechanisms of human disease.
We describe here a software tool, called FIMO (Find Individual Motif Occurrences, pronounced fēmō), that carries out in an efficient, statistically rigorous fashion one of the core functions required for any motif-based sequence analysis: scanning a collection of DNA or protein sequences for occurrences of one or more motifs. FIMO is by no means the first motif scanning method; however, many publicly available motif scanners are either not currently maintained or lack some of FIMO's features. Table 1 summarizes the differences between FIMO and eight currently available motif scanners. Furthermore, as part of the MEME Suite (Bailey et al., 2009), FIMO can be used seamlessly in conjunction with a variety of complementary motif-based sequence analysis tools.
Table 1.
Comparison of motif search functionality
| Method | Scans DNA | Scans proteins | Supports custom backgrounds | Reports P-values | Performs multiple testing correction | Source code freely available | Web accessible | GFF/WIG output | XML/HTML output |
|---|---|---|---|---|---|---|---|---|---|
| MotifScanner | ✓ | ✓ | ✓ | ✓ | ✓ | ||||
| MotifViz | ✓ | ✓ | ✓ | ✓ | ✓ | ||||
| STORM | ✓ | ✓ | ✓ | ||||||
| TRED | ✓ | ✓ | ✓ | ||||||
| RSAT | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||
| Patser | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||
| PoSSuMsearch | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| MATCH | ✓ | ✓ | ✓ | ||||||
| FIMO | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
References for the motif scanning algorithms are provided in the supplement. Note that FIMO only supports zero-order custom backgrounds.
Note that the MEME Suite provides two other motif scanning algorithms that are useful in different scenarios. MAST (Bailey and Gribskov, 1998) searches with one or more DNA or protein motifs against a database composed of relatively short sequences, e.g. proteins or candidate regulatory regions, assigning a single score to each target sequence assuming that every motif occurs exactly once in the sequence. MCAST (Bailey and Noble, 2003), in contrast, uses a hidden Markov model to search DNA sequences for regions that are enriched with occurrences of one or more of the given motifs. Thus, MCAST is designed to scan chromosomes to detect cis-regulatory modules containing a known collection of cofactor motifs. Compared with MAST and MCAST, FIMO is simpler and more general. FIMO only assigns scores to individual motif occurrences; it makes no attempt to assign scores to joint occurrences of motifs, to sequence regions or to complete sequences. FIMO is thus a general-purpose tool for identifying individual candidate binding sites or protein motifs.
2 IMPLEMENTATION
FIMO takes as input one or more fixed-length motifs, represented as position-specific frequency matrices. These motifs can be generated from the MEME motif discovery algorithm, extracted from an existing motif database or created by hand using a simple text format. The program computes a log-likelihood ratio score (often referred to incorrectly as a ‘log-odds score’) for each motif with respect to each sequence position and converts these scores to P-values using dynamic programming (Staden, 1994), assuming a zero-order null model in which sequences are generated at random with user-specified per-letter background frequencies. Finally, FIMO employs a bootstrap method (Storey, 2002) to estimate false discovery rates (FDRs). Because the FDR is not monotonic relative to the P-value, FIMO instead reports for each P-value a corresponding q-value, which is defined as the minimal FDR threshold at which the P-value is deemed significant (Storey, 2003).
FIMO produces as output a ranked list of motif occurrences, each with an associated log-likelihood ratio score, P-value and q-value. This list is represented in multiple ways: as an HTML report, as an XML file in CisML format (Haverty and Weng, 2004), as a plain text file and as tab-delimited files in formats suitable for input to the UCSC Genome Browser (.gff and .wig).
The FIMO web server allows the user to upload one or more motifs and then search either a user-supplied sequence file or one of 3102 single and multiorganism DNA and protein databases from Ensembl and Genbank. Search results are stored online, and the user is notified of their availability via email.
3 EXAMPLE
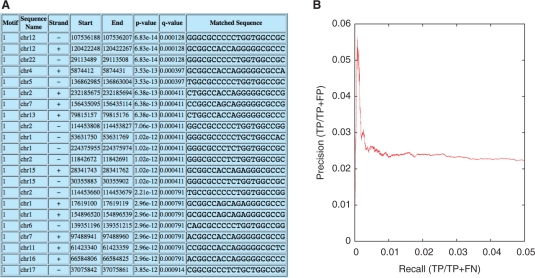
To demonstrate FIMO's functionality, we searched the human genome with a motif for CTCF, a highly conserved zinc finger DNA-binding protein that exhibits diverse regulatory functions and that plays a major role in the global organization of the chromatin architecture of the human genome (Phillips and Corces, 2009). Figure 1 shows the FIMO HTML output for the top-scoring predicted occurrences of the motif, and a precision–recall curve comparing the predicted CTCF binding sites with a gold standard derived from a ChIP-seq experiment (see Supplementary Material for details). Overall, FIMO identified 8647 candidate binding sites with q < 0.05. The precision–recall curve suggests that the top of the list is enriched with sites that overlap ChIP-seq peaks. Note that the absolute precision is low, presumably for two reasons: first, a single motif lacks sufficient information to reliably scan an entire eukaryotic genome with high precision; second, FIMO identifies many bona fide CTCF binding sites that are not active in the particular cell type in which the ChIP-seq experiment was carried out. Scanning the entire human genome took 30 min 10 s of wall clock time on an Intel Xeon 2.2 GHz CPU, equivalent to scanning 3.5 Mp/s.
Fig. 1.
Using FIMO to identify candidate CTCF binding sites in the human genome. (A) Sample FIMO HTML output, showing the locations of the top-scoring occurrences of the CTCF motif in the human genome. (B) A precision-recall curve created by comparing FIMO's ranked list of CTCF sites with a gold standard derived from a ChIP-seq experiment.
Funding: This work was supported by National Institutes of Health award 2 R01 RR021692.
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Bailey T.L., Noble W.S. Searching for statistically significant regulatory modules. Bioinformatics. 2003;19(Suppl. 2):ii16–ii25. doi: 10.1093/bioinformatics/btg1054. [DOI] [PubMed] [Google Scholar]
- Bailey T.L., Gribskov M. Combining evidence using p-values: Application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- Bailey T., et al. MEME suite: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverty P.M., Weng Z. CisML: an XML-based format for sequence motif detection software. Bioinformatics. 2004;20:1815–1817. doi: 10.1093/bioinformatics/bth162. [DOI] [PubMed] [Google Scholar]
- Phillips J.E., Corces V.G. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Searching for motifs in nucleic acid sequences. Methods Mol. Biol. 1994;25:93–102. doi: 10.1385/0-89603-276-0:93. [DOI] [PubMed] [Google Scholar]
- Storey J.D. A direct approach to false discovery rates. J. R. Stat. Soci. 2002;64:479–498. [Google Scholar]
- Storey J.D. The positive false discovery rate: a bayesian interpretation and the q-value. Ann. Stat. 2003;31:2013–2035. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.