Abstract
Gene fusions involving members of the RAF family of protein kinases have recently been identified as characteristic aberrations of low-grade astrocytomas, the most common tumors of the central nervous system in children. While it has been shown that these fusions cause constitutive activation of the ERK/MAPK pathway, very little is known about their formation. Here, we present a detailed analysis of RAF gene fusion breakpoints from a well-characterized cohort of 43 low-grade astrocytomas. Our findings show that the rearrangements that generate these RAF gene fusions may be simple or complex and that both inserted nucleotides and microhomology are common at the DNA breakpoints. Furthermore, we identify novel enrichment of microhomologous sequences in the regions immediately flanking the breakpoints. We thus provide evidence that the tandem duplications responsible for these fusions are generated by microhomology-mediated break-induced replication (MMBIR). Although MMBIR has previously been implicated in the pathogenesis of other diseases and the evolution of eukaryotic genomes, we demonstrate here that the proposed details of MMBIR are consistent with a recurrent rearrangement in cancer. Our analysis of repetitive elements, Z-DNA and sequence motifs in the fusion partners identified significant enrichment of the human minisatellite conserved sequence/χ-like element at one side of the breakpoint. Therefore, in addition to furthering our understanding of low-grade astrocytomas, this study provides insights into the molecular mechanistic details of MMBIR and the sequence of events that occur in the formation of genomic rearrangements.
Genomic rearrangements are genetic aberrations in which the gross structure of one or several chromosomes has been altered. These events may be copy number neutral, as is the case for balanced translocations and inversions, or result in chromosomal segments being lost or gained, as in deletions and duplications, respectively (Gu et al. 2008). Genomic rearrangements affecting the germline play a role in the creation of genetic variation, the development of functionally divergent genes, and, ultimately, speciation (Feuk et al. 2006; Fan et al. 2008; Kitano et al. 2009). In addition to their evolutionary significance, germline genomic rearrangements are responsible for a group of human diseases known collectively as genomic disorders (Stankiewicz and Lupski 2010), which includes a number of cancer-predisposition syndromes. The role of genomic rearrangements in carcinogenesis is not restricted to these predisposition syndromes, with somatically acquired changes to chromosome structure present in a wide range of cancers (Stratton et al. 2009). Some of these rearrangements are believed to be early, perhaps even initiating, events (Greaves and Wiemels 2003), while others are linked to tumor progression (Maher et al. 2006).
One possible consequence of genomic rearrangements is the creation of in-frame gene fusions. Characteristic fusions arising from chromosome translocations are found in leukemias, lymphomas, and sarcomas (Mitelman et al. 2007). Gene fusions have also been identified in several adult carcinomas, such as prostate cancer (Tomlins et al. 2005), adenoid cystic carcinomas (Persson et al. 2009), and small-cell lung cancer (Pleasance et al. 2010). In addition to these neoplasms, two gene fusions, KIAA1549–BRAF at 7q34 and SRGAP3–RAF1 at 3p25, have recently been identified in low-grade astrocytoma, the most common form of brain tumor in children (Jones et al. 2008, 2009; Forshew et al. 2009; Sievert et al. 2009).
We believe that these RAF gene fusions represent a powerful model for investigating genomic rearrangements for several reasons. Firstly, they occur in the majority of pilocytic astrocytomas, the most common subgroup of low-grade astrocytomas (Forshew et al. 2009; Lawson et al. 2010). Secondly, they are formed by tandem duplications rather than translocations. Tandem duplications have previously been identified as important rearrangements in several specific tumors (Nakao et al. 1996; Fenstermaker et al. 1998). However, recent next-generation sequencing studies suggest that tandem duplications may be of widespread significance as they were found to be both common in lung cancers (Campbell et al. 2008) and the major cause of non-amplified in-frame gene fusions in breast cancer (Stephens et al. 2009). Thirdly, the discovery of two fusion genes on separate chromosomes allows us to directly compare different regions of the genome, while the existence of five known KIAA1549–BRAF fusion variants facilitates the analysis and identification of local factors. Finally, pilocytic astrocytomas appear to have a relatively stable genome, exhibiting few additional copy number changes (Jones et al. 2006). Therefore, RAF fusions in low-grade astrocytomas may also be used as a model for investigating the mechanisms that cause germline tandem duplications and events in carcinogenesis that occur prior to the onset of genomic instability.
Here, we present a detailed analysis of 43 low-grade astrocytomas with RAF gene fusions in which we identify common properties of their breakpoints, investigate local and global factors to explain their relative frequencies, and discuss possible mechanisms for their formation.
Results
Relative frequencies of RAF gene fusion variants cannot be explained by intron size alone
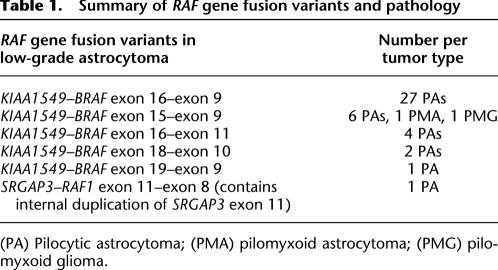
Our tumor cohort of 43 pediatric low-grade astrocytomas includes all known KIAA1549–BRAF variants (Tatevossian et al. 2010) and one of the two reported SRGAP3–RAF1 fusion genes (Table 1; Supplemental Table S1; Forshew et al. 2009). One variant of the BRAF fusions (KIAA1549–BRAF exon 16–exon 9) was found to occur more frequently than the others (27/42), a finding that is concordant with previous studies (Jones et al. 2008; Sievert et al. 2009; Lawson et al. 2010). One potential explanation for this disparity in fusion frequency is intron size. The introns associated with the overrepresented fusion variant, KIAA1549 intron 16 (8869 bp) and BRAF intron 8 (6723 bp), are the largest of all introns involved in the formation of KIAA1549–BRAF fusions. However, this alone cannot explain the discrepancy as the observed frequencies of KIAA1549–BRAF fusion variants are still significantly different from the expected distribution even when intron sizes are taken into account (P = 0.021, χ2-test).
Table 1.
Summary of RAF gene fusion variants and pathology
(PA) Pilocytic astrocytoma; (PMA) pilomyxoid astrocytoma; (PMG) pilomyxoid glioma.
RAF fusion breakpoints are not clustered within introns
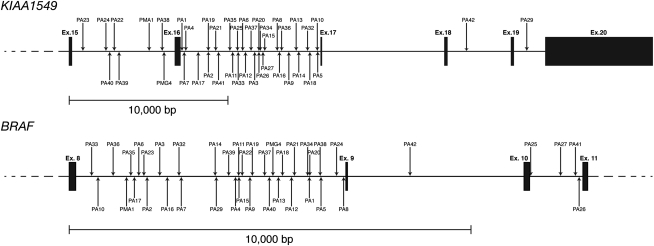
We used PCR and sequencing analyses to map the exact intronic positions of the DNA breakpoints for 42 samples. Despite numerous attempts using different primers and PCR conditions, the breakpoint of an additional tumor sample, PA28, could not be identified. The distribution of breakpoints from 41 tumors with KIAA1549–BRAF fusions is depicted in Figure 1. No discernible clustering of breakpoints was identified within any of the introns (Supplemental Fig. S1).
Figure 1.
Distribution of DNA breakpoints for 41 KIAA1549–BRAF gene fusions.
Sequence alignment identifies distinct categories of fusion breakpoint
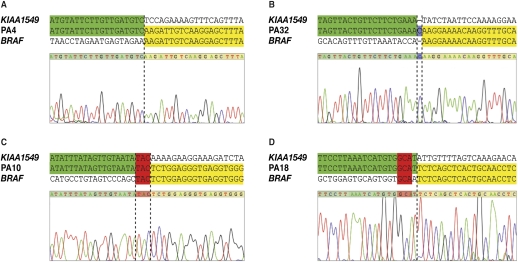
In 40 samples the fusions appeared to have been produced by simple rearrangements, whereas the remaining two (PA27 and PA30) were created by complex rearrangements, as indicated by the presence of large insertions. Sequence alignment of the simple breakpoints revealed the existence of three distinct categories. In 6/40 simple rearrangements, we observed a seamless transition from one sequence to the next, with a clearly defined breakpoint (Fig. 2A). The second category (n = 6) is defined by the presence of short inserted (also known as non-templated) sequences at the breakpoint that match neither reference sequence (Fig. 2B). We ensured that these insertions were not, in fact, germline SNPs or in-dels by sequencing the unrearranged alleles of KIAA1549 and BRAF. The final category, which includes the majority of tumors (n = 28), is characterized by breakpoint microhomology—the presence of one or more base pairs of sequence homology at the breakpoint junction that could be assigned to either gene (Fig. 2C). In our tumor cohort, insertions and breakpoint microhomology ranged from 1 to 6 bp and 1 to 5 bp in length, respectively. The sequence alignment for each tumor sample is shown in Supplemental Figure S2.
Figure 2.
Categories of simple RAF gene fusion breakpoints. Fusion sequences aligned with KIAA1549 (green) and BRAF (yellow). (Blue) Nucleotides that align to neither reference gene (insertions); (red) nucleotides that align to both reference genes (microhomology). (A) Seamless transition with defined breakpoint. (B) Insertion. (C) Breakpoint microhomology. (D) Flanking microhomology only.
Breakpoint and flanking microhomology are significantly enriched, whereas large regions of homology are uncommon
To determine whether the observed microhomology could have arisen by chance, we randomly generated five sets of 500 breakpoints within KIAA1549 intron 16 and BRAF intron 8. Only 643/2500 (25.7%) simulated breakpoints exhibited microhomology, with a mean of 0.36 bp per junction, whereas 28/40 (70%) simple RAF gene fusions contained breakpoint microhomology, with a mean of 1.78 bp per junction. Therefore, the distribution of microhomology in the simulated breakpoints differed significantly from the observed distribution in the 40 fusions caused by simple rearrangements (P = 3.8 × 10−15, Mann-Whitney U Test) (Supplemental Fig. S3).
In addition, several samples exhibited flanking microhomology, which we defined as the presence of ≥3 consecutive homologous nucleotides within ±10 bp of, but not overlapping, the breakpoint (Fig. 2D). Flanking microhomology was found at 3/6 breakpoints with insertions, 5/28 of those with breakpoint microhomology, and 3/6 with neither insertions nor breakpoint microhomology. Although flanking microhomology was not significantly enriched in all of the fusions caused by simple rearrangements (11/40, 27.5%) relative to the five control sets, we would not expect enrichment in those samples that possess breakpoint microhomology of equivalent size as both types of microhomology most likely perform the same function of mediating interactions between the fusion partners. Therefore, after excluding those samples with ≥3 nucleotides of breakpoint microhomology from the tumor and control sets, flanking microhomology was found to be significantly enriched in the observed breakpoints (11/28, 39.3%) compared to four of the five sets of simulated breakpoints (P < 0.05, Fisher's exact test; odds ratio 1.7–2.3). As with the insertions, we sequenced the normal KIAA1549 and BRAF alleles to confirm that these regions were not breakpoint microhomology masked by germline SNPs or in-dels.
We analyzed the reference genome sequence (GRCh37) within ±2 kb of each simple breakpoint for the presence of larger regions of homology (>100 bp) between KIAA1549 and BRAF. All the regions identified were caused by Alu repeats. It has previously been suggested that Alu repeats within 300 bp of the breakpoints may mediate recombination (Bailey et al. 2003). However, this was the case in only 5/40 samples with simple rearrangements (Supplemental Table S2). Moreover, there are no segmental duplications (regions of >1 kb in size with ≥95% homology) in KIAA1549, BRAF, SRGAP3, or RAF1.
Complex rearrangements create a subset of RAF gene fusions
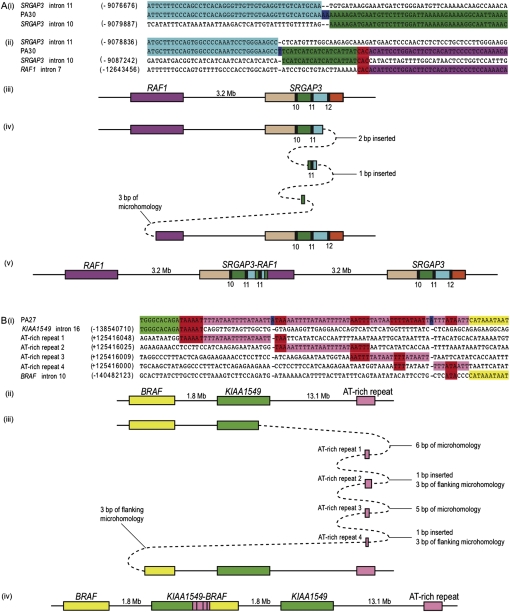
The two RAF gene fusions caused by complex rearrangements were KIAA1549–BRAF exon 16–exon 11 (PA27) and SRGAP3–RAF1 exon 11–exon 8 (PA30). Our initial characterization of the SRGAP3–RAF1 fusion identified an internal tandem duplication of SRGAP3 exon 11, which enables the resultant fusion gene to be in-frame (Forshew et al. 2009). We successfully sequenced both DNA breakpoints and the entire 1063-bp insertion. The majority of the insertion mapped to a contiguous sequence starting within SRGAP3 intron 10 and ending in SRGAP3 intron 11, which differed from the reference genome by only three SNPs (rs2271205, rs35647176, and rs11131157) and one in-del (rs35966925). Insertions were present at both the SRGAP3 intron 11–intron 10 breakpoint (2 bp, AA) and the SRGAP3–RAF1 intron 11–intron 7 breakpoint (20 bp, TTCATCATCATCATCATTAT). We hypothesized that the latter insertion had not been created by the random addition of nucleotides but was instead generated from a DNA template, with the most likely candidate a 19-bp region within the SRGAP3 intron 10 (Fig. 3A).
Figure 3.
Breakpoint analyses of two complex rearrangements. The figure is not to scale. In both panels, microhomology (red); insertions (blue). Genes are depicted by their orientation on the minus strand. (A) Complex rearrangement in PA30. (Purple) RAF1; (beige) SRGAP3 intron 9; (green) intron 10; (light blue) intron 11; (orange) intron 12. Exons are numbered and represented by black rectangles. (i) Sequence alignment showing the SRGAP3 intron 11–intron 10 breakpoint. (ii) Sequence alignment for the SRGAP3–RAF1 breakpoint. (iii) Schematic representation of the region on chromosome 3p25 prior to duplication. (iv) The three template switching events that may have given rise to the complex breakpoint in PA30. (v) Observed complex rearrangement. (B) Complex rearrangement in PA27. (Light green) KIAA1549; (yellow) BRAF; (pink) AT-rich region. (i) Sequence alignment showing the KIAA1549–BRAF breakpoint in PA27. (ii) Schematic representation of the region on chromosome 7q34 prior to duplication. (iii) The five template switching events that could have generated the complex breakpoint in PA27. (iv) Observed complex rearrangement.
The KIAA1549–BRAF complex rearrangement in PA27 contains a 67-bp insertion that could not be mapped to a single genomic locus. We were able to identify several ways in which this fusion could have been created, one of which is depicted in Figure 3B.
Repetitive elements and non-B DNA conformations are not significantly enriched in the fusion partners
Repetitive elements, fragile sites, and regions of non-B DNA have all been associated with the formation of genomic rearrangements (Arlt et al. 2006; Bacolla et al. 2006; Wells 2007; de Smith et al. 2008). There are no known common or rare fragile sites at 7q34 or 3p25 (Rebhan et al. 1997). We analyzed the presence of both repetitive elements and Z-DNA in KIAA1549, BRAF, SRGAP3, and RAF1 and compared their distribution to 1000 control genes. After correction for multiple testing, none of the calculated empirical P-values were significant at the 5% level (Supplemental Table S3).
In addition to this analysis, we compared the distribution of these elements within the relevant introns of KIAA1549 and BRAF (Supplemental Table S4). The majority of repetitive elements tested were observed at higher frequencies in introns other than KIAA1549 intron 16 and BRAF intron 8 and so cannot explain the prevalence of the KIAA1549–BRAF exon 16–exon 9 fusion variant. The only exceptions were L2 repeats and long terminal repeats (LTRs), which were observed exclusively in KIAA1549 intron 16 and BRAF intron 8.
The human minisatellite conserved sequence/χ-like element is significantly enriched at the KIAA1549 side of the breakpoint
A number of common motifs have previously been shown to be significantly enriched at the breakpoints of genomic rearrangements (Abeysinghe et al. 2003). We identified 39 motifs that are associated with gene rearrangements, mutations, DNA cleavage, replication, and site-specific recombination and so could potentially be involved in the formation of RAF gene rearrangements in low-grade astrocytomas (Supplemental Table S5). For the 41 tumors with KIAA1549–BRAF fusions, we tested for enrichment of a given motif in the region ±25 bp of the breakpoint at: (i) the KIAA1549 side of the breakpoint only, (ii) the BRAF side only, (iii) either side, and (iv) both sides. Only one motif, the human minisatellite conserved sequence/χ-like element, was found to be significantly enriched (P = 0.040, Fisher's exact test). This motif was overrepresented at the KIAA1549 side of the breakpoint only, occurring in 4/41 (9.8%) tumor samples (PA7, PA15, PA17, and PA39) compared to 52/5000 (1.0%) 50-bp control sequences.
Discussion
Although the significance of genomic rearrangements is widely appreciated and, in some cases, their effects largely understood, our knowledge of the mechanisms that create them remains incomplete. Our results not only provide evidence for the involvement of specific processes in the formation of RAF gene fusions in low-grade astrocytomas, but they also allow greater insight into the general underlying mechanisms.
Until recently, most of the proposed mechanisms have focused on the incorrect repair of DNA double-strand breaks (DSBs) (Hastings et al. 2009b). The two main pathways that carry out DSB repair in eukaryotes are homologous recombination and non-homologous DNA end joining (NHEJ). Although homologous recombination is believed to be less error-prone than NHEJ (Khanna and Jackson 2001), errors in either of these processes can cause genomic rearrangements.
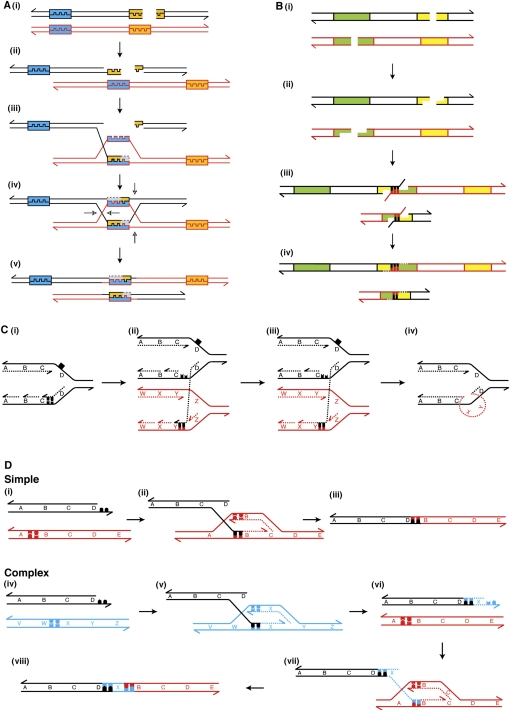
Homologous recombination repairs DSBs by using another section of DNA, most commonly the corresponding allele on the sister chromatid (Kadyk and Hartwell 1992), as a template. If homologous recombination occurs between two paralogous sequences of DNA, instead of between two alleles, genomic rearrangements are created. This process is known as non-allelic homologous recombination (NAHR) (Fig. 4A). It is estimated that NAHR requires at least 200 bp of homology between the invading strand and the template (Rubnitz and Subramani 1984; Reiter et al. 1998), which is the case in only 5/40 simple rearrangements among our cases. Furthermore, segmental duplications and breakpoint clustering, which are both commonly associated with NAHR, are absent. Therefore, we are confident that NAHR is not the predominant mechanism of RAF gene fusion formation in low-grade astrocytomas.
Figure 4.
Schematic representations of postulated mechanisms for the formation of genomic rearrangements. In all panels, 3′ ends are represented by half arrows. (A) Non-allelic homologous recombination. (i) A DSB occurs. (ii) The DNA ends are resected by exonucleases leaving 3′ single-stranded DNA (ssDNA) overhangs. (iii) One of the 3′ ssDNA ends invades a paralogous sequence (blue and orange boxes) and is extended by DNA synthesis. (iv) Strand invasion and extension of the second DNA end occurs, resulting in the formation of a double Holliday junction. (v) The Holliday junctions are resolved in the opposite sense producing a reciprocal duplication and deletion. (B) Non-homologous DNA end joining. (i) Two DSBs occur in sister chromatids or homologous chromosomes. (ii) The Artemis–DNA-dependent protein kinase catalytic subunit (DNA-PKcs) complex resects the DNA ends. (iii) Incorrect DNA ends are brought together by the Ku protein, and may anneal through microhomology. The Artemis–DNA–PKcs complex cleaves the flaps. (iv) DNA polymerase λ or μ (POLL or POLM) adds nucleotides to fill the gaps, and the NHEJ1–XRCC4–DNA ligase IV (LIG4) complex ligates the DNA ends, producing a reciprocal deletion and duplication. (C) Fork stalling template switching. (i) A replication fork stalls after encountering a DNA lesion (black square) on the template strand. (ii) The lagging strand from the stalled replication fork anneals to the template strand of another replication fork using microhomology. (iii) The invading strand is either extended by DNA synthesis or ligated to the 5′ end of the preceding Okazaki fragment. (iv) The FoSTeS model predicts that the lagging strand returns to the original template, allowing normal DNA replication to resume. Prior to this occurring, the lagging strand may invade multiple different replication forks (not shown). (D) Microhomology-mediated break-induced replication. (i–iii) Formation of a simple rearrangement by MMBIR. (i) An unpaired DSB end undergoes 5′ resection. (ii) The 3′ overhang uses microhomology to anneal to a non-paralogous section of DNA. (iii) No subsequent template switching events occur. (iv–viii) Formation of a complex rearrangement by MMBIR. (iv,v) Strand invasion occurs as in the creation of a simple rearrangement. (vi) The extended end dissociates as the low-processivity replication fork collapses. (vii) The 3′ end anneals to a second template. For tandem duplications this is upstream of the original break. (viii) Further template switching events may occur (not shown) until ultimately a fully processive replication fork is formed that goes to completion or collides with a replication fork moving in the opposite direction.
Unlike NAHR, NHEJ does not require a template and so is the favored mechanism of DNA DSB repair during G0, G1, and early S phases (Takata et al. 1998). However, NHEJ repair pathways can also operate in G2 and late S phases, and play a crucial role in repairing physiological DSBs such as those incurred during V(D)J recombination (Grawunder et al. 1997; Wilson et al. 1997; Wu et al. 2008). NHEJ primarily causes genomic rearrangements by ligating DNA ends from different DSBs together (Fig. 4B). Therefore, NHEJ does not require large regions of homology, may use breakpoint microhomology, and can create insertions due to the random addition of nucleotides by DNA polymerase μ (POLM) (Lieber 2008). Although these characteristics are consistent with the observed attributes of the simple breakpoints, it is extremely unlikely that NHEJ could have generated the complex rearrangements as this would require more than three DSBs being repaired incorrectly in the same region. Therefore, a more parsimonious explanation is provided by the recently proposed replication-based mechanisms.
The first of these replication-based mechanisms to be proposed was fork stalling and template switching (FoSTeS) (Lee et al. 2007), which is based on a suggested mechanism of gene amplification in Escherichia coli (Slack et al. 2006). In this model a genomic rearrangement is created by a replication fork stalling and the nascent lagging strand dissociating and invading one or multiple adjacent replication forks (Fig. 4C). Although such a mechanism is theoretically capable of producing both simple and complex rearrangements and would explain the formation of copy number changes following inhibition of replication (Arlt et al. 2009), FoSTeS has yet to be validated experimentally, and so several significant questions concerning its molecular details remain unanswered. Importantly, it has been shown that the induction of fork stalling events in yeast does not result in the formation of segmental duplications (Payen et al. 2008).
An alternative replication-based model is break-induced replication (BIR). This mechanism was first proposed to have evolved in yeast as a means of maintaining ploidy following DNA breakage (Morrow et al. 1997) but has now been implicated in telomere maintenance and genomic rearrangement formation in both humans and yeast (McEachern and Haber 2006; Bauters et al. 2008; Deem et al. 2008). Unlike FoSTeS, BIR requires an unpaired DSB end, which may be produced by the collapse of a replication fork, the erosion of telomeres, or the separation of two DSB ends. Classical BIR requires large regions of homology, comparable to those necessary for NAHR; however, it has recently been proposed that an alternate Rad51-independent pathway may exist that could use microhomology to anneal single-stranded DNA (Hastings et al. 2009a). This process is known as microhomology-mediated break-induced replication (MMBIR) (Fig. 4D).
Although MMBIR is only speculative at present, with no direct experimental evidence in humans or yeast, our findings are almost entirely consistent with such a model. Of the 40 RAF gene fusions formed by simple genomic rearrangements, 34 (85%) contained either breakpoint or flanking microhomology that could mediate MMBIR. In addition to this, simple rearrangements account for the vast majority of RAF gene fusions (95%), which is similar to studies of BIR in yeast, where template switching was observed in only 20% of cases (Smith et al. 2007). Unlike other studies that have reported large interspersed duplications and deletions (Lee et al. 2007; Vissers et al. 2007), the complex rearrangements we detected were <2 kb in length and directly adjacent to the breakpoint. Although it is possible that the interspersed copy number changes observed by other groups could occur by MMBIR (Hastings et al. 2009a), the experimental evidence from BIR in yeast has shown that most template switching events occur up to 10 kb from the initial site of strand invasion (Smith et al. 2007), a characteristic that is shared by our findings. Furthermore, it has previously been suggested that MMBIR can occur during mitosis and generate rearrangements ranging from several megabases to a few hundred base pairs in length (Zhang et al. 2009). Therefore, of the currently proposed mechanisms, we believe that MMBIR is most likely to be responsible for the formation of RAF gene fusions in low-grade astrocytomas.
Our study is not only the first to illustrate that MMBIR is consistent with a specific recurrent rearrangement in cancer, it also has implications concerning the mechanism itself. The first of these is our discovery that flanking microhomology is enriched at the DNA breakpoints. Several studies have previously demonstrated that breakpoint microhomology is overrepresented at the junctions of pathogenic genomic rearrangements, including those associated with cancer (Bignell et al. 2007; Vissers et al. 2009). However, none, to our knowledge, have investigated the presence of microhomology at sites that do not directly overlap the breakpoint. Therefore, it is possible that in many cases the reported extent of microhomology between junction sequences in genomic rearrangements is an underestimate. Even our definition may prove too conservative because it fails to consider microhomology from alternative alignments (Supplemental Fig. S4). In order for flanking microhomology to mediate MMBIR, it must be present on the unpaired DSB end. Of the 11 simple rearrangements exhibiting flanking microhomology, eight samples had it on the KIAA1549 side, two had it on the BRAF side, and one had a region on each side. By using this principle, other studies may be able to verify whether replication fork collapse consistently occurs at one side of the breakpoint.
The only motif that we found to be significantly enriched at the fusion breakpoints was the human minisatellite conserved sequence/χ-like element. This motif was present in the region ±25 bp of the KIAA1549 junction in only four tumor samples and so clearly cannot be an absolute requirement for the creation of these tandem duplications. Despite this, it is still possible that, in a minority of cases, the χ-like element plays a role in RAF gene fusion formation. The χ element was initially identified as a mediator of prokaryotic recombination, specifically altering the activity of the RecBCD enzyme (Lam et al. 1974; Dixon and Kowalczykowski 1993). However, it has now been identified as a recombination hotspot involved in the formation of many eukaryotic genomic rearrangements, including several oncogenic translocations (Krowczynska et al. 1990; Jaeger et al. 1993; Lopez-Correa et al. 2001; Abeysinghe et al. 2003). It is possible that this is due to the χ-like element acting as a recognition site for V(D)J recombinase (Wyatt et al. 1992) and so increases the likelihood of NHEJ occurring. This could, therefore, indicate that NHEJ is responsible for the simple rearrangements we observed or, conversely, that χ-like elements may also mediate MMBIR.
Another observation that furthers our understanding of MMBIR is the presence of inserted nucleotides at the breakpoints of complex rearrangements. Small insertions are often considered to be a defining characteristic of NHEJ, as DNA polymerase μ is capable of template-independent synthesis under physiological conditions (Gu et al. 2007). However, the presence of inserted nucleotides at both of the SRGAP3–RAF1 breakpoints, along with similar findings in a previous study (Zhang et al. 2009), could either suggest the involvement of DNA polymerase μ (POLM) in MMBIR or be indicative of additional template switching events. In the case of the latter, adjacent nucleotides may have been incorporated using another template but are not identified as insertions due to breakpoint microhomology. It is, therefore, possible that some or all of the rearrangements that we designated as simple with small insertions may, in fact, be complex.
Several other factors further complicate the identification of complex rearrangements. PA28, the single sample in which we were unable to identify the fusion breakpoint, may contain a complex rearrangement. Our failure to amplify and sequence the junction here may be due to the presence of a large insertion. Another possibility, which could affect a greater number of samples, is that we may have been unable to identify interspersed duplications or deletions. To reduce the likelihood of this, we analyzed the region of gain and its neighboring segments for gains or losses in 38 samples using the Affymetrix 6.0 data from our previous investigation (Forshew et al. 2009). We did not identify any potential interspersed duplications or deletions using this approach; however, we cannot conclusively eliminate this possibility as the template for inserted sequences could theoretically come from anywhere in the genome, and very small regions of copy number change may not be detectable using the Affymetrix 6.0 array.
In addition to investigating the mechanisms capable of creating RAF gene fusions, we aimed to explain their relative frequencies by analyzing the local DNA sequence. Using this approach, we were unable to identify significant enrichment of repetitive elements or Z-DNA in KIAA1549, BRAF, SRGAP3, or RAF1 that could account for the specificity of fusion partners. This is not particularly surprising as either the promoter or a specific protein domain of KIAA1549 and SRGAP3 may be necessary for the associated RAF gene fusion to provide a growth advantage (Tatevossian et al. 2010). Our intronic analysis indicates that, of the repetitive elements tested, only L2 repeats and LTRs could potentially account for the discrepancy in KIAA1549–BRAF fusion variant frequency; however, the significance of the contribution made by these elements remains to be established.
It is also important to note that a sequence-based explanation for the uneven distribution of RAF gene fusion variants may not exist. Several other factors could explain this disparity, with one such possibility being variation in the local chromatin architecture. A previous study identified depletion of histone H1 and hyperacetylation of histone H3 within intron 5 of the RUNX1 gene (Stuardo et al. 2009). All of the chromosome 21 breakpoints associated with the RUNX1–RUNX1T1 (formerly known as AML1-ETO) fusion gene, which is created by a t(8;21) translocation and commonly associated with acute myeloid leukemia (Miyoshi et al. 1991), are restricted to this intron. Similar histone modifications could result in an open chromatin conformation specifically within KIAA1549 intron 16 and BRAF intron 8. Stuardo et al. (2009) also found that the same histone modifications were present in a hematopoietic cell line but absent in a cervical carcinoma cell line. Therefore, local chromatin architecture could provide an explanation for the specificity of KIAA1549–BRAF and SRGAP3–RAF1 fusions to low-grade astrocytomas.
In addition to local chromatin configuration, it may also prove useful to consider the chromatin architecture of the entire duplicated region. All of the putative mechanisms we have discussed above require interaction between the two halves of the breakpoint. There is currently some debate as to whether this spatial proximity occurs before or after DNA breakage; however, several recent studies have provided evidence in support of the “contact-first” model (Lever and Sheer 2010). If this is the case for the RAF gene fusions found in low-grade astrocytomas, it is possible that investigating the interaction between KIAA1549 and BRAF using fluorescent in situ hybridization (FISH) and chromatin conformation capture methodologies may prove informative in identifying the cell of origin for low-grade astrocytomas.
It is also of interest to consider the possibility that the interaction between KIAA1549 and BRAF may be transient, with their spatial proximity either restricted to a specific stage of development or induced by cell signaling pathways. A recent example of the latter is that androgen signaling has been shown to induce spatial proximity between TMPRSS2 and ERG, two genes that fuse together in ∼50% of prostate cancers (Lin et al. 2009; Mani et al. 2009). Alternatively, spatial proximity could be restricted to a certain stage of the cell cycle. Investigating this could, in turn, help confirm that MMBIR is the mechanism of fusion formation as one would predict the fusion partners to interact within S phase, possibly with both genes being replicated in the same replication focus.
In conclusion, while it is currently not possible to observe directly the sequence of events involved in the formation of genomic rearrangements due to technological limitations, we have, through detailed analysis of the breakpoint sequences, been able to identify key features of the underlying mechanism. Furthermore, our findings provide a framework for future investigations into the generation of both cancer-specific gene fusions and genomic variation.
Methods
Tumor tissue and control samples
Low-grade astrocytomas (WHO Grade I-II) from 43 patients aged 1–20 yr were studied. Of these samples, 33 were included in our previous study (Forshew et al. 2009) in which they are denoted by the same identifiers used here. Clinical information and molecular analysis for individual tumors can be found in Supplemental Table S1. Tissue samples were obtained at surgery prior to adjuvant therapy. Samples were fully anonymized to the researchers, and access to tissue and clinical data were in accordance with Institutional Review Board and MREC guidelines: St Jude Children's Research Hospital (USA) XPD07-107/IRB and Tissue Resource Request No 07-007; Newcastle (UK) REC ref No 2002/112; Blizard Institute of Cell and Molecular Science (UK) ICMS/PR/09/77. Pooled control DNA was obtained from the blood of 21 male volunteers.
Extraction and preparation of nucleic acid
Tumor DNA was extracted from fresh frozen tissue using the QIAamp DNA mini kit (QIAGEN). RNA was extracted using TRIzol (Invitrogen) and eluted into RNAse-free water using the RNeasy mini kit (QIAGEN). cDNA was synthesized using random hexamers and the SuperScript First-Strand cDNA synthesis system (Invitrogen). Genomic DNA was whole genome amplified using the REPLI-g whole genome amplification kit (QIAGEN). Control DNA was extracted from each individual blood sample using the Chemagen magnetic bead blood purification system (Chemagen AG).
Detection of fusion breakpoints in cDNA and genomic DNA
The cDNA of all tumor samples was screened by PCR for the presence of RAF gene fusions prior to their admission into the study. For detection of the DNA breakpoints, primers were designed at ∼500-bp intervals throughout the appropriate introns of KIAA1549, BRAF, SRGAP3, and RAF1 (Supplemental Table S6). Tumors with the same fusion variant had their whole genome amplified DNA pooled and were analyzed by PCR using all relevant primer combinations. This process was then repeated for the pooled control DNA to allow the identification of non-specific products. Primer combinations that produced tumor-specific products were tested on individual tumor samples, and the resulting products were analyzed in both directions by direct Sanger sequencing on either a 3100 Genetic Analyzer or a 3730 DNA Analyzer (Applied Biosystems). Results were analyzed using Sequence Scanner Software v1.0 (Applied Biosystems). For those tumors where the breakpoints failed to amplify, we repeated the aforementioned assay using the Extensor Long Range PCR Kit (Thermo Fisher Scientific) instead of Platinum Taq DNA polymerase (Invitrogen).
Bioinformatics analyses
The assembly of the human genome used was GRCh37. The statistical significance of fusion variant frequency was tested using a χ2-test. The intronic distribution of breakpoints was analyzed as follows: For each intron, we randomly distributed as many simulated breakpoints throughout the intron as we observed in our data set. By repeating this process and indexing the breakpoints, we established 95% confidence intervals for the expected location of successive breakpoints. Additionally, we carried out a Kolmogorov-Smirnov test against the null hypothesis that the locations are drawn from a uniform distribution across the intron.
To test for the enrichment of microhomology, five control sets of 500 simulated breakpoint pairs, with one breakpoint randomly situated in BRAF intron 8 and the other located in KIAA1549 intron 16, were generated, and, for each breakpoint pair, the extent of microhomology was determined. The statistical significance of differences between the simulated and observed sample distributions was tested by a Mann-Whitney U-test. The significance of flanking microhomology was tested by a Fisher's exact test.
The Human Genome Segmental Duplication Database (Cheung et al. 2003) (http://projects.tcag.ca/humandup/) was used to check KIAA1549, BRAF, SRGAP3, and RAF1 for segmental duplications. The search for large regions of homology was conducted using BLASTN 2.2.22+ (Altschul et al. 1997) (http://blast.ncbi.nlm.nih.gov/).
A list of 1000 randomly selected, non-overlapping, autosomal human genes between 100 kb and 250 kb in length was generated using the BioMart tool on the Ensembl genome browser (http://www.ensembl.org/biomart/). Analysis of repetitive elements within the control genes and fusion partners was carried out using the table browser tool on the UCSC genome browser and the RepeatMasker software (Smit et al. 1996) (http://www.repeatmasker.org). Zhunt online (Ho et al. 1986) (http://bioinfo.cgrb.oregonstate.edu/zDNA/) was used to detect possible regions of Z-DNA. Empirical P-values, with Bonferroni correction, were used to test the differences in the number of repetitive elements and Z-DNA within the fusion partners relative to the set of control genes. Empirical P-values were calculated by (r + 1)/(n + 1), where n is the number of replicate samples and r is the number of replicates with a test statistic greater than the actual value (North et al. 2002).
The analysis of common motifs, on either strand, within ±25 bp of fusion breakpoints was carried out using fuzznuc, which is available as part of the European Molecular Biology Open Software Suite (EMBOSS) (Rice et al. 2000). A set of 10,000 DNA sequences of 50 bp in length was generated from the list of 1000 control genes and tested for the same common motifs with fuzznuc. This was subdivided into two sets of 5000 sequences that were used as separate control sets for KIAA1549 and BRAF. The statistical significance of motif enrichment at the breakpoints was tested using a Fisher's exact test with Bonferroni correction at the 5% significance level.
Acknowledgments
This work was supported by the Samantha Dickson Brain Tumour Trust, Cancer Research UK London Research Institute, Cancer Research UK program grant C5321/A8318, and the American Lebanese Syrian Associated Charities. We thank the Equipment Park of the Cancer Research UK London Research Institute for assistance with sequencing.
Footnotes
[Supplemental material is available for this article. The sequence data from this study have been submitted to the European Nucleotide Archive (http://www.ebi.ac.uk/ena/) under accession nos. FR799511-FR799553.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.115782.110.
References
- Abeysinghe SS, Chuzhanova N, Krawczak M, Ball EV, Cooper DN 2003. Translocation and gross deletion breakpoints in human inherited disease and cancer I: Nucleotide composition and recombination-associated motifs. Hum Mutat 22: 229–244 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt MF, Durkin SG, Ragland RL, Glover TW 2006. Common fragile sites as targets for chromosome rearrangements. DNA Repair (Amst) 5: 1126–1135 [DOI] [PubMed] [Google Scholar]
- Arlt MF, Mulle JG, Schaibley VM, Ragland RL, Durkin SG, Warren ST, Glover TW 2009. Replication stress induces genome-wide copy number changes in human cells that resemble polymorphic and pathogenic variants. Am J Hum Genet 84: 339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacolla A, Wojciechowska M, Kosmider B, Larson JE, Wells RD 2006. The involvement of non-B DNA structures in gross chromosomal rearrangements. DNA Repair (Amst) 5: 1161–1170 [DOI] [PubMed] [Google Scholar]
- Bailey JA, Liu G, Eichler EE 2003. An Alu transposition model for the origin and expansion of human segmental duplications. Am J Hum Genet 73: 823–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauters M, Van Esch H, Friez MJ, Boespflug-Tanguy O, Zenker M, Vianna-Morgante AM, Rosenberg C, Ignatius J, Raynaud M, Hollanders K, et al. 2008. Nonrecurrent MECP2 duplications mediated by genomic architecture-driven DNA breaks and break-induced replication repair. Genome Res 18: 847–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell GR, Santarius T, Pole JC, Butler AP, Perry J, Pleasance E, Greenman C, Menzies A, Taylor S, Edkins S, et al. 2007. Architectures of somatic genomic rearrangement in human cancer amplicons at sequence-level resolution. Genome Res 17: 1296–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PJ, Stephens PJ, Pleasance ED, O'Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, et al. 2008. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet 40: 722–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung J, Estivill X, Khaja R, MacDonald JR, Lau K, Tsui LC, Scherer SW 2003. Genome-wide detection of segmental duplications and potential assembly errors in the human genome sequence. Genome Biol 4: R25 doi: 10.1186/gb-2003-4-4-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deem A, Barker K, Vanhulle K, Downing B, Vayl A, Malkova A 2008. Defective break-induced replication leads to half-crossovers in Saccharomyces cerevisiae. Genetics 179: 1845–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Smith AJ, Walters RG, Coin LJ, Steinfeld I, Yakhini Z, Sladek R, Froguel P, Blakemore AI 2008. Small deletion variants have stable breakpoints commonly associated with Alu elements. PLoS ONE 3: e3104 doi: 10.1371/journal.pone.0003104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DA, Kowalczykowski SC 1993. The recombination hotspot χ is a regulatory sequence that acts by attenuating the nuclease activity of the E. coli RecBCD enzyme. Cell 73: 87–96 [DOI] [PubMed] [Google Scholar]
- Fan C, Chen Y, Long M 2008. Recurrent tandem gene duplication gave rise to functionally divergent genes in Drosophila. Mol Biol Evol 25: 1451–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenstermaker RA, Ciesielski MJ, Castiglia GJ 1998. Tandem duplication of the epidermal growth factor receptor tyrosine kinase and calcium internalization domains in A-172 glioma cells. Oncogene 16: 3435–3443 [DOI] [PubMed] [Google Scholar]
- Feuk L, Carson AR, Scherer SW 2006. Structural variation in the human genome. Nat Rev Genet 7: 85–97 [DOI] [PubMed] [Google Scholar]
- Forshew T, Tatevossian RG, Lawson AR, Ma J, Neale G, Ogunkolade BW, Jones TA, Aarum J, Dalton J, Bailey S, et al. 2009. Activation of the ERK/MAPK pathway: A signature genetic defect in posterior fossa pilocytic astrocytomas. J Pathol 218: 172–181 [DOI] [PubMed] [Google Scholar]
- Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, Mann M, Lieber MR 1997. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature 388: 492–495 [DOI] [PubMed] [Google Scholar]
- Greaves MF, Wiemels J 2003. Origins of chromosome translocations in childhood leukaemia. Nat Rev Cancer 3: 639–649 [DOI] [PubMed] [Google Scholar]
- Gu J, Lu H, Tippin B, Shimazaki N, Goodman MF, Lieber MR 2007. XRCC4:DNA ligase IV can ligate incompatible DNA ends and can ligate across gaps. EMBO J 26: 1010–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Zhang F, Lupski JR 2008. Mechanisms for human genomic rearrangements. Pathogenetics 1: 4 doi: 10.1186/1755-8417-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR 2009a. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet 5: e1000327 doi: 10.1371/journal.pgen.1000327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G 2009b. Mechanisms of change in gene copy number. Nat Rev Genet 10: 551–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho PS, Ellison MJ, Quigley GJ, Rich A 1986. A computer aided thermodynamic approach for predicting the formation of Z-DNA in naturally occurring sequences. EMBO J 5: 2737–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger U, Purtscher B, Karth GD, Knapp S, Mannhalter C, Lechner K 1993. Mechanism of the chromosomal translocation t(14;18) in lymphoma: Detection of a 45-Kd breakpoint binding protein. Blood 81: 1833–1840 [PubMed] [Google Scholar]
- Jones DT, Ichimura K, Liu L, Pearson DM, Plant K, Collins VP 2006. Genomic analysis of pilocytic astrocytomas at 0.97 Mb resolution shows an increasing tendency toward chromosomal copy number change with age. J Neuropathol Exp Neurol 65: 1049–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Kocialkowski S, Liu L, Pearson DM, Backlund LM, Ichimura K, Collins VP 2008. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res 68: 8673–8677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Kocialkowski S, Liu L, Pearson DM, Ichimura K, Collins VP 2009. Oncogenic RAF1 rearrangement and a novel BRAF mutation as alternatives to KIAA1549:BRAF fusion in activating the MAPK pathway in pilocytic astrocytoma. Oncogene 28: 2119–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk LC, Hartwell LH 1992. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132: 387–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna KK, Jackson SP 2001. DNA double-strand breaks: Signaling, repair and the cancer connection. Nat Genet 27: 247–254 [DOI] [PubMed] [Google Scholar]
- Kitano J, Ross JA, Mori S, Kume M, Jones FC, Chan YF, Absher DM, Grimwood J, Schmutz J, Myers RM, et al. 2009. A role for a neo-sex chromosome in stickleback speciation. Nature 461: 1079–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krowczynska AM, Rudders RA, Krontiris TG 1990. The human minisatellite consensus at breakpoints of oncogene translocations. Nucleic Acids Res 18: 1121–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam ST, Stahl MM, McMilin KD, Stahl FW 1974. Rec-mediated recombinational hot spot activity in bacteriophage lambda. II. A mutation which causes hot spot activity. Genetics 77: 425–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson AR, Tatevossian RG, Phipps KP, Picker SR, Michalski A, Sheer D, Jacques TS, Forshew T 2010. RAF gene fusions are specific to pilocytic astrocytoma in a broad paediatric brain tumour cohort. Acta Neuropathol 120: 271–273 [DOI] [PubMed] [Google Scholar]
- Lee JA, Carvalho CM, Lupski JR 2007. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131: 1235–1247 [DOI] [PubMed] [Google Scholar]
- Lever E, Sheer D 2010. The role of nuclear organization in cancer. J Pathol 220: 114–125 [DOI] [PubMed] [Google Scholar]
- Lieber MR 2008. The mechanism of human nonhomologous DNA end joining. J Biol Chem 283: 1–5 [DOI] [PubMed] [Google Scholar]
- Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, Zhang J, Rose DW, Fu XD, Glass CK, et al. 2009. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell 139: 1069–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Correa C, Dorschner M, Brems H, Lazaro C, Clementi M, Upadhyaya M, Dooijes D, Moog U, Kehrer-Sawatzki H, Rutkowski JL, et al. 2001. Recombination hotspot in NF1 microdeletion patients. Hum Mol Genet 10: 1387–1392 [DOI] [PubMed] [Google Scholar]
- Maher EA, Brennan C, Wen PY, Durso L, Ligon KL, Richardson A, Khatry D, Feng B, Sinha R, Louis DN, et al. 2006. Marked genomic differences characterize primary and secondary glioblastoma subtypes and identify two distinct molecular and clinical secondary glioblastoma entities. Cancer Res 66: 11502–11513 [DOI] [PubMed] [Google Scholar]
- Mani RS, Tomlins SA, Callahan K, Ghosh A, Nyati MK, Varambally S, Palanisamy N, Chinnaiyan AM 2009. Induced chromosomal proximity and gene fusions in prostate cancer. Science 326: 1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern MJ, Haber JE 2006. Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem 75: 111–135 [DOI] [PubMed] [Google Scholar]
- Mitelman F, Johansson B, Mertens F 2007. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer 7: 233–245 [DOI] [PubMed] [Google Scholar]
- Miyoshi H, Shimizu K, Kozu T, Maseki N, Kaneko Y, Ohki M 1991. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc Natl Acad Sci 88: 10431–10434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow DM, Connelly C, Hieter P 1997. “Break copy” duplication: A model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics 147: 371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao M, Yokota S, Iwai T, Kaneko H, Horiike S, Kashima K, Sonoda Y, Fujimoto T, Misawa S 1996. Internal tandem duplication of the flt3 gene found in acute myeloid leukemia. Leukemia 10: 1911–1918 [PubMed] [Google Scholar]
- North BV, Curtis D, Sham PC 2002. A note on the calculation of empirical P values from Monte Carlo procedures. Am J Hum Genet 71: 439–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payen C, Koszul R, Dujon B, Fischer G 2008. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet 4: e1000175 doi: 10.1371/journal.pgen.1000175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson M, Andren Y, Mark J, Horlings HM, Persson F, Stenman G 2009. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci 106: 18740–18744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Stephens PJ, O'Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, et al. 2010. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 463: 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D 1997. GeneCards: Integrating information about genes, proteins and diseases. Trends Genet 13: 163. [DOI] [PubMed] [Google Scholar]
- Reiter LT, Hastings PJ, Nelis E, De Jonghe P, Van Broeckhoven C, Lupski JR 1998. Human meiotic recombination products revealed by sequencing a hotspot for homologous strand exchange in multiple HNPP deletion patients. Am J Hum Genet 62: 1023–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A 2000. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet 16: 276–277 [DOI] [PubMed] [Google Scholar]
- Rubnitz J, Subramani S 1984. The minimum amount of homology required for homologous recombination in mammalian cells. Mol Cell Biol 4: 2253–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievert AJ, Jackson EM, Gai X, Hakonarson H, Judkins AR, Resnick AC, Sutton LN, Storm PB, Shaikh TH, Biegel JA 2009. Duplication of 7q34 in pediatric low-grade astrocytomas detected by high-density single-nucleotide polymorphism-based genotype arrays results in a novel BRAF fusion gene. Brain Pathol 19: 449–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack A, Thornton PC, Magner DB, Rosenberg SM, Hastings PJ 2006. On the mechanism of gene amplification induced under stress in Escherichia coli. PLoS Genet 2: e48 doi: 10.1371/journal.pgen.0020048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P 1996. RepeatMasker Open-3.0. http://www.repeatmasker.org/
- Smith CE, Llorente B, Symington LS 2007. Template switching during break-induced replication. Nature 447: 102–105 [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR 2010. Structural variation in the human genome and its role in disease. Annu Rev Med 61: 437–455 [DOI] [PubMed] [Google Scholar]
- Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, et al. 2009. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 462: 1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA 2009. The cancer genome. Nature 458: 719–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuardo M, Martinez M, Hidalgo K, Montecino M, Javed A, Lian JB, Stein GS, Stein JL, Gutierrez SE 2009. Altered chromatin modifications in AML1/RUNX1 breakpoint regions involved in (8;21) translocation. J Cell Physiol 218: 343–349 [DOI] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A, Takeda S 1998. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J 17: 5497–5508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatevossian RG, Lawson AR, Forshew T, Hindley GF, Ellison DW, Sheer D 2010. MAPK pathway activation and the origins of pediatric low-grade astrocytomas. J Cell Physiol 222: 509–514 [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. 2005. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310: 644–648 [DOI] [PubMed] [Google Scholar]
- Vissers LE, Stankiewicz P, Yatsenko SA, Crawford E, Creswick H, Proud VK, de Vries BB, Pfundt R, Marcelis CL, Zackowski J, et al. 2007. Complex chromosome 17p rearrangements associated with low-copy repeats in two patients with congenital anomalies. Hum Genet 121: 697–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers LE, Bhatt SS, Janssen IM, Xia Z, Lalani SR, Pfundt R, Derwinska K, de Vries BB, Gilissen C, Hoischen A, et al. 2009. Rare pathogenic microdeletions and tandem duplications are microhomology-mediated and stimulated by local genomic architecture. Hum Mol Genet 18: 3579–3593 [DOI] [PubMed] [Google Scholar]
- Wells RD 2007. Non-B DNA conformations, mutagenesis and disease. Trends Biochem Sci 32: 271–278 [DOI] [PubMed] [Google Scholar]
- Wilson TE, Grawunder U, Lieber MR 1997. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature 388: 495–498 [DOI] [PubMed] [Google Scholar]
- Wu W, Wang M, Singh SK, Mussfeldt T, Iliakis G 2008. Repair of radiation induced DNA double strand breaks by backup NHEJ is enhanced in G2. DNA Repair (Amst) 7: 329–338 [DOI] [PubMed] [Google Scholar]
- Wyatt RT, Rudders RA, Zelenetz A, Delellis RA, Krontiris TG 1992. BCL2 oncogene translocation is mediated by a chi-like consensus. J Exp Med 175: 1575–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Khajavi M, Connolly AM, Towne CF, Batish SD, Lupski JR 2009. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat Genet 41: 849–853 [DOI] [PMC free article] [PubMed] [Google Scholar]