Abstract
Reciprocal chromosome translocations are often not exactly reciprocal. Most familiar are deletions at the breakpoints, up to megabases in extent. We describe here the opposite phenomenon—duplication of tens or hundreds of kilobases at the breakpoint junction, so that the same sequence is present on both products of a translocation. When the products of the translocation are mapped on the genome, they overlap. We report several of these “overlapping-breakpoint” duplications in breast cancer cell lines HCC1187, HCC1806, and DU4475. These lines also had deletions and essentially balanced translocations. In HCC1187 and HCC1806, we identified five cases of duplication ranging between 46 kb and 200 kb, with the partner chromosome showing deletions between 29 bp and 31 Mb. DU4475 had a duplication of at least 200 kb. Breakpoints were mapped using array painting, i.e., hybridization of chromosomes isolated by flow cytometry to custom oligonucleotide microarrays. Duplications were verified by fluorescent in situ hybridization (FISH), PCR on isolated chromosomes, and cloning of breakpoints. We propose that these duplications are the counterpart of deletions and that they are produced at a replication bubble, comprising two replication forks with the duplicated sequence in between. Both copies of the duplicated sequence would go to one daughter cell, on different products of the translocation, while the other daughter cell would show deletion. These duplications may have been overlooked because they may be missed by FISH and array-CGH and may be interpreted as insertions by paired-end sequencing. Such duplications may therefore be quite frequent.
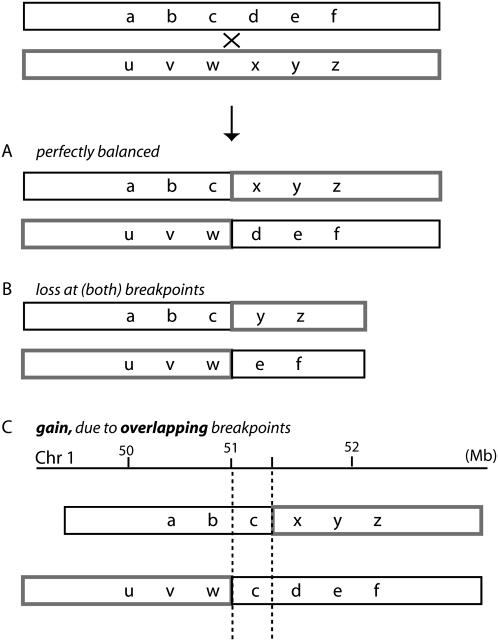
It has been recognized for some time that reciprocal chromosome translocations are often not perfectly reciprocal, i.e., there is some additional rearrangement of the genome around the breakpoint junctions. This is of more than academic interest since the additional rearrangement often determines the genetic consequences of the translocations. This applies both to neoplastic and constitutional translocations. While some additional rearrangements involve only a few base pairs to a few hundred base pairs (Shimizu et al. 1992; Gajecka et al. 2008), many of them involve tens to thousands of kilobases. Most familiar are deletions at the breakpoints, illustrated in Figure 1B. In neoplasia, for example, around one-fifth to one-third of the t(9;22) translocations of chronic myeloid leukemia show deletion of at least 100 kb and up to several megabases (Sinclair et al. 2000): While the breakpoint junction on the der(22) product always creates the classic BCR-ABL gene fusion (Sinclair et al. 2000), the der(9) product can have breakpoints up to several megabases away. Similarly, constitutional reciprocal translocations that are associated with a phenotype frequently have megabase deletions at the breakpoints, which probably often determine their phenotypic consequences (Ciccone et al. 2005; Gribble et al. 2005; De Gregori et al. 2007).
Figure 1.
Breakpoint complexity at apparently balanced translocations. (A) Perfectly balanced rearrangement with no net gain or loss of material at the junction. (B) Deletion of material at the breakpoints of one or both translocation partners. (C) Duplication of material due to the same sequence being present on both products of a translocation—the breakpoints overlap on a genomic map (shown schematically for chromosome 1 as an example). Breakpoints are indicated with a dashed line, and the sequence in between is common to both translocation products.
We describe here the opposite phenomenon—duplication of tens or hundreds of kilobases of sequence so that the same sequence is present on both products of a translocation. When the translocation products are mapped on the genome, they overlap (Fig. 1C). We could find only three previous reports of such duplications at a translocation breakpoint. Cox et al. (2003) described a 650-kb duplication in a constitutional translocation, Stankiewicz et al. (2001) described a 250-kb duplication at the (4;19) evolutionary reciprocal translocation in Gorilla gorilla, and Koszul et al. (2004) described a 115-kb duplication in a model yeast experiment.
While analyzing the chromosome translocation breakpoints of four breast cancer cell lines (Howarth et al. 2008; this study), we found several examples of apparently balanced translocations where the breakpoints on the two product chromosomes appeared to overlap by tens or hundreds of kilobases; i.e., the same sequence was present on both products of the translocation (Fig. 1C). We present here detailed characterization of these duplications.
Results
We first characterized two examples, each from a different cell line, using several independent methods, to confirm that the apparent duplication was, indeed, present, with the two translocation products carrying tens of kilobases of common sequence.
Duplication at a t(4;6) reciprocal translocation in HCC1806
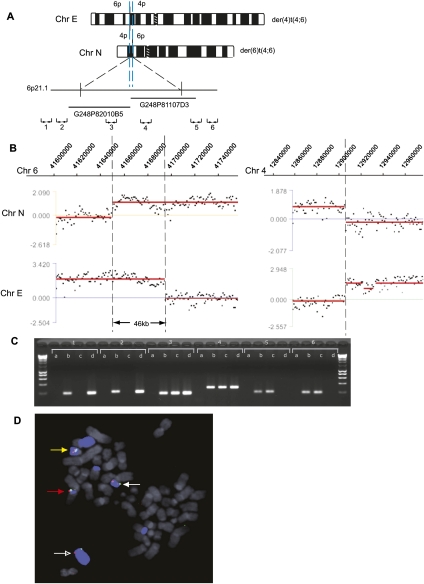
The breast cancer cell line HCC1806 has a reciprocal translocation between chromosomes 4 and 6, t(4;6)(p15.33;p21.1) (Fig. 2; Table 1; Howarth et al. 2008). Breakpoints were initially determined by “array painting,” i.e., isolating chromosomes by flow cytometry and hybridizing them to DNA microarrays (Howarth et al. 2008). The two product chromosomes, the der(6)t(4;6) and der(4)t(4;6) (meaning, respectively, the products that retain the chromosome 6 and chromosome 4 centromeres), were hybridized to custom oligonucleotide arrays with an average probe spacing of 150 to 170 bp. As previously reported (Howarth et al. 2008), this showed breakpoints on chromosome 6p21.1 at 41.65 Mb on the der(6)t(4;6) and 41.695 Mb on the der(4)t(4;6), implying that the 46 kb of sequence between these points was present on both products (Fig. 2B; Table 1).
Figure 2.
HCC1806 t(4;6) duplication at a breakpoint. (A) Schematic representation of the products (Chr E and Chr N) of the reciprocal translocation between chromosomes 4 and 6 in HCC1806. The duplicated region of Chr 6 is shown between blue dashed lines, and the approximate location of fosmids used in FISH mapping is shown. The approximate location of chromosome 6 PCR primer pairs (1–6) is also shown. (B) Hybridization of Chr N and Chr E to a custom NimbleGen oligonucleotide array covering specified regions on chromosome 6 (left) and chromosome 4 (right). Breakpoints are indicated with a broken line. (C) Breakpoint mapping by PCR on flow-sorted chromosomes. PCR results with primer pairs 1–6 are shown. Lanes are labeled a (negative water control), b (HCC1806 genomic DNA), c (Chr N), and d (Chr E). (D) Breakpoint mapping by FISH using fosmids G248P81107D3 (shown in red) and G248P82010B5 (shown in green). Chromosome 6 is shown in blue. The der(4)t(1;6;4) is indicated with a red arrow (Chr B), the der(4)t(4;6) with a white arrow (Chr E), the der(6)t(4;6) with an open white arrow (Chr N), and the del (6) with a yellow arrow (Chr j). Other pieces of Chr 6 are the der(10)t(6;10) (Chr V), the der(14)t(6;14) (Chr Z), and the der(6)t(1;6) (Chr F) (Howarth et al. 2008).
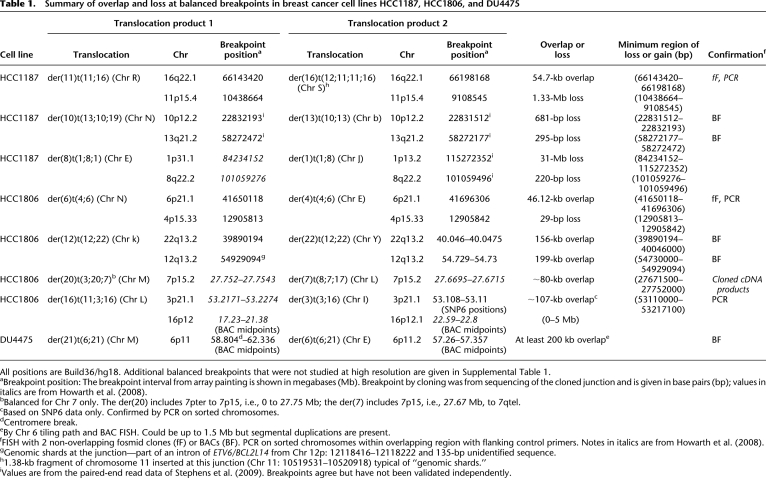
Table 1.
Summary of overlap and loss at balanced breakpoints in breast cancer cell lines HCC1187, HCC1806, and DU4475
All positions are Build36/hg18. Additional balanced breakpoints that were not studied at high resolution are given in Supplemental Table 1.
aBreakpoint position: The breakpoint interval from array painting is shown in megabases (Mb). Breakpoint by cloning was from sequencing of the cloned junction and is given in base pairs (bp); values in italics are from Howarth et al. (2008).
bBalanced for Chr 7 only. The der(20) includes 7pter to 7p15, i.e., 0 to 27.75 Mb; the der(7) includes 7p15, i.e., 27.67 Mb, to 7qtel.
cBased on snp6 data only. Confirmed by PCR on sorted chromosomes.
dCentromere break.
eBy Chr 6 tiling path and BAC FISH. Could be up to 1.5 Mb but segmental duplications are present.
fFISH with 2 non-overlapping fosmid clones (fF) or BACs (BF). PCR on sorted chromosomes within overlapping region with flanking control primers. Notes in italics are from Howarth et al. (2008).
gGenomic shards at the junction—part of an intron of ETV6/BCL2L14 from Chr 12p: 12118416–12118222 and 135-bp unidentified sequence.
h1.38-kb fragment of chromosome 11 inserted at this junction (Chr 11: 10519531–10520918) typical of “genomic shards.”
iValues are from the paired-end read data of Stephens et al. (2009). Breakpoints agree but have not been validated independently.
We confirmed the position of the breakpoints by cloning: The chromosome 4 breakpoints were mapped in the same way (see below), and the junctions were amplified by long-range PCR. From these junctions, the breakpoints on chromosome 6 were at 41.650118 Mb on the der(6) and 41.696306 Mb on the der(4), in excellent agreement with the array mapping (Fig. 2B; Table 1; for junction sequences, see Supplemental Table 3).
The presence of the duplicated sequences on both products was confirmed independently by PCR and fluorescent in situ hybridization (FISH). PCR was performed on the isolated chromosomes, using a series of primer pairs spaced across the region. The region of duplication, but not the flanking regions, was present on both product chromosomes (Fig. 2C). Fosmids within the region of duplication, G248P82010B5 and G248P81107D3, were positive by FISH on both the der(6)t(4;6) and the der(4)t(4;6) (Fig. 2D).
The region of duplication is a copy number gain, so would, in principle, be detectable by array comparative genomic hybridization (array-CGH) of sufficiently high resolution. There are published array-CGH data for the cell lines used in this study from Bignell et al. (2010) (http://www.sanger.ac.uk/genetics/CGP) on the Affymetrix SNP6 array, which are consistent with the presence of the duplication, but the gain is at the limit of resolution (Supplemental Fig. 1A).
Collectively, these observations demonstrate that the duplicated sequence is, indeed, present on both products of the translocation, although the individual approaches might leave some doubt. For example, the apparent overlap of mapped breakpoints in the array hybridization could, in principle, arise from errors in mapping the hybridization, e.g., in translation between genome assemblies. However, the cloned junction sequences and PCR marker results rule out such a mistake, and 46 kb does not correspond to the differences between recent assemblies. Hybridization to an array can be cross-hybridization of a closely related sequence, but there are no segmental duplications in this region (http://www.genome.ucsc.edu) (Kent et al. 2002). Similarly, the fosmid FISH shows there are sequences from each fosmid present on each product, but it does not confirm the true extent of the common sequence. The fosmids actually share 409 bp of sequence at 41.669959–41.670368 Mb, but as hybridization of less than a few kilobases would not be detected, and a breakpoint at 41.67 Mb would be incompatible with both the array painting and junction cloning, there must be some duplication.
In contrast to this duplication on chromosome 6, the breakpoints on chromosome 4 were within 29 bp of each other (Fig. 2B; Table 1). They were mapped similarly, by array painting to custom arrays (Howarth et al. 2008), and cloning, and the breaks were at 12.905842 Mb on the der(4) and 12.905813 Mb on the der(6), resulting in loss of 29 bp.
Knowledge of the duplication was essential for determining the genetic consequences of the rearrangement. The der(4) has a break downstream from FOXP4 so that it carries an intact copy of FOXP4, while the der(6) break falls in the second intron of FOXP4, retaining the 3′ end of the gene. There does not, however, appear to be a gene broken on chromosome 4 that would form a gene fusion. Real time RT-PCR analysis did not reveal any striking difference in the expression of FOXP4 in this cell line compared to other breast cancer cell lines (data not shown).
Origin of duplicated sequences
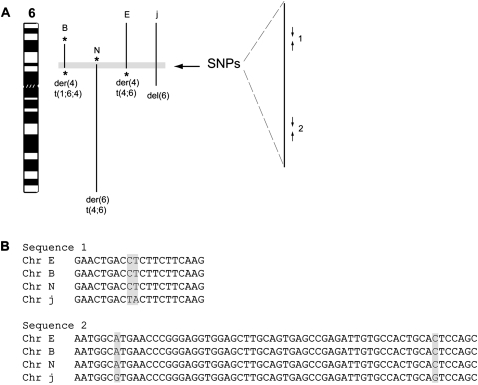
The two copies of the same sequence on chromosome 6 could, in principle, arise from two different copies of the chromosome; elsewhere (Alsop et al. 2008) we have argued that recombination may occur between two copies of the same chromosome. To test this, we chose SNPs (single nucleotide polymorphisms) that were heterozygous in this cell line, within the region on chromosome 6 that is common to the two translocation products (Fig. 3A). The sequences were the same on the two product chromosomes, showing that the two copies came from the same parent chromosome, while the other allele of the SNPs was detected on the unrelated copy of this region, on the del(6) (chromosome j in Fig. 3B).
Figure 3.
Sequencing of SNPs at the duplicated chromosome 6 junction in HCC1806 reveals products of the reciprocal translocation between chromosomes 4 and 6 have the same haplotype. (A) An ideogram of chromosome 6 is shown, with abnormal chromosome 6 segments in HCC1806 indicated with black lines, labeled with their short chromosome names, chromosome B, etc. They are the der(4)t(1;6;4) (Chr B), the der(6)t(4;6) (Chr N), the der(4)t(4;6) (Chr E), and the del(6) (Chr j) (Howarth et al. 2008). Balanced breaks are indicated by *. The 46-kb region of chromosome 6 common to all chromosomes is highlighted with a gray box and shown expanded to the right. The approximate location of PCR primer pairs 1 and 2 is shown (giving PCR products of 250 bp and 350 bp, respectively; not to scale). (B) Examples of sequences from primer pairs 1 and 2 from the different flow-sorted chromosomes. SNPs are highlighted with gray boxes.
This further argues against the detected duplication being an artifact of segmental duplication, since the sequence around the heterozygous SNPs on the two product chromosomes was unique and identical.
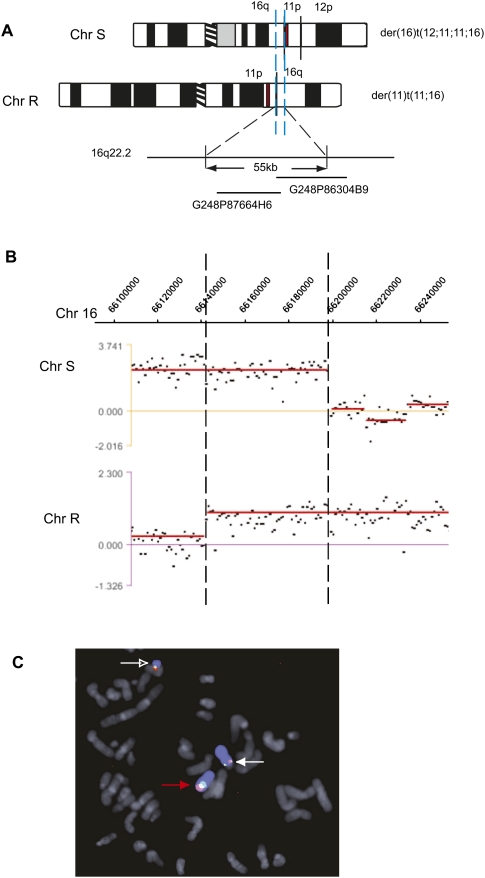
Duplication at a t(11;16) reciprocal translocation in HCC1187
Using similar approaches, we showed that the reciprocal translocation between chromosomes 11 and 16 in HCC1187 results in a gain of 55 kb of chromosome 16 (Fig. 4; Table 1). This translocation occurred between chromosome 16 and one of two copies of a previous 11;12 translocation, resulting in products der(16)t(12;11;11;16) and der(11)t(11;16) (Howarth et al. 2008). Array painting on custom arrays showed breakpoints on chromosome 16q22.1 at ∼66.143 Mb on the der(11)t(11;16) and 66.198 Mb on the der(16)t(12;11;11;16), implying that the 55 kb between these points was present on both products (Fig. 4B; Howarth et al. 2008).
Figure 4.
HCC1187 t(11;16) duplication at a breakpoint. (A) Schematic representation of the products of the reciprocal translocation between chromosomes 11 and 16 in HCC1187 (Chr S and Chr R). The duplicated region of Chr 16 is shown between blue dashed lines. The approximate location of fosmids used in FISH mapping is shown. A 1.3-kb duplicated piece of chromosome 11 is shown with a red bar. (B) Hybridization of Chr S and Chr R to a custom NimbleGen oligonucleotide array covering a specified region on chromosome 16. Breakpoints are indicated with a broken line. (C) Breakpoint mapping by FISH using fosmids G248P87664H6 (shown in red) and G248P86304B9 (shown in green). Chromosome 16 is shown in blue. Normal chromosome 16 is indicated with a red arrow, the der (16) with a white arrow (Chr S), and the der(11) with an open white arrow (Chr R) (confirming single-color FISH shown in Howarth et al. 2008).
Both breakpoint junction sequences were obtained by mapping the chromosome 11 breaks on custom arrays (see below) and cloning the junctions by PCR. The breakpoints on chromosome 16 were at 66,143,420 bp and 66,198,168 bp, precisely consistent with the array painting (Table 1). Junction sequences for the der(16) (junction 4 in Supplemental Table 3) are in exact agreement with recent paired-end read data for HCC1187 (Stephens et al. 2009).
The presence of sequence on both translocation products was confirmed by FISH, refining our previous data (Howarth et al. 2008). Fosmids G248P87664H6 and G248P86304B9 map largely or partially within the region of duplication, and both were positive by FISH on both products of the translocation (Fig. 4C). They have a small region, 6 kb, of common sequence.
The expected copy number increase in the 55-kb region of duplication was again at the limit of resolution of the SNP6 array-CGH of Bignell et al. (2010), but the data were consistent with the gain—in comparison with the matched normal lymphoblastoid line, which is available for HCC1187 (Supplemental Fig. 1F).
In contrast, there was net loss of 1.33 Mb at the breakpoints on chromosome 11p15.4. The breakpoints were both mapped using custom oligonucleotide arrays, to 9.1085–9.109 Mb and 10.435–10.438 Mb (Supplemental Fig. 2B; Howarth et al. 2008). The cloned junctions were at 10,438,664 bp and 9,108,545 bp (Table 1). Inserted into the der(16) junction was a small (1.38 kb) fragment of chromosome 11 from 10.519531 Mb to 10.520918 Mb (Table 1; Supplemental Fig. 2C; Supplemental Table 3), 80 kb away from the breakpoint, typical of the small fragments of distant sequence that are often found inserted in junctions, named “genomic shards” (Bignell et al. 2007; Hastings et al. 2009a). The resulting loss was clear in the SNP6 array-CGH data from Bignell et al. (2010) (Supplemental Fig. 1G).
Again, the duplication alters the consequences for the genes at the translocation junctions. On the der(16), the translocation creates a fusion gene: the break on chromosome 16 is in the second intron of CTCF, retaining the first two untranslated exons, and it is joined to chromosome 11, 38 kb upstream of SCUBE2, at 9.108545 Mb. (The 1.38 kb shard of chromosome 11 is inserted at this junction.) This would be expected to fuse the second exon of CTCF to the first splice acceptor in SCUBE2, and, indeed, the predicted CTCF-SCUBE2 fusion was detected by RT-PCR, fusing exon 2 of CTCF to exon 2 of SCUBE2 (S Newman, KD Howarth, and PAW Edwards, unpubl.). However, the fusion is predicted to be out-of-frame. On the other product chromosome, the chromosome 16 break is not within CTCF but 10.5 kb upstream, leaving an intact copy of CTCF. This is joined to chromosome 11 at 10.438664 Mb, in intron 1 of AMPD3. No fusion is created as the genes in question are transcribed in opposite directions.
Further examples of duplication in HCC1806
There were three further examples of duplication at balanced breaks in HCC1806, which we characterized in less detail (Table 1), bringing the total identified in HCC1806 alone to four.
The reciprocal translocation between chromosomes 12 and 22 had a duplication of 199 kb on chromosome 12 and a duplication of 156 kb on chromosome 22 (Table 1). This was detected by array painting on high-resolution oligonucleotide arrays (Table 1; Supplemental Fig. 3B). The breakpoint junction of the der(12)t(12;22)(q13.2;q13.2) was cloned, and breaks were at chr22:39890194 bp and chr12:54929094 bp (Table 1). Sequencing revealed a genomic shard, inserted at the 12;22 junction, of 135 bp of unidentified sequence joined to 195 bp from 43 Mb away on chromosome 12 at 12.118416–12.118222 Mb (from an intron of ETV6/BCL2L14) (Supplemental Table 3). We were unable to clone the junction on the der(22), so it may have undergone additional rearrangement. The duplication on chromosome 12 was verified by FISH. Two non-overlapping BACs, separated by ∼40 kb, were both positive on both translocation products (Supplemental Fig. 3C). The signal from BAC RP11-620A9 was weaker on the der(22) (Chr Y). This is consistent with a breakpoint at ∼54.729 Mb on chromosome 12, leaving only 36 kb of sequence present on this product, compared to the entire BAC (187 kb), which is positive on the der(12) (Chr k). Similarly, on chromosome 22, two adjacent PACS, with an overlap of only 99 bp, were both positive on both translocation products (Supplemental Fig. 3D). Again, the SNP6-array-CGH is consistent with gain of the duplicated region on both chromosomes 12 and 22, but it is not well resolved (Supplemental Fig. 1B,C).
This duplication could also be important at the gene level, especially as the breaks are close to both ERBB3 on chromosome 12 and EP300 (p300) on chromosome 22. The der(12)t(12;22)(q13.2;q13.2) has breaks in ANKRD52 and EP300, whereas the der(22)t(12;22)(q13.2;q13.2) breaks are in the intergenic region ∼30 kb 5′ to ERBB3 and in ZC3H7B. ANKRD52 and EP300 are transcribed in opposite directions so there is no fusion predicted. There is the potential for a fusion product between ZC3H7B and ERBB3, but we were unable to detect transcripts by RT-PCR. We were unable to clone this junction, suggesting possible further rearrangement.
The third example in HCC1806 was found by inspection of Bignell et al.'s (2010) array-CGH data, which showed a copy number increase on chromosome 3 at the t(3;16)(p21.1;p12.1) reciprocal breakpoint in HCC1806 (Supplemental Fig. 4), suggesting that there might be duplication at this breakpoint as well. The copy number increase extended from 53.108–53.110 Mb to 53.217–53.220 Mb; ∼107 kb in length (Supplemental Fig. 4). The latter breakpoint agrees with the breakpoint (53.22 Mb) obtained by array painting results of product der(16)t(11;3;16) (Chr L) on oligonucleotide arrays (Howarth et al. 2008); the other breakpoint was not determined by array painting. Two non-overlapping fosmids within the region were both positive on both products of the translocation (Supplemental Fig. 4C), and PCR on flow-sorted chromosomes with multiple primer pairs confirmed that sequence is present on both products (Supplemental Fig. 4C).
The breaks on chromosome 16 were not mapped precisely. They were not well-resolved by array painting on BAC arrays because of segmental duplications in this region, and custom oligonucleotide arrays were not attempted.
The fourth example in HCC1806, reported previously (Howarth et al. 2008), was a balanced but non-reciprocal translocation of chromosome 7: The pieces of chromosome 7 in the two chromosomes der(20)t(3;20;7) and der(7)t(8;7;17) were almost balanced, with duplication at the breakpoints of 80 kb by array painting (Supplemental Fig. 5; Table 1). The SNP6-array-CGH of Bignell et al. (2010) was consistent with the gain of this region (Supplemental Fig. 1E). The 20;7 junction creates an expressed TAX1BP1-AHCY fusion, which has been verified at the cDNA level previously (Howarth et al. 2008).
Duplication at the reciprocal t(6;21) in DU4475
We identified an example in a third breast cancer cell line, DU4475, with a duplication of at least 200 kb on chromosome 6, in a reciprocal t(6;21)(p11;q21) translocation. Hybridization to a chromosome 6 tiling path BAC array gave positive hybridization for both products of the translocation to probes between 57.2 Mb and the centromere, which starts at 58.8 Mb, over 1.5 Mb (Supplemental Fig. 6B). However, this region contains segmental duplications (Supplemental Fig. 6B), which could give false-positive hybridization, so the full extent of the duplication could not be confirmed. We could confidently identify by FISH a duplication of at least 220 kb between 57.42 and 57.64 Mb, the innermost ends of PACs RP3-422B11 and RP3-401D24, which do not contain duplicated regions (Supplemental Fig. 6B; Table 1; Supplemental Table 4). Two further BAC probes, RP11-343D24 and RP11-136G2, containing 41 kb and 109 kb of duplicated sequence, respectively, were also positive on both translocation products, so the duplication may extend over the whole >1.5-Mb interval.
Junction sequences
Sequences through the junctions showed 1 to 8 bp of microhomology between the joined sequences in six of seven cases (Supplemental Table 3)—consistent with microhomology-mediated mechanisms of joining, including MMEJ (microhomology-mediated end joining) and MMBIR (microhomology-mediated break-induced replication) (Hastings et al. 2009b). Fragments of DNA from elsewhere in the genome (genomic shards) (Bignell et al. 2007) were inserted at two of the junctions (Supplemental Table 3).
Discussion
We have described an unexpected type of complexity at translocation breakpoints: “overlapping-breakpoint” duplications, where tens to hundreds—maybe thousands—of kilobases of sequence are present on both products of the translocation, so that, when the products are mapped on the genome, they overlap. We found only three previously described examples of such duplications: a constitutional reciprocal translocation in which 650 kb of chromosome X was present on both products (Cox et al. 2003), an evolutionary (4;19) reciprocal translocation in G. gorilla, in which 250 kb of Gorilla chromosome 19 was present on both products of the translocation (Stankiewicz et al. 2001), and a 115-kb duplication in a model yeast experiment (Koszul et al. 2004). Smaller duplications at reciprocal breakpoints have been noted previously, but much smaller than the tens of kilobases found here. For example, in four cases of MALT lymphoma, duplications ranging from 105 bp to 3.3 kb were observed at reciprocal breakpoints (Liu et al. 2004); duplications of ∼250 bp have been found in chronic myeloid leukemia (Litz et al. 1993), and a 337-bp duplication of chromosome 22 was seen at a balanced t(1;22) breakpoint in a phenotypically normal individual (Gajecka et al. 2008). Microduplication of a few base pairs has also been described (Gajecka et al. 2006), which is presumably just a signature of the joining mechanism.
In principle, these apparent duplications at reciprocal translocations could have arisen when two independent unbalanced chromosome translocations happened to have breakpoints in approximately the same place. However, this is very unlikely to have been the case where the breakpoints on one chromosome are essentially at the same point. In particular, in the t(4;6)(p15.33;p21.1) in HCC1806, while the breakpoints on chromosome 6 overlapped by 46 kb, the breakpoints on chromosome 4 were balanced to within 29 bp (29 bp are lost) (Table 1). On the other hand, the apparent duplication on chromosome 7 in the der(20)t(3;20;7) and der(7)t(8;7;17) could be fortuitous, if these were unrelated rearrangements.
How frequent are these duplications at breakpoints?
It remains to be seen how widespread these overlapping-breakpoint duplications are. They could be quite common, since these duplications would often have been missed by classical breakpoint mapping, as discussed below. They may alternatively be a rearrangement architecture specific to a particular kind of genetic instability found in epithelial cancer, just as unbalanced translocations are typical of epithelial cancers but not hemopoietic neoplasms. They may be specific to particular cases—as described for other breast cancer cell lines with particular patterns of rearrangement, such as HCC38, which contains more than 100 tandem duplications (Stephens et al. 2009).
Estimating the proportion of translocations that showed these duplications in our cell lines was not straightforward, because not all breakpoints were mapped precisely, and we chose some breakpoints to study because we suspected that there was duplication. Identifying duplications usually required hybridization of both products of a translocation to custom oligonucleotide arrays, and this was not applied comprehensively. Subsequently, five of the breakpoints that we had not mapped to high resolution were provisionally identified in the paired-end-read sequencing data of Stephens et al. (2009). These additional breakpoints revealed deletions at the junctions ranging from 220 bp to 31 Mb (Supplemental Table 1). This gave results for 14 broken chromosomes out of a total of 21 in HCC1187 and HCC1806 (each reciprocal translocation involves two broken chromosomes, while translocations that appear to be balanced for only one chromosome contribute only one). Six (43% of 14; 29% of 21) had duplication (∼46 kb to 200 kb), three had large deletions (∼1 Mb to 31 Mb), and five had very small deletions (29 bp to 681 bp) at the junctions, with seven uncharacterized (Table 1; Supplemental Table 1).
Our analysis of DU4475 was only at 0.2- or 1-Mb resolution, but this uncovered the one large duplication—of between 220 kb and >1.5 Mb—on chromosome 6, in a 6;21 reciprocal translocation, and a deletion of 38 Mb on chromosome 10, in a 10;21 reciprocal translocation.
In summary, our data suggest that duplications (and, equally, deletions) occurred in around perhaps one-quarter to one-half of participating chromosomes in these breast cancer cell lines. One-quarter would be the average predicted by the following model.
Possible mechanisms of formation
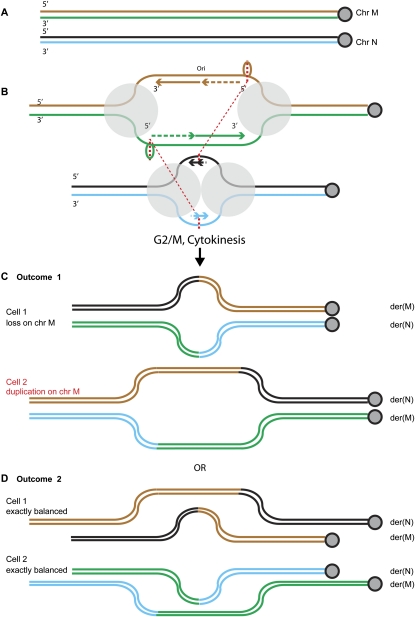
We suggest that these duplications might arise from replication bubbles (Fig. 5). The duplications could form in the same event as the more familiar situation in which material is lost at junctions of reciprocal translocations (Sinclair et al. 2000; Gribble et al. 2005; De Gregori et al. 2007). In a replication bubble, the segment of DNA between two replication forks is present in two copies but remains connected to flanking unreplicated sequence. This is an obvious source of duplication or deletion: If both copies of the segment end up in one daughter cell, this cell has duplication, while the other daughter cell will have loss of the segment (Fig. 5).
Figure 5.
Replication bubbles as a source of duplications and deletions at reciprocal translocation junctions. (A) Two normal chromosomes, M and N. Each DNA strand is shown as a thin line, centromere shown to the right. (B) Replication bubbles form, with the new strands shown. The polymerase complexes are shown as gray circles, the trombone loops at the lagging strand polymerase as ovals. For simplicity, the bubble on chromosome N is shown before significant synthesis has occurred, but it could be like the bubble on chromosome M, in which case, there are four possible outcomes. Breaks or template switching occurs at the thicker dotted red lines, perhaps at the single-stranded trombone or primer loops adjacent to the lagging strand polymerases, and reciprocal joining, i.e., translocation, occurs as shown by the thinner dotted lines, between them and breaks on chromosome N. Replication proceeds and the chromatids separate to daughter cells. According to which chromatids end up in the same cell, two outcomes are possible. (C) Outcome 1: Daughter cell 1 inherits a translocation with loss of the segment of chromosome M between the two breakpoints. Daughter cell 2 inherits two copies of this segment, to give a duplication at the breakpoint. (D) Outcome 2: Both cells receive exactly balanced products.
The length of gained or lost material is consistent with this model, as the distance between neighboring replication origins is estimated to be around the size of the duplications we have found. Lucas et al. (2007) obtained a range of 5–400 kb, average 60 kb, and presumably, when adjacent bubbles coalesce, larger bubbles are created.
The rearrangement event may occur simultaneously at the two forks, or sequentially—but the two forks, which started from a single origin, may be attached to each other, making it more likely that both forks are affected together. For simplicity, Figure 5 shows a case in which the breaks on chromosome N are not separated, so that there is no loss or gain of material from chromosome N in either daughter cell. However, loss or gain could equally well occur on both participating chromosomes M and N, if the replication bubble on chromosome N has completed significant synthesis. Thus, duplication on one participating chromosome could be accompanied by duplication or deletion on the other, as we have observed. Variations on the mechanism, with rearrangements between opposite ends of a single replication bubble, can also give simple interstitial deletions and tandem duplications (Supplemental Fig. 7).
One argument for this model is the occurrence of both deletions and duplications, sometimes in the same translocation (Table 1). Superficially, it might seem that deletions do not require a mechanism to explain them—they could result from erosion of material at broken ends during repair—but it seems unlikely that so much—tens to thousands of kilobases—would be discarded this way. Deletions require mechanistic explanation just as duplications do.
How does this model relate to existing models of translocation?
The distinguishing feature of this model (Fig. 5) is the focus on replication bubbles—i.e., a pair of replication forks—rather than individual replication forks, which have been the focus of recent discussion of structural change in the human genome (for review, see Hastings et al. 2009b).
Our model does not specify the events that join a pair of replication forks to another pair (or a simple break in another chromosome). Various ways of initiating rearrangement can be imagined. For example, replication forks comprise unwound template strands, where single-strand damage can interrupt synthesis (Hastings et al. 2009b); and single-strand Okazaki fragments may be vulnerable structures, because they have to be primed and concatenated (Dixon 2009).
However, a likely mechanism for joining the forks would be MMBIR (microhomology-mediated break-induced replication) (Hastings et al. 2009a), a development of FoSTeS (fork stalling and template switching) (Lee et al. 2007). These mechanisms focus on template switching during replication—in contrast to other models based on repair of strand breaks (Hastings et al. 2009b). They invoke the rescue of individual stalled replication forks by switching the template during strand synthesis or resynthesis to an unrelated template that has only a few base pairs of homology. This results in joining of a strand to an unrelated sequence; it explains the insertion of genomic shards (small fragments of genome from elsewhere in the genome) (Bignell et al. 2007), which are often found in the junctions, and are presumed to be the result of abortive attempts to use another template (Hastings et al. 2009b); and it specifies that junctions show microhomology between the joined sequences of typically 2 to 5 bp. Our translocations were consistent with this pattern, showing microhomology of 1 to 8 bp at the junctions, and, in two cases, typical genomic shards (Supplemental Table 3).
It is difficult to see how the MMBIR/FoSTeS mechanism (Hastings et al. 2009a,b), acting at a single replication fork, could generate these duplications, because it would tend to give unbalanced translocations rather than reciprocal translocations (i.e., it would generate a single chromosome product rather than a pair). Its simple consequences are a linear concatenation of fragments of sequence—such as duplications, deletions, and unbalanced translocations (Hastings et al. 2009b). Reciprocal translocation would require some balancing event that generates a second “mirror-image” structure. Invoking a coordinated event involving two replication forks naturally provides the reciprocal translocation event, with deletions or duplications in the observed size range.
Other genome rearrangements could well share this mechanism, since rearrangement of replication bubbles can give tandem duplications and deletions (Supplemental Fig. 7). However, there is no obvious diagnostic feature of our mechanism that could be used to test this. We can only say that a wide range of genome rearrangements seem to share joining mechanisms with our duplications, with microhomology and occasional genomic shards, but, again, this is not diagnostic. In breast cancers, Stephens et al. (2009) found that 15% of rearrangements showed non-templated sequence and a further 4% contained genomic shards. Microhomology is usual at the junctions of rearrangements in leukemia translocations (Nickoloff et al. 2008), constitutional rearrangements (Gajecka et al. 2008), and at many non-recurrent CNV (copy number variant) breakpoint junctions, including tandem duplications (Hastings et al. 2009b; Vissers et al. 2009).
An alternative scenario to our model would be that a duplication occurs first, and translocation second: either the duplication would be a tandem duplication, and the junction between the copies would then be a hotspot for subsequent translocation formation; or, as suggested for the duplication at the evolutionary translocation in the Gorilla genome, the duplicated copy would be inserted into another chromosome, followed by reciprocal translocation between the two copies (Stankiewicz et al. 2001). However, how and why translocation should tend to occur specifically at the duplications would remain unexplained. Tandem duplications of tens to hundreds of kilobases are quite common (Jones et al. 2008; Stephens et al. 2009). We looked for evidence of a preexisting tandem duplication in our first example, the t(11;16)(p15.4;q22.1) of HCC1187. The der(16)t(12;11;11;16) and the der(11)t(11;16) are of the same parental origin as the “normal” chromosome 16 (S Newman, KD Howarth, and PAW Edwards, unpubl.). The “normal” copy of chromosome 16 did not have a tandem duplication.
SINEs, LINEs, and Alu elements have been implicated in chromosomal translocation in human tumors (Onno et al. 1992; Rudiger et al. 1995; Jeffs et al. 1998; Hill et al. 2000). We found no evidence for such repetitive elements at the breakpoints. It therefore seems unlikely that the rearrangements are mediated by homologous recombination.
Origin of duplicated sequences
We showed that both copies of the chromosome 6 duplication in the HCC1806 t(4;6)(p15.33;p21.1) had the same parental origin. This is consistent both with our proposed mechanism and a tandem duplication followed by translocation. It does, however, rule out a mechanism involving an unequal exchange with the homologous chromosome, which we have invoked in the formation of some homozygous deletions (Alsop et al. 2008).
Consequences for the analysis of chromosome translocations
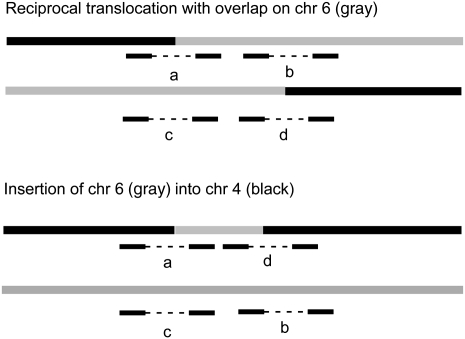
These overlapping-breakpoint duplications are likely to lead to errors in mapping of translocations. In classical FISH, translocation breakpoints have often been located by finding a probe that gives a “split signal,” i.e., that is positive on both products of a reciprocal translocation. In fact, if there are sequences common to both translocation products, the breakpoints could be outside such a “split probe.” As we have shown, array CGH will not usually detect the small duplications (Supplemental Fig. 1), so it would not usually provide warning of extra complexity at the breakpoint. The new “paired-end read” methods for identifying translocations may also misinterpret these duplications (Campbell et al. 2008) because a map of the genome has to be assembled from junctions and copy number: The duplicated material would be indistinguishable from an insertion (Fig. 6). For example, in HCC1806, the duplicated piece of chromosome 6 could be misinterpreted as an insertion of an extra copy of this piece into chromosome 4 (Fig. 6).
Figure 6.
“Overlapping-breakpoint” duplications could be interpreted as an insertion by “paired-end read” methods. The duplication at the breakpoint on chromosome 6 of the t(4;6) in HCC1806 is shown in gray. Paired-end read fragments are shown as black bars joined with a dotted line. The same junction sequences (a, b, c, and d) would be generated by a reciprocal translocation with an “overlapping-breakpoint” duplication (above) or by an insertion of chromosome 6 (gray) into chromosome 4 (black) (below).
The method we used to analyze translocations, “array painting,” was particularly suited to discovering the duplications. Chromosomes are separated by flow cytometry and then hybridized to high-resolution arrays (Fiegler et al. 2003; Howarth et al. 2008), so each breakpoint was analyzed separately. As Figure 2B shows, this is almost uniquely suited to showing the presence of sequences on both products of the translocation.
In most cases, knowledge of the duplication at balanced breaks was necessary for the correct interpretation of gene targets. In addition to our previous analysis (Howarth et al. 2008), we found a number of genes broken and fused. Translocations occurred at FOXP4, EP300, AMPD3, CTCF, ANKRD52, ZC3H7B, TAX1BP1 (described previously), and RFT1, and a fusion was formed between CTCF and SCUBE2. In addition to genes broken, the duplications resulted in an extra copy of a number of interesting genes, including PRKCD, RANGAP1, and ERBB3.
Conclusion
In conclusion, the duplication of material onto both products of a balanced translocation adds to the expanding range of complexities encountered at translocation breakpoints. They may be quite frequent and, for technical reasons, may have been overlooked. These duplications alter the apparent genetic consequences of translocations and are a potential pitfall for both classical FISH-based translocation mapping and current methods based on sequencing through junctions. Their structure suggests that translocations could arise at replication bubbles.
Methods
All genomic positions are relative to human genome Build NCBI36/hg18.
Cell culture
Breast cancer cell lines HCC1187 and HCC1806 (Gazdar et al. 1998) were obtained from ATCC, and DU4475 (Langlois et al. 1979) was from Professor M.J. O'Hare (LICR/UCL Breast Cancer Laboratory, University College Medical School, London, UK). All cell lines were grown in RPMI 1640 medium.
Fluorescence in situ hybridization
Metaphase chromosome preparation and FISH methods were as described previously (Pole et al. 2006). Chromosome paints were kindly supplied by Professor M. Ferguson-Smith (Department of Veterinary Medicine, University of Cambridge, UK). BAC/PAC probes and fosmids used in FISH experiments were obtained from BACPAC Resources. Clone positions are listed in Supplemental Table 4 according to Human Genome Sequence NCBI Build 36. All BACs, PACs, and fosmids were checked by hybridization to normal metaphase chromosomes.
Array painting and PCR and SNP detection on sorted chromosomes
Array painting to custom oligonucleotide arrays from NimbleGen Systems and chromosome 6 tiling path BAC arrays was as described (Howarth et al. 2008). Briefly, metaphase chromosome suspensions were prepared and separated by standard methods on the basis of size and GC:AT content. Typically, 5000 copies of each chromosome were collected, amplified using phi29 DNA polymerase (GenomiPhi kit; GE Healthcare) and the DNA hybridized to genomic DNA arrays essentially as for array CGH. The same chromosome fractions were also used to test for the presence of sequences by PCR and for the detection of heterozygous SNPs. PCR reactions were carried out in a 25-μL reaction volume containing 15 ng of GenomiPhi-amplified DNA, 0.4 μM each primer, 400 μM each dNTP, HotMaster Taq buffer (containing 2.5 mM MgCl2) and 1.25 units of HotMaster Taq DNA polymerase (5 PRIME). Amplification conditions were: 95°C for 30 sec, 30 cycles of 95°C for 30 sec, 58°C for 30 sec, 72°C for 1 min, followed by a final extension at 72°C for 10 min. PCR primers are given in Supplemental Table 2.
Allelic variants were determined by sequencing PCR products in both directions.
Junction sequencing
PCR primers (Supplemental Table 2) were designed at ∼1–2-kb intervals on each side of the estimated breakpoints and used in trial-and-error combinations to amplify junction fragments by long-range PCR using Elongase (Invitrogen). PCR reactions were carried out in a 50-μL reaction volume containing 100 ng of genomic DNA, 200 μM each dNTP, 0.2 μM each primer, Elongase buffer A and B (1.7 mM MgCl2) and 1 μL of Elongase enzyme mix. Cycling conditions were: hot start at 94°C for 30 sec, followed by 35 cycles of 94°C for 30 sec, 58°C for 30 sec, 68°C for 10 min. Amplification products were analyzed on a 1% TBE agarose gel. Products were cloned using the TOPO-XL and S.N.A.P. UV-Free gel purification kit (Invitrogen) and sequenced in both directions.
Acknowledgments
We thank Bee Ling Ng and Nigel Carter, Wellcome Trust Sanger Institute, for chromosome sorting; Koichi Ichimura and V. Peter Collins for chromosome 6 tiling path arrays; and Lucy Raymond and Susanna Cooke for helpful discussions. This work was supported by Cancer Research UK, Breast Cancer Campaign, Breast Cancer Research Trust, UK Medical Research Council (studentship for S.N.), and the Wellcome Trust.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.114116.110.
References
- Alsop AE, Taylor K, Zhang J, Gabra H, Paige AJ, Edwards PA 2008. Homozygous deletions may be markers of nearby heterozygous mutations: The complex deletion at FRA16D in the HCT116 colon cancer cell line removes exons of WWOX. Genes Chromosomes Cancer 47: 437–447 [DOI] [PubMed] [Google Scholar]
- Bignell GR, Santarius T, Pole JC, Butler AP, Perry J, Pleasance E, Greenman C, Menzies A, Taylor S, Edkins S, et al. 2007. Architectures of somatic genomic rearrangement in human cancer amplicons at sequence-level resolution. Genome Res 17: 1296–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell GR, Greenman CD, Davies H, Butler AP, Edkins S, Andrews JM, Buck G, Chen L, Beare D, Latimer C, et al. 2010. Signatures of mutation and selection in the cancer genome. Nature 463: 893–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PJ, Stephens PJ, Pleasance ED, O'Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, et al. 2008. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet 40: 722–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccone R, Giorda R, Gregato G, Guerrini R, Giglio S, Carrozzo R, Bonaglia MC, Priolo E, Lagana C, Tenconi R, et al. 2005. Reciprocal translocations: A trap for cytogenetists? Hum Genet 117: 571–582 [DOI] [PubMed] [Google Scholar]
- Cox JJ, Holden ST, Dee S, Burbridge JI, Raymond FL 2003. Identification of a 650 kb duplication at the X chromosome breakpoint in a patient with 46,X,t(X;8)(q28;q12) and non-syndromic mental retardation. J Med Genet 40: 169–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gregori M, Ciccone R, Magini P, Pramparo T, Gimelli S, Messa J, Novara F, Vetro A, Rossi E, Maraschio P, et al. 2007. Cryptic deletions are a common finding in “balanced” reciprocal and complex chromosome rearrangements: A study of 59 patients. J Med Genet 44: 750–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon NE 2009. DNA replication: Prime-time looping. Nature 462: 854–855 [DOI] [PubMed] [Google Scholar]
- Fiegler H, Gribble SM, Burford DC, Carr P, Prigmore E, Porter KM, Clegg S, Crolla JA, Dennis NR, Jacobs P, et al. 2003. Array painting: A method for the rapid analysis of aberrant chromosomes using DNA microarrays. J Med Genet 40: 664–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajecka M, Pavlicek A, Glotzbach CD, Ballif BC, Jarmuz M, Jurka J, Shaffer LG 2006. Identification of sequence motifs at the breakpoint junctions in three t(1;9)(p36.3;q34) and delineation of mechanisms involved in generating balanced translocations. Hum Genet 120: 519–526 [DOI] [PubMed] [Google Scholar]
- Gajecka M, Gentles AJ, Tsai A, Chitayat D, Mackay KL, Glotzbach CD, Lieber MR, Shaffer LG 2008. Unexpected complexity at breakpoint junctions in phenotypically normal individuals and mechanisms involved in generating balanced translocations t(1;22)(p36;q13). Genome Res 18: 1733–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdar AF, Kurvari V, Virmani A, Gollahon L, Sakaguchi M, Westerfield M, Kodagoda D, Stasny V, Cunningham HT, Wistuba II, et al. 1998. Characterization of paired tumor and non-tumor cell lines established from patients with breast cancer. Int J Cancer 78: 766–774 [DOI] [PubMed] [Google Scholar]
- Gribble SM, Prigmore E, Burford DC, Porter KM, Ng BL, Douglas EJ, Fiegler H, Carr P, Kalaitzopoulos D, Clegg S, et al. 2005. The complex nature of constitutional de novo apparently balanced translocations in patients presenting with abnormal phenotypes. J Med Genet 42: 8–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR 2009a. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet 5: e1000327 doi: 10.1371/journal.pgen.1000327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G 2009b. Mechanisms of change in gene copy number. Nat Rev Genet 10: 551–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill AS, Foot NJ, Chaplin TL, Young BD 2000. The most frequent constitutional translocation in humans, the t(11;22)(q23;q11) is due to a highly specific alu-mediated recombination. Hum Mol Genet 9: 1525–1532 [DOI] [PubMed] [Google Scholar]
- Howarth KD, Blood KA, Ng BL, Beavis JC, Chua Y, Cooke SL, Raby S, Ichimura K, Collins VP, Carter NP, et al. 2008. Array painting reveals a high frequency of balanced translocations in breast cancer cell lines that break in cancer-relevant genes. Oncogene 27: 3345–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffs AR, Benjes SM, Smith TL, Sowerby SJ, Morris CM 1998. The BCR gene recombines preferentially with Alu elements in complex BCR-ABL translocations of chronic myeloid leukaemia. Hum Mol Genet 7: 767–776 [DOI] [PubMed] [Google Scholar]
- Jones DT, Kocialkowski S, Liu L, Pearson DM, Backlund LM, Ichimura K, Collins VP 2008. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res 68: 8673–8677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D 2002. The human genome browser at UCSC. Genome Res 12: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R, Caburet S, Dujon B, Fischer G 2004. Eucaryotic genome evolution through the spontaneous duplication of large chromosomal segments. EMBO 23: 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois AJ, Holder WD Jr, Iglehart JD, Nelson-Rees WA, Wells SA Jr, Bolognesi DP 1979. Morphological and biochemical properties of a new human breast cancer cell line. Cancer Res 39: 2604–2613 [PubMed] [Google Scholar]
- Lee JA, Carvalho CM, Lupski JR 2007. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131: 1235–1247 [DOI] [PubMed] [Google Scholar]
- Litz CE, McClure JS, Copenhaver CM, Brunning RD 1993. Duplication of small segments within the major breakpoint cluster region in chronic myelogenous leukemia. Blood 81: 1567–1572 [PubMed] [Google Scholar]
- Liu H, Hamoudi RA, Ye H, Ruskone-Fourmestraux A, Dogan A, Isaacson PG, Du MQ 2004. t(11;18)(q21;q21) of mucosa-associated lymphoid tissue lymphoma results from illegitimate non-homologous end joining following double strand breaks. Br J Haematol 125: 318–329 [DOI] [PubMed] [Google Scholar]
- Lucas I, Palakodeti A, Jiang Y, Young DJ, Jiang N, Fernald AA, Le Beau MM 2007. High-throughput mapping of origins of replication in human cells. EMBO Rep 8: 770–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickoloff JA, De Haro LP, Wray J, Hromas R 2008. Mechanisms of leukemia translocations. Curr Opin Hematol 15: 338–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onno M, Nakamura T, Hillova J, Hill M 1992. Rearrangement of the human tre oncogene by homologous recombination between Alu repeats of nucleotide sequences from two different chromosomes. Oncogene 7: 2519–2523 [PubMed] [Google Scholar]
- Pole JC, Courtay-Cahen C, Garcia MJ, Blood KA, Cooke SL, Alsop AE, Tse DM, Caldas C, Edwards PA 2006. High-resolution analysis of chromosome rearrangements on 8p in breast, colon and pancreatic cancer reveals a complex pattern of loss, gain and translocation. Oncogene 25: 5693–5706 [DOI] [PubMed] [Google Scholar]
- Rudiger NS, Gregersen N, Kielland-Brandt MC 1995. One short well conserved region of Alu-sequences is involved in human gene rearrangements and has homology with prokaryotic chi. Nucleic Acids Res 23: 256–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Miyoshi H, Kozu T, Nagata J, Enomoto K, Maseki N, Kaneko Y, Ohki M 1992. Consistent disruption of the AML1 gene occurs within a single intron in the t(8;21) chromosomal translocation. Cancer Res 52: 6945–6948 [PubMed] [Google Scholar]
- Sinclair PB, Nacheva EP, Leversha M, Telford N, Chang J, Reid A, Bench A, Champion K, Huntly B, Green AR 2000. Large deletions at the t(9;22) breakpoint are common and may identify a poor-prognosis subgroup of patients with chronic myeloid leukemia. Blood 95: 738–743 [PubMed] [Google Scholar]
- Stankiewicz P, Park SS, Inoue K, Lupski JR 2001. The evolutionary chromosome translocation 4;19 in Gorilla gorilla is associated with microduplication of the chromosome fragment syntenic to sequences surrounding the human proximal CMT1A-REP. Genome Res 11: 1205–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, et al. 2009. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 462: 1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers LE, Bhatt SS, Janssen IM, Xia Z, Lalani SR, Pfundt R, Derwinska K, de Vries BB, Gilissen C, Hoischen A, et al. 2009. Rare pathogenic microdeletions and tandem duplications are microhomology-mediated and stimulated by local genomic architecture. Hum Mol Genet 18: 3579–3593 [DOI] [PubMed] [Google Scholar]