Abstract
Insertions occur when a segment of one chromosome is translocated and inserted into a new region of the same chromosome or a non-homologous chromosome. We report 71 cases with unbalanced insertions identified using array CGH and FISH in 4909 cases referred to our laboratory for array CGH and found to have copy-number abnormalities. Although the majority of insertions were non-recurrent, several recurrent unbalanced insertions were detected, including three der(Y)ins(Y;18)(q?11.2;p11.32p11.32)pat inherited from parents carrying an unbalanced insertion. The clinical significance of these recurrent rearrangements is unclear, although the small size, limited gene content, and inheritance pattern of each suggests that the phenotypic consequences may be benign. Cryptic, submicroscopic duplications were observed at or near the insertion sites in two patients, further confounding the clinical interpretation of these insertions. Using FISH, linear amplification, and array CGH, we identified a 126-kb duplicated region from 19p13.3 inserted into MECP2 at Xq28 in a patient with symptoms of Rett syndrome. Our results demonstrate that although the interpretation of most non-recurrent insertions is unclear without high-resolution insertion site characterization, the potential for an otherwise benign duplication to result in a clinically relevant outcome through the disruption of a gene necessitates the use of FISH to determine whether copy-number gains detected by array CGH represent tandem duplications or unbalanced insertions. Further follow-up testing using techniques such as linear amplification or sequencing should be used to determine gene involvement at the insertion site after FISH has identified the presence of an insertion.
Insertions occur when a segment of one chromosome is translocated and inserted into an interstitial region of another non-homologous chromosome (interchromosomal) (Abuelo et al. 1988; Van Hemel and Eussen 2000) or into a different region of the same chromosome (intrachromosomal) (Madan and Menko 1992). Insertions may occur in a direct fashion, in which the inserted segment maintains its orientation with respect to the centromere, or may be inverted. Estimates of the incidence of insertions range from 1:10,000 to 1:80,000 live births by cytogenetic techniques (Van Hemel and Eussen 2000).
Many factors affect the phenotypic consequences of insertions, including the size and gene content of the inserted segment, which may cause functional aneusomy of a dosage-sensitive gene(s); the orientation of the insertion; position effects exerted on genes in the inserted segment and/or at the site of insertion; disruption of a gene by the insertion; and the presence or absence of additional alterations around the breakpoints of the insertion (Baptista et al. 2005, 2008). Although balanced insertions are less likely to have clinical consequences, malsegregation of a balanced insertion present in a carrier parent can result in an unbalanced form of the rearrangement, with either partial trisomy or partial monosomy, in progeny (Fogu et al. 2007).
Here, we report the identification and characterization of insertions in 71 probands in a diagnostic setting by array CGH and FISH. Our results demonstrate that although the interpretation of most non-recurrent insertions is unclear without high-resolution insertion site characterization, follow-up techniques such as FISH and linear amplification coupled with array CGH may determine whether copy-number gains detected by array CGH represent tandem duplications or unbalanced insertions. High-resolution insertion-site characterization may also determine gene involvement at the insertion site.
Results
During the study period from March 2004 to February 2010, we tested more than 40,000 patients in our laboratory by array CGH and reported 8861 copy-number alterations in 7441 patients. Of these alterations, 4648 involved copy-number gain, whereas 4213 involved loss of material. Metaphase FISH analysis was performed on a total of 3884 copy-number gains and 1644 losses, and parental FISH was performed for 1982 copy-number gains and 1759 losses. In total, 5643 abnormalities (3884 gains and 1759 losses) in 4909 patients were further investigated by performing FISH analysis and/or obtaining parental samples after array CGH identified a copy-number alteration potentially resulting from an unbalanced insertion. Unbalanced insertions were identified in a total of 71 of the 4909 probands tested (Table 1; Supplemental Table 1). Of the 71 unbalanced insertions, 50 were detected by oligonucleotide array as copy-number gains out of 1724 patients with copy-number gains detected by oligonucleotide array and FISH analysis. Thus, based on our population of patients, ∼2.9% of copy-number gains detected by oligonucleotide array CGH were found to represent unbalanced insertions when subjected to follow-up FISH analysis.
Table 1.
Thirty-six probands with unbalanced insertions
(DD) developmental delay; (DF) dysmorphic features; (FAS) fetal alcohol syndrome; (MCA) multiple congenital anomalies; (MR) mental retardation.
aOnly FISH was performed on this specimen.
Of the 71 insertions identified, 55 (77%) were interchromosomal and 16 (23%) were intrachromosomal. Parental samples were obtained for 36 probands, and, after FISH analysis, 30 (84%) insertions were found to be inherited from a carrier parent. The remaining six (16%) insertions were apparently de novo, although paternity was not confirmed in these cases. Of the 30 inherited insertions, 22 (73%) probands with unbalanced insertions inherited their rearrangement directly from an unbalanced carrier parent, whereas eight unbalanced insertions (27%) were the result of abnormal segregation or recombination in a parent with a balanced rearrangement.
Balanced parental insertional translocations
Of the eight balanced insertions detected in parental samples (Table 2), two were reciprocal insertional translocations, one interchromosomal and the other intrachromosomal. Of the remaining six one-way, insertional translocations, five were interchromosomal and one was intrachromosomal. The reciprocal insertions observed in the parents of probands 26 and 32 resulted in partial monosomy and partial trisomy of two different regions of the genome in the respective probands, either by malsegregation of a reciprocal interchromosomal insertion in the case of proband 32 or by meiotic recombination between the derivative and normal homologs in the case of the intrachromosomal insertion in proband 26. Of the remaining six insertions inherited from balanced carrier parents (probands 9, 10, 11, 13, 21, 26), four were observed by array CGH as gains in the proband, and two were observed as apparent deletions.
Table 2.
Eight parents with balanced insertional translocations
Inherited unbalanced insertions may represent benign structural variation
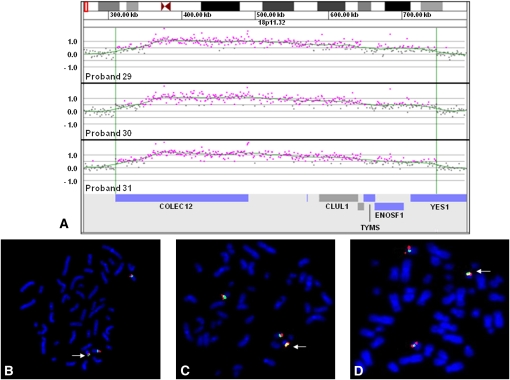
Of 36 unbalanced insertions in which parental samples were tested, 22 were inherited from an unbalanced parent (Table 1). For most cases, no clinical information was obtained for the parents who were carriers of an unbalanced insertion. In three probands (probands 29–31), array CGH detected a gain of a 437-kb region of 18p11.32 that contains six genes from the NCBI reference sequence (RefSeq) database: COLEC12, CETN1, CLUL1, TYMS, ENOSF1, and YES1 (Fig. 1A). The breakpoints within 18p11.32 appeared identical among all three probands, and this was confirmed by additional high-resolution array CGH using a 2.1M-feature oligonucleotide array, which further refined the duplicated segments to chr18:309,785–747,102 (hg18 genome assembly). FISH analysis performed on each proband determined that the duplicated segment was inserted into the pericentromeric region of the Y chromosome, at approximately Yp11.2 (Fig. 1B–D). The indications for diagnostic array study were varied among probands 29–31: dysmorphic features and multiple congenital anomalies, coloboma of the iris, and failure to thrive, respectively. In all three cases, the clinically normal fathers also carried apparently identical unbalanced insertions. The father of proband 29 had mild growth delay during childhood and transverse palmar creases but grew to be 6 feet 1 inches and had no other remarkable features. Based on a parental report of a great-grandparent of proband 29 who lived in the same state as proband 31 and shared the same last name, probands 29 and 31 may have shared a paternal ancestor, at least three generations back, but no relation to this lineage could be established for proband 30.
Figure 1.
Characterization of recurrent der(Y)ins(Y;18)(?p11.2;p11.32p11.32) by oligonucleotide array CGH and FISH. (A) Oligonucleotide microarray results showing identical single-copy gains of 340 probes from 18p11.32, ∼437 kb in size (chr18:309,785–747,102 based on UCSC 2006 hg 18 assembly), in three probands. Probes are ordered on the x-axis according to physical mapping positions, with the most distal 18p11.32 probes to the left and the most proximal 18p11.32 probes to the right. Values along the y-axis represent log2 ratios of patient:control signal intensities. Genes in the interval are shown as blue and gray bars below. (B) Metaphase FISH results showing insertion of RP11-720L2 (red) from the duplicated region on chromosome 18 in proband 29 into the pericentromeric region of the Y chromosome (arrow). (Green) A centromere probe for chromosome 18 and a Yp11.31 probe specific to SRY (RP11-400O10). (C) Metaphase FISH results showing insertion of RP11-133D9 (red) from the duplicated region on chromosome 18 into the pericentromeric region of the Y chromosome in proband 30 (arrow). (Green) Centromere probes for chromosome 18 and the Y chromosome. (D) Metaphase FISH results showing the insertion of RP11-720L2 (red) into the pericentromeric region of the Y chromosome in proband 31 (arrow). (Green) Centromere probes for chromosome 18 and the Y chromosome.
Insertions with complex breakpoints
An insertion of an interstitial segment of Xp22.33 into Xq28 was observed in two male patients in our cohort (probands 16 and 17). In proband 16, array CGH detected a 238-kb gain of material from the pseudoautosomal region at Xp22.33/Yp11.32, resulting in partial trisomy of PPP2R3B and terminating just distal to the SHOX locus at chrX:249,940–488,150. In proband 17, array CGH detected a 370-kb gain of the pseudoautosomal region including SHOX at chrX:300,092–669,611 (Supplemental Fig. 1A). Array CGH also detected ∼33-kb gains of Xq28 in both unrelated probands at chrX:154,396,893–154,429,972 (Supplemental Fig. 1B), partially overlapping TMLHE. FISH analysis performed on both samples showed the insertion of the Xp22.33/Yp11.32 region into Xq28 (Supplemental Fig. 1C,D). Several similar duplications of Xq28 have been identified in control samples and recorded in the Database of Genomic Variants (http://projects.tcag.ca/variation/). A second FISH experiment using one probe corresponding to the inserted segment and a second probe mapping to the location of the Xq28 gain showed that the insertion and gain (duplication) occurred in close proximity (Supplemental Fig. 1E,F), within the resolution of metaphase FISH using BAC clones (80–200 kb) (Shaffer et al. 2001). Maternal FISH in both cases confirmed the presence of an unbalanced der(X)ins(X)(q28p22.33p22.33) in the mother of each male proband. In addition, array CGH performed on the mother of proband 16 identified the duplication of Xq28 identical to the alteration observed in the child. No array CGH was performed on the mother of proband 17.
A duplication at the apparent insertion site was also identified in proband 1, who had a de novo unbalanced der(1)ins(1;X)(p36.32;q22.2q22.2)dup(1)(p36.32p36.32). Array CGH detected two gains: a 310-kb gain of Xq22.2 at approximately chrX:102,675,779–102,986,217, including the PLP1 locus, and a 729-kb gain within 1p36.32 at chr1:3,323,541–4,052,757 (Supplemental Fig. 2A,B). FISH analysis visualized the insertion of the Xq22.2 region into 1p36 (Supplemental Fig. 2C). A second FISH experiment using probes from both the Xq22.2 and the 1p36.32 duplicated regions (RP11-832L2 and RP11-893K17, respectively) confirmed that the 1p36.32 duplication detected by array CGH lies near the insertion site (Supplemental Fig. 2D). Metaphase FISH analysis was performed on both parental samples using a probe specific to the Xq22.2 region and failed to identify a rearrangement Xq22.2 in either parent. In addition, interphase FISH analysis was performed on both parental samples using a probe specific to the duplicated region at 1p36.32 and failed to identify a duplication of this region in either parent. Thus, the rearrangements observed in the proband were determined to be apparently de novo in origin.
Insertion site resolution using linear amplification and PCR identifies gene disruption
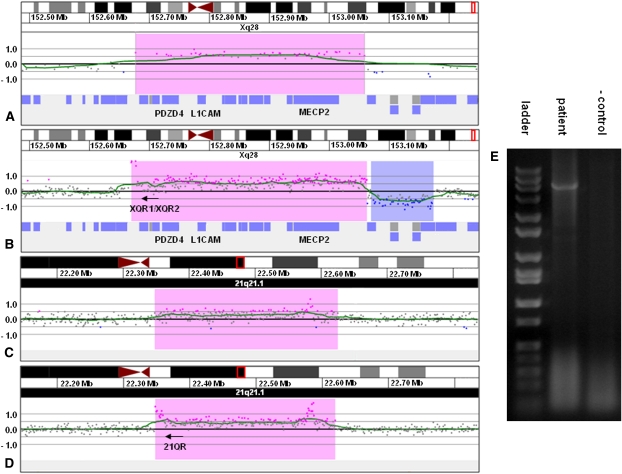
Two de novo insertions were further investigated to elucidate the precise site of insertion and determine the pathogenicity of the insertion. In proband 2, array CGH detected a 382-kb gain of Xq28 containing 14 genes including L1CAM, AVPR2, and MECP2 (Fig. 2A) and a 231-kb duplication of 21q21.1 containing no genes. Linear amplification performed with primers XQR1 and XQR2 located just inside the proximal breakpoint of the Xq28 gain and subsequent array CGH of the amplified product indicated that the Xq28 segment was inserted in an inverted orientation at the proximal boundary of the 21q21.1 duplication (Fig. 2B,C). PCR performed with primers XQR2 and 21QR located on each side of the suspected junction (Fig. 2D) showed a junction fragment and confirmed the insertion site in 21q21.1 (Fig. 2E).
Figure 2.
Characterization of der(21)ins(21;X)(q21.1;q28q28)dup(21)(q21.1q21.1) by oligonucleotide array CGH, linear amplification, and PCR. (A) 135k feature oligonucleotide microarray results showing a single-copy gain of 48 probes, ∼382 kb in size (chrX:152,676,843–153,058,941 based on the UCSC 2006 hg18 assembly), from Xq28 in proband 2. Probes are ordered on the x-axis with the most proximal Xq28 probes to the left and the most distal Xq28 probes to the right. Values along the y-axis represent log2 ratios of patient:control signal intensities. (B) 2.1M feature oligonucleotide microarray results showing the same duplication as in A after linear amplification with primers XQR1 and XQR2. Successful amplification is evidenced by the elevated log ratios of probes in the proximal portion of the duplicated region. (C) 2.1M feature oligonucleotide microarray results showing a single-copy gain of 210 probes, ∼272 kb in size (chr21:22,347,877–22,623,043 based on the UCSC 2006 hg18 assembly), from 21q21.1 in proband 2. (D) 2.1 M feature oligonucleotide microarray results showing the same duplication as in C after linear amplification with the primers from B. The elevated log ratios in the proximal portion of the duplicated segment are indicative of the insertion of Xq22.2 sequence into 21q21.1, which allowed for continuous amplification across the breakpoint. The cluster of elevated probes that can be seen in the distal portion of the duplicated segment was also present in the unamplified sample and probably represents a CNV or artifact. (E) Gel electrophoresis of PCR amplicon produced with primers XQR1 and 21QR confirming the insertion site detected by linear amplification.
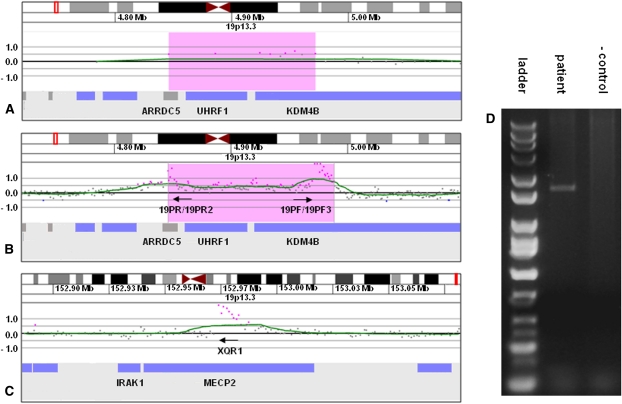
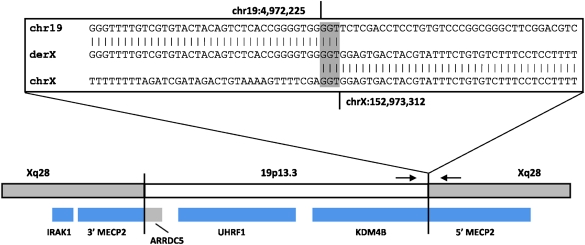
In proband 6, array CGH detected a 126-kb gain of 19p13.3 that contained three genes—ARRDC5, UHRF1, and KDM4B (JMJD2B) (Fig. 3A). Linear amplification using primers 19PF, 19PF3, 19PR, and 19PR2 was followed by high-resolution array CGH of the amplified products and showed that the site of insertion was between chrX:152,971,845 and chrX:152,973,394, within a large intron of MECP2 (Fig. 3B,C). PCR was performed using primers 19PF and XQR1 located on each side of the suspected junction (Fig. 3D) and resulted in a junction fragment, confirming that MECP2 was disrupted by the insertion. Subsequently, using several primers specific to this region, the site of insertion on chromosome X was narrowed to an 83-bp region between chrX:152,973,327 and chrX:152,973,409. Sequencing performed on this product identified the junction between the proximal side of the gain of chromosome 19 and the insertion site on the X chromosome (Fig. 4).
Figure 3.
Characterization of der(X)ins(X;19)(q28;p13.3p13.3) by oligonucleotide array CGH, linear amplification, and PCR. (A) 135k-feature oligonucleotide microarray results showing a single-copy gain of 9 probes, ∼126 kb in size (chr19:4,845,920–4,971,768 based on the UCSC 2006 hg 18 assembly), from 19p13.3 in proband 6. Probes are ordered along the x-axis with the most distal 19p13.3 probes to the left and the most proximal 19p13.3 probes to the right. Values along the y-axis represent log2 ratios of patient:control signal intensities. (B) 2.1M feature oligonucleotide microarray analysis showing the same duplication as in A after linear amplification with primers 19PF, 19PF3, 19PR, and 19PR2. Successful amplification is evidenced by the elevated log ratios of probes at the proximal and distal edges of the duplicated region. (C) 2.1M feature oligonucleotide microarray analysis showing linear amplification from B extending across the insertion junction into intron 2 of MECP2. (D) Gel electrophoresis of PCR amplicon produced with primers 19PF and XQR1 confirming the insertion site detected by linear amplification.
Figure 4.
Diagram representing the insertion site in the der(X) resulting from insertion of chromosome 19 material into Xq28 in proband 6. (Gray shaded area within the inset box) The 3-nucleotide span of microhomology shared at the distal insertion breakpoint. (Small black arrows) The positions of primers used to generate the PCR fragment and obtain the junction sequence. Genes are displayed as blue and gray bars below, and show the disruption of MECP2 as well as the two possible fusion gene products that could result from the insertion.
Discussion
Estimated incidence of insertions
The incidence of insertions in the population can be estimated by comparing their detection rate in our laboratory to that of a genomic disorder with a well-established incidence, such as 7q11.23 microduplication syndrome, which has an incidence of ∼1:13,000 to 1:20,000 in the general population (Van der Aa et al. 2009). The lack of a clearly recognizable set of clinical features associated with the disease and the likelihood of FISH analysis being performed to visualize the abnormality after detection by microarray analysis make 7q11.23 microduplications a suitable genomic disorder to use for comparison with insertions. Whole-genome oligonucleotide arrays can be expected to yield a higher incidence of insertions because of their high-density coverage; therefore, only cases with alterations detected by these array types (SignatureChipOS V1.1 or V2, introduced in November 2007) were used for this incidence calculation. In total, 50 patients with unbalanced insertions and 13 patients with duplications of 7q11.23 were identified by oligonucleotide array. Thus, based on its relative frequency in our experience, the incidence of insertions can be estimated to be ∼1:3380 to 1:5200. This is a higher incidence compared to previous estimates of 1:10,000 to 1:80,000, likely owing to the fact that previous estimates were based on insertions detectable by cytogenetic techniques rather than array CGH and FISH (Van Hemel and Eussen 2000). The use of higher-resolution techniques, with the ability to consistently detect insertions smaller than 50 kb, would likely yield still higher incidence estimates.
Clinical interpretation of unbalanced insertions detected in carrier parents
In our cohort, 81% of insertions of known inheritance were inherited from a carrier parent, and 65% of insertions of known inheritance were inherited directly from a parent who carried an identical unbalanced rearrangement. The interpretation of the clinical significance of inherited insertions should always be approached with care. While the large number of apparently normal parents carrying unbalanced insertions may suggest that these partial trisomies that impact the dosage of a small number of genes can be tolerated with little phenotypic consequence and may represent benign structural variation, a number of these seemingly benign rearrangements may unmask a recessive mutation not present in the parent by disruption of a gene at the insertion site or act in conjunction with another alteration or genetic factor to contribute to a multifactorial condition displayed in the proband. In addition, variable expressivity cannot be excluded and could account for an abnormal proband born to an apparently normal carrier parent (Sharp 2009).
Examples of a recurrent rearrangement involving insertion of 6p25.3 into 3p13 have been previously reported in four individuals with idiopathic ID/DD. In addition, identical unbalanced rearrangements were identified in the phenotypically normal mothers of three out of four of these probands (Kang et al. 2010). We detected a similar alteration of unknown inheritance in proband 61 of our cohort, supporting the conclusion that this insertion represents a recurrent structural variation within the human population that may be benign. The recurrent der(Y)ins(Y;18) observed in our cohort is likely another example of an insertion persisting as a benign structural variant in the population. The relatively small size of the trisomic region and the absence of genes associated with known genomic disorders in the inserted segment and at the insertion site suggest that it makes little contribution to the phenotypes of the probands. Our results support two possible modes by which this rearrangement may have arisen. First, this rearrangement may have arisen in a common ancestor shared by the families involved in this study and been subsequently transmitted through many generations. In this scenario, this insertion could represent an uncharacterized Y-chromosome variant that arose in recent human history (Hammer 1994; Jobling and Tyler-Smith 1995; Jobling et al. 2007; Karafet et al. 2008). Alternatively, because there is no known relationship between the family of proband 30 and those of probands 29 and 31, the observed recurrent rearrangements may represent two independent events, potentially mediated by genomic architecture present in the regions involved in the insertion. Low-copy repeats were not observed at the breakpoints of the inserted segments in these probands; however, that does not exclude the presence of shorter repetitive sequences that may mediate these rearrangements. Higher-resolution characterization of the insertion breakpoints would be required to determine the extent of the homology that is present.
Risks associated with carriers of balanced insertions
Balanced interchromosomal insertions present in carrier parents have a 50% risk of malsegregation during meiosis resulting in partial monosomy or partial trisomy of the inserted segment. In our cohort, six probands inherited an unbalanced insertion from a parent who carried a one-way, balanced insertion: Four of these cases were observed by array CGH as a partial trisomy in the proband, and two were detected as partial monosomy. Two cases had both partial monosomy and partial trisomy of different regions resulting from malsegregation of a two-way, reciprocal insertional translocation in the parent (proband 32) or recombination between the rearranged chromosome and the normal homolog (proband 26). The risk of unbalanced offspring associated with balanced insertion carriers highlights the need for parental FISH studies when an unbalanced insertion is identified by array CGH and FISH in a proband. In addition, parental FISH studies are crucial to determine the origin of interstitial losses detected by array CGH because deletions resulting from malsegregation of balanced insertions cannot be distinguished from typical interstitial deletions until parental FISH analysis is performed.
Risks associated with female carriers of X-chromosome insertions
Although X-chromosome insertions may produce no or only mild effects in carrier females, presumably because of X inactivation, an abnormal phenotype may result when the insertion is transmitted to a male. The two unbalanced insertions of segments of the pseudoautosomal region at Xp22.33 into Xq28 that were observed in male patients (subjects 16 and 17) referred for developmental delay and multiple congenital anomalies, respectively, illustrate these potential risks. Deletions of, and distal to, SHOX have been well described (Robertson et al. 2000; Benito-Sanz et al. 2005; Schneider et al. 2005). The impact of duplications including, and distal to, SHOX are less clear, although they have been linked to tall stature (Thomas et al. 2009). The unbalanced insertion in proband 17 would be expected to result in functional trisomy of SHOX, because two copies of the gene are expressed from the pseudoautosomal region in both male and female carriers, whereas X inactivation of the copy of the gene inserted into Xq28 on the rearranged X would prevent full functional trisomy in the mother. Although the association between a duplication of SHOX and the phenotype of proband 17 is unclear, the resulting expected dosage differences of SHOX expression in female and male carriers caused by the insertion suggests that the rearrangement potentially influences the phenotype of the proband and demonstrates the complex inheritance patterns associated with insertions involving the X chromosome. Although not confirmed in this study, it is also possible that these insertions cause disruption of a gene at the insertion site that results in the phenotypes observed in probands 16 and 17.
Disrupted genes at insertion sites
While the prevalence of pathogenic insertions that disrupt clinically significant genes is poorly understood because of the difficulty associated with the characterization of the precise insertion site in most cases, the rearrangement identified in proband 6 demonstrates the potential significance of this type of insertion. The insertion of a 126-kb segment of 19p13.3 into MECP2 is expected to disrupt its transcription and result in haploinsufficiency in this female patient. Although the majority of Rett syndrome cases are caused by mutations within MECP2 (Renieri et al. 2003), disruption of MECP2 resulting from this insertion likely explains the clinical suspicion of Rett syndrome in this patient. The discovery that this gain of 19p13.3 material represents an unbalanced insertion instead of a tandem duplication and that the rearrangement likely results in haploinsufficiency of a gene known to be associated with human disease affects the relevance of the array CGH results.
Mechanisms of insertion formation
Non-allelic homologous recombination (NAHR), homologous recombination, and non-homologous end joining (NHEJ) have been implicated as mechanisms of chromosomal translocations (Gu et al. 2008) and may be similarly implicated in the creation of insertions. Although NAHR may explain the formation of some insertions, stretches of homology of adequate length to mediate homologous recombination were not present at the breakpoints of those re-examined in our cohort (data not shown). In addition, the creation of large-scale insertion-site imbalances is difficult to associate with either of these mechanisms. This implicates another mechanism in the formation of non-recurrent insertions in which long stretches of breakpoint homology are not observed and large insertion-site alterations are present.
Recently, replication-based mechanisms have been proposed to explain the occurrence of non-recurrent and complex genomic rearrangements (Ballif et al. 2004; Lee et al. 2007; Bauters et al. 2008). Specifically, variations of break-induced replication (BIR) termed microhomology/microsatellite-induced replication (MMIR) and microhomology-mediated break-induced replication (MMBIR), which, instead of requiring large tracts of sequence identity to mediate recombination and repair (Reiter et al. 1998), are mediated by very short spans of microhomology, have been proposed to generate intra- and interchromosomal rearrangements (Payen et al. 2008; Hastings et al. 2009). In these models, 3′ overhangs generated at collapsed replication forks, caused by a break in the template strand, invade regions of exposed single-stranded DNA at potentially distant locations in the genome based on microhomology. Subsequent cycles of replication fork collapse, strand invasion, and replication initiation can occur at multiple sites to create complex rearrangements with various deletions, duplications, and insertions. These models could account for the breakpoint diversity and insertion site complexities observed in our cohort and have been implicated in the formation of segmental duplications in other organisms (Payen et al. 2008).
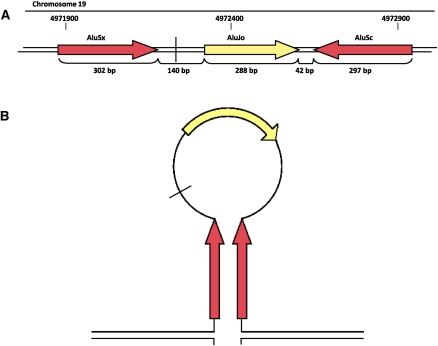
Additionally, breakpoint regions of many insertions may be associated with inverted repetitive sequences (such as Alu elements), which have been shown to encourage genomic rearrangement (VanHulle et al. 2007) and could mediate insertions based on these proposed mechanisms by promoting secondary structures. In proband 6, the proximal breakpoint of the inserted segment on chromosome 19 is in close proximity to two inverted Alu repeats which could mediate the formation of a stem–loop structure (Fig. 5) and expose single-stranded DNA as a target for strand invasion. This explanation is further supported by the propensity of the MECP2 region to incur double-strand breaks (DSBs), which have been shown to initiate BIR (Bauters et al. 2008). Breakpoint sequence analysis of additional insertions may help to elucidate the contributions of both homology-dependent and microhomology-dependent mechanisms to the formation of insertions.
Figure 5.
(A) Diagram outlining the repetitive sequences around the proximal breakpoint of the copy-number gain on chromosome 19. (Vertical black line) The position of the insertion breakpoint. Members of AluS families (red boxes) and AluJ families (yellow boxes) of repetitive elements. (B) Diagram of potential stem–loop structure mediated by inverted AluS repeats present at the breakpoint.
Conclusion
Our results demonstrate the surprising prevalence of familial unbalanced insertions and the potential risks of pathogenic insertions. The characterization of 2.9% of copy-number gains detected by oligonucleotide array CGH as unbalanced insertions emphasizes the importance of performing FISH after array CGH to determine the nature of copy-number gains and losses detected by microarray and visualize the chromosomes to assess the need for parental follow-up studies and allow provision of accurate genetic counseling. The ability of high-density oligonucleotide array CGH to detect small insertions and cryptic breakpoint alterations makes it a valuable tool in the characterization of insertions, although the clinical significance of most unbalanced insertions detected by array CGH and FISH remains uncertain without more precise insertion-site characterization.
Methods
Array comparative genomic hybridization
Microarray analysis was performed as previously described for BAC (Ballif et al. 2008a) and oligo-based (Ballif et al. 2008b) arrays. Microarray coverage and criteria for reporting abnormalities are detailed in the Supplemental Methods.
FISH
Copy-number gains detected by array CGH were further analyzed with metaphase FISH using one or more BAC clones indicated to be abnormal by array CGH to determine whether they represented a tandem or insertional duplication (Traylor et al. 2009). Parental samples were obtained for copy-number losses detected by array CGH, and metaphase FISH was performed to determine whether the parent was a carrier of a balanced insertion. The insertion in proband 20 was characterized using only FISH without array CGH. This patient was studied because of known karyotypic findings.
Linear amplification and polymerase chain reaction (PCR)
Linear amplification was performed across the breakpoints of two insertions to resolve the site of insertion more accurately. Fifty-microliter linear amplification reactions were performed on probands 2 and 6 with the Failsafe PCR System (Epicentre Biotechnologies) using Premix D and custom primers specific to the breakpoints of the insertion as determined by array CGH (Table 3). Custom primers were designed using the Primer3 software (http://primer3.sourceforge.net/). Linear amplifications were conducted with an initial denaturation of 2 min at 94°C followed by 10 cycles of denaturation for 10 sec at 94°C, annealing for 30 sec at 62°C, and elongation for 20 min at 68°C; 10 cycles of denaturation for 15 sec at 94°C, annealing for 30 sec at 62°C, and elongation for 20 min at 68°C, with an additional 20 sec of elongation added per cycle; and a final elongation step for 1 min at 68°C. Linear amplification products were then purified using the QuickStep 2 PCR Purification System (EdgeBio), labeled with Cy3 or Cy5 dyes using a Roche-NimbleGen labeling kit according to the manufacturer's instructions and hybridized to 2.1M-feature arrays as previously described to determine the site of insertion in each proband. PCR was performed to confirm the site of insertion in each proband using the Failsafe PCR System with Premix E and primers designed on each side of the insertion breakpoints (Table 3). PCR reactions were initially denatured for 2 min at 94°C followed by 28 cycles of denaturation for 30 sec at 94°C, annealing for 30 sec at 64°C, elongation for 5 min at 68°C, and final extension for 1 min at 68°C. Sequencing of the PCR-amplified junction fragment isolated from proband 6 was performed by SeqWright, Inc.
Table 3.
Primers used for linear amplification (LA) and PCR in proband 2
Gel electrophoresis
Gel electrophoresis of amplified products was performed by running 3 μL of amplified product at 120 V for 45 min in a 1% agarose gel containing ethidium bromide. Ten microliters of all purpose hi-lo DNA marker (Bionexus) and a negative amplification control using reference female DNA as template were run alongside each amplified sample.
Acknowledgments
We gratefully acknowledge Aaron Theisen (Signature Genomic Laboratories, Spokane, WA) for his editing of the manuscript; Sara Minier, Victoria Cawich, Caitlin Valentin, Ryan Traylor, and all of the laboratory staff at Signature Genomics for their assistance in conducting the array CGH and FISH experiments; and Dr. Paul Mark, Anne Spencer, Jennifer Eichmeyer, and Stacie Rosenthal for their assistance in contacting the families of patients involved in this study.
Footnotes
[Supplemental material is available for this article. The microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/). A full list of accession numbers can be found in Supplemental Table 2.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.114579.110.
References
- Abuelo DN, Barsel-Bowers G, Richardson A 1988. Insertional translocations: Report of two new families and review of the literature. Am J Med Genet 31: 319–329 [DOI] [PubMed] [Google Scholar]
- Ballif BC, Wakui K, Gajecka M, Shaffer LG 2004. Translocation breakpoint mapping and sequence analysis in three monosomy 1p36 subjects with der(1)t(1;1)(p36;q44) suggest mechanisms for telomere capture in stabilizing de novo terminal rearrangements. Hum Genet 114: 198–206 [DOI] [PubMed] [Google Scholar]
- Ballif BC, Theisen A, Coppinger J, Gowans GC, Hersh JH, Madan-Khetarpal S, Schmidt KR, Tervo R, Escobar LF, Friedrich CA, et al. 2008a. Expanding the clinical phenotype of the 3q29 microdeletion syndrome and characterization of the reciprocal microduplication. Mol Cytogenet 1: 8 doi: 10.1186/1755-8166-1-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballif BC, Theisen A, McDonald-McGinn DM, Zackai EH, Hersh JH, Bejjani BA, Shaffer LG 2008b. Identification of a previously unrecognized microdeletion syndrome of 16q11.2q12.2. Clin Genet 74: 469–475 [DOI] [PubMed] [Google Scholar]
- Baptista J, Prigmore E, Gribble SM, Jacobs PA, Carter NP, Crolla JA 2005. Molecular cytogenetic analyses of breakpoints in apparently balanced reciprocal translocations carried by phenotypically normal individuals. Eur J Hum Genet 13: 1205–1212 [DOI] [PubMed] [Google Scholar]
- Baptista J, Mercer C, Prigmore E, Gribble SM, Carter NP, Maloney V, Thomas NS, Jacobs PA, Crolla JA 2008. Breakpoint mapping and array CGH in translocations: Comparison of a phenotypically normal and an abnormal cohort. Am J Hum Genet 82: 927–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauters M, Van Esch H, Friez MJ, Boespflug-Tanguy O, Zenker M, Vianna-Morgante AM, Rosenberg C, Ignatius J, Raynaud M, Hollanders K, et al. 2008. Nonrecurrent MECP2 duplications mediated by genomic architecture-driven DNA breaks and break-induced replication repair. Genome Res 18: 847–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito-Sanz S, Thomas NS, Huber C, Gorbenko del Blanco D, Aza-Carmona M, Crolla JA, Maloney V, Rappold G, Argente J, Campos-Barros A, et al. 2005. A novel class of Pseudoautosomal region 1 deletions downstream of SHOX is associated with Leri-Weill dyschondrosteosis. Am J Hum Genet 77: 533–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogu G, Bandiera P, Cambosu F, Carta AR, Pilo L, Serra G, Soro G, Tondi M, Tusacciu G, Montella A 2007. Pure partial trisomy of 6p12.1–p22.1 secondary to a familial 12/6 insertion in two malformed babies. Eur J Med Genet 50: 103–111 [DOI] [PubMed] [Google Scholar]
- Gu W, Zhang F, Lupski JR 2008. Mechanisms for human genomic rearrangements. Pathogenetics 1: 4 doi: 10.1186/1755-8417-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer MF 1994. A recent insertion of an alu element on the Y chromosome is a useful marker for human population studies. Mol Biol Evol 11: 749–761 [DOI] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR 2009. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet 5: e1000327 doi: 10.1371/journal.pgen.1000327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobling MA, Tyler-Smith C 1995. Fathers and sons: the Y chromosome and human evolution. Trends Genet 11: 449–456 [DOI] [PubMed] [Google Scholar]
- Jobling MA, Lo IC, Turner DJ, Bowden GR, Lee AC, Xue Y, Carvalho-Silva D, Hurles ME, Adams SM, Chang YM, et al. 2007. Structural variation on the short arm of the human Y chromosome: recurrent multigene deletions encompassing Amelogenin Y. Hum Mol Genet 16: 307–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SH, Shaw C, Ou Z, Eng PA, Cooper ML, Pursley AN, Sahoo T, Bacino CA, Chinault AC, Stankiewicz P, et al. 2010. Insertional translocation detected using FISH confirmation of array-comparative genomic hybridization (aCGH) results. Am J Med Genet A 152A: 1111–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF 2008. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res 18: 830–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Carvalho CM, Lupski JR 2007. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131: 1235–1247 [DOI] [PubMed] [Google Scholar]
- Madan K, Menko FH 1992. Intrachromosomal insertions: a case report and a review. Hum Genet 89: 1–9 [DOI] [PubMed] [Google Scholar]
- Payen C, Koszul R, Dujon B, Fischer G 2008. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet 4: e1000175 doi: 10.1371/journal.pgen.1000175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter LT, Hastings PJ, Nelis E, De Jonghe P, Van Broeckhoven C, Lupski JR 1998. Human meiotic recombination products revealed by sequencing a hotspot for homologous strand exchange in multiple HNPP deletion patients. Am J Hum Genet 62: 1023–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renieri A, Meloni I, Longo I, Ariani F, Mari F, Pescucci C, Cambi F 2003. Rett syndrome: the complex nature of a monogenic disease. J Mol Med 81: 346–354 [DOI] [PubMed] [Google Scholar]
- Robertson SP, Shears DJ, Oei P, Winter RM, Scambler PJ, Aftimos S, Savarirayan R 2000. Homozygous deletion of SHOX in a mentally retarded male with Langer mesomelic dysplasia. J Med Genet 37: 959–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider KU, Sabherwal N, Jantz K, Roth R, Muncke N, Blum WF, Cutler GB Jr, Rappold G 2005. Identification of a major recombination hotspot in patients with short stature and SHOX deficiency. Am J Hum Genet 77: 89–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer LG, Ledbetter DH, Lupski JR 2001. Molecular cytogenetics of contiguous gene syndromes: Mechanisms and consequences of gene dosage imbalance. In Metabolic and molecular basis of inherited disease (ed. Scriver CR et al. ), pp. 1291–1324 McGraw Hill, New York [Google Scholar]
- Sharp AJ 2009. Emerging themes and new challenges in defining the role of structural variation in human disease. Hum Mutat 30: 135–144 [DOI] [PubMed] [Google Scholar]
- Thomas NS, Harvey JF, Bunyan DJ, Rankin J, Grigelioniene G, Bruno DL, Tan TY, Tomkins S, Hastings R 2009. Clinical and molecular characterization of duplications encompassing the human SHOX gene reveal a variable effect on stature. Am J Med Genet A 149A: 1407–1414 [DOI] [PubMed] [Google Scholar]
- Traylor RN, Fan Z, Hudson B, Rosenfeld JA, Shaffer LG, Torchia BS, Ballif BC 2009. Microdeletion of 6q16.1 encompassing EPHA7 in a child with mild neurological abnormalities and dysmorphic features: case report. Mol Cytogenet 2: 17 doi: 10.1186/1755-8166-2-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Aa N, Rooms L, Vandeweyer G, van den Ende J, Reyniers E, Fichera M, Romano C, Delle Chiaie B, Mortier G, Menten B, et al. 2009. Fourteen new cases contribute to the characterization of the 7q11.23 microduplication syndrome. Eur J Med Genet 52: 94–100 [DOI] [PubMed] [Google Scholar]
- Van Hemel JO, Eussen HJ 2000. Interchromosomal insertions. Identification of five cases and a review. Hum Genet 107: 415–432 [DOI] [PubMed] [Google Scholar]
- VanHulle K, Lemoine FJ, Narayanan V, Downing B, Hull K, McCullough C, Bellinger M, Lobachev K, Petes TD, Malkova A 2007. Inverted DNA repeats channel repair of distant double-strand breaks into chromatid fusions and chromosomal rearrangements. Mol Cell Biol 27: 2601–2614 [DOI] [PMC free article] [PubMed] [Google Scholar]