Abstract
Expression levels of many human genes are under the genetic control of expression quantitative trait loci (eQTLs). Despite technological advances, the precise molecular mechanisms underlying most eQTLs remain elusive. Here, we use deep mRNA sequencing of two CEU individuals to investigate those mechanisms, with particular focus on the role of splicing control loci (sQTLs). We identify a large number of genes that are differentially spliced between the two samples and associate many of those differences with nearby single nucleotide polymorphisms (SNPs). Subsequently, we investigate the potential effect of splicing SNPs on eQTL control in general. We find a significant enrichment of alternative splicing (AS) events within a set of highly confident eQTL targets discovered in previous studies, suggesting a role of AS in regulating overall gene expression levels. Next, we demonstrate high correlation between the levels of mature (exonic) and unprocessed (intronic) RNA, implying that ∼75% of eQTL target variance can be explained by control at the level of transcription, but that the remaining 25% may be regulated co- or post-transcriptionally. We focus on eQTL targets with discordant mRNA and pre-mRNA expression patterns and use four examples: USMG5, MMAB, MRPL43, and OAS1, to dissect the exact downstream effects of the associated genetic variants.
Expression quantitative trait loci (eQTLs), defined as loci in which genetic variation is associated with changes in the expression levels of target genes, have been studied extensively over the past decade and shown to be widespread in human populations. Particular attention has been paid to their possible role in complex genetic disorders (Emilsson et al. 2008; Nica et al. 2010). Numerous studies have correlated gene and transcript isoform expression levels with nearby, cis-acting genetic markers (Morley et al. 2004; Cheung et al. 2005; Stranger et al. 2005; Pastinen et al. 2006; Dixon et al. 2007; Göring et al. 2007; Kwan et al. 2008; Montgomery et al. 2010; Pickrell et al. 2010). We generally assume that causative polymorphisms, which are in linkage disequilibrium with the associated markers, are responsible for regulating transcript levels. However, in a vast majority of cases, the actual regulatory mechanisms have not been elucidated. In particular, it is not known if eQTLs act preferentially at the level of transcription or at the level of mRNA processing or stability (Gilad et al. 2008; Veyrieras et al. 2008).
There exist only isolated instances in which the molecular mechanisms of regulatory single nucleotide polymorphisms (SNPs) have been identified. For example, a SNP in the CHI3L2 promoter has been implicated in recruiting variable amounts of RNA polymerase, suggesting regulation at the level of transcription (Cheung et al. 2005). In a contrasting case, a SNP affecting a polyadenylation site of IRF5 has been shown to affect the length of the 3′ untranslated regions (UTRs) (Graham et al. 2007), which, in turn, results in variable levels of the entire protein-coding region of the gene, presumably by affecting the stability of the entire mRNA molecule. Additional evidence from computational analyses shows an excess of SNPs associated with eQTLs both in the promoter and in the 3′ regions of genes, suggesting that SNPs affecting mRNA stability may, in general, post-transcriptionally regulate gene expression levels (Veyrieras et al. 2008; Pickrell et al. 2010). Since expression control using 3′ UTRs has also been observed in other model systems (Sandberg et al. 2008), it has been proposed that SNPs altering microRNA binding sites may be a common mechanism of genetic control—however, no evidence currently exists for this mechanism. Finally, for several genes, splicing regulatory SNPs that exert control on the type of isoform as well as the total amount of mRNA produced have been identified, providing further evidence for cotranscriptional regulation of eQTL targets (Field et al. 2005; Pinyol et al. 2007; Kwan et al. 2008). However, with the exception of individual examples, the overall proportion of eQTL targets regulated at the level of transcription compared co- and post-transcriptionally could not be determined until now.
Recently, next-generation RNA sequencing (RNA-seq) has been shown to be an effective way of simultaneously profiling both gene expression levels and the types of isoforms that are being expressed (Marioni et al. 2008; Wang et al. 2008). Two independent groups have shown that RNA-seq is capable of identifying eQTLs and providing insights into their regulatory mechanisms (Montgomery et al. 2010; Pickrell et al. 2010). Those two studies have estimated that there are 839 and 939 eQTLs in Northern European and Nigerian populations, respectively, demonstrating the power of this approach. In our study, we extend those findings by providing the first estimate of the proportion of eQTLs acting transcriptionally versus co- or post-transcriptionally and by describing in detail three typical mechanisms underlying post-transcriptional regulation by eQTLs. We use ultra-deep RNA sequencing data from two healthy, unrelated HapMap individuals of European descent (NA12891 and NA12892) to characterize all alternative splicing (AS) events, to dissect in depth the number of previously identified eQTLs and to describe the differences in the expressed transcripts at single-base resolution. Specifically, using our unique RNA preparation method, we contrast results obtained from exonic (mostly processed mRNA) and intronic (mostly unspliced pre-mRNA) sequences to draw conclusions regarding the relative prevalence of various modes of regulation. We then focus our analysis on four genes with discordant mRNA and pre-mRNA expression patterns to illustrate how SNPs can affect gene and isoform expression by affecting splice-site strength and usage or mRNA stability.
Results
We sequenced total, poly(A)-enriched RNA from two unrelated HapMap CEU lymphoblast cell lines. Although, compared to previous population-wide studies, our approach uses only two individuals, this allows us to obtain very high sequencing depths, with true “single-base” resolution in deconvoluting transcript structures. Below, we first describe the potential biases present within RNA sequencing data. We then characterize global AS events, with particular attention on those associated with genetic variation in the vicinity of exon junctions. Finally, we investigate the potential involvement of splicing and other regulatory mechanisms as underlying causes of eQTL action. In order to ensure that the differences observed between our two samples are truly genetic and not caused by random sample-to-sample variation, we focus the latter part of our analysis on genes known to be genetically controlled from independent studies.
Detection of biases across transcripts
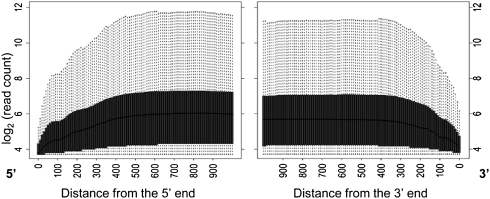
Whole-transcript profiling of RNA expression data suffers from uneven coverage of transcripts due to variation in GC content, sample preparation, and fragmentation of the template. In our earlier analysis of exon microarray data (Bemmo et al. 2008), we noted a strong 5′ versus 3′ bias, resulting in frequent saturation of signal at the 5′-end and reduction of signal at the 3′-end of genes. Since RNA sequencing library construction uses several steps that could result in similar biases [e.g., poly(A) selection and random priming], we investigated whether sequencing-based transcript profiling results in coverage bias across genes. As expected, we found reduced coverage at the 3′ region (Fig. 1). This is the effect of reduced likelihood of random priming during first-strand cDNA synthesis at the edge of the transcript (Bemmo et al. 2008). However, contrary to expectations, we did not find a loss of coverage of long transcripts that may be caused by RNA degradation combined with poly(A) selection. The coverage does not decrease with distance from the 3′ end of the molecules, even for transcripts longer than 10 kb (data not shown). However, we did observe a decrease of coverage at the 5′ end of genes. Such an effect is predicted to result from fragmentation of RNA molecules (Wang et al. 2009). As a result of these “edge” biases, our data set may have reduced power to detect AS events occurring near the termini of the gene, such as alternative poly(A) sites, and alternative promoter usage. However, data generated using this RNA sequencing protocol is well suited for estimating whole gene expression level and detecting AS events involving internal exons.
Figure 1.
Sequencing coverage as a function of the position across a transcript in NA12892. For all genes longer than 1000 nt and expressed above a nominal background (≥10 reads), the number of mapping reads at each position of the transcript is plotted against the distance to the transcript ends. Data points have been averaged within 10-nt bins.
Detection of alternatively spliced variants and sQTLs
We searched for alternative splicing of cassette exons by identifying exons that are skipped in a subset of the transcripts and where the two adjacent flanking exons are constitutively included. These cassette exons were then filtered for those that show statistically significant differential splicing between the two individuals used in our study, which was assessed using a Fisher's exact test on a 2 × 2 contingency table. The list of all cassette exons is provided in Supplemental Table 1.
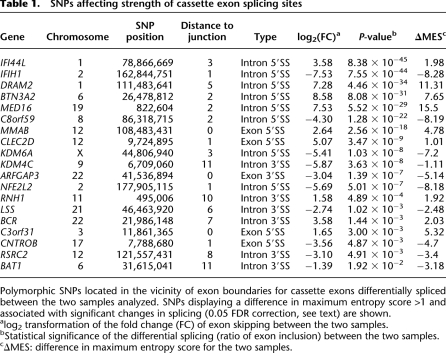
Since we are interested in AS events that are genetically regulated, we compiled a list of all the polymorphic SNPs in our two samples within 200 bp of the splice junctions of these cassette exons. We then analyzed their potential effect on the choice of splicing site. AS events with proximal polymorphic SNPs show a twofold enrichment for the most significant changes (using a nominal P < 0.01 cutoff) in splicing between the two samples (Supplemental Fig. 1), suggesting that these SNPs are at least partly responsible for the splicing differences observed between the two samples. We further assessed the potential of these SNPs to modify exonic splicing enhancer (ESE) motifs (Cartegni et al. 2003), as well as the strength of the resulting splicing site, by using a maximum entropy-based scoring method (Supplemental Table 2; Yeo and Burge 2004). In order to determine whether these computational predictions can be used to infer the effect of the SNP on splicing, we tested whether the levels of exon inclusion correlated with these metrics. We found no correlation between exon inclusion levels and the strength of splicing enhancer binding sites (data not shown), suggesting that ESE predictions themselves are a poor indicator of the effect of SNPs on splicing patterns. However, we found excellent agreement between the maximum entropy (MES) scores and the levels of exon inclusion. We first selected all SNPs predicted to result in |ΔMES| > 1 between the two samples. Next, in order to select the confident subset of cassette exons that were differentially spliced between samples, we ranked the cassette exons by their P-value and carried out a false discovery rate (FDR) correction (Benjamini-Hochberg). For all 19 cassette exons that passed the 0.05 FDR correction, the direction of the splicing change (exon inclusion level) agreed with the theoretical prediction of splice site strength (100% concordance, sign test P = 1.9 × 10−6). The list of cassette exons, the corresponding SNPs, and their predicted effect on splice sites is shown in Table 1. As expected, this level of concordance decreases with relaxed statistical significance levels and ΔMES cutoffs. However, this approach illustrates that the combination of RNA-seq data and theoretical predictions can be used to identify sQTLs, even with only two samples. Three of the 19 AS events have been observed and validated in our previous study (Coulombe-Huntington et al. 2009). Importantly, the remainder are novel, underscoring the utility of this approach as an alternative to population-based designs, and its ability to identify the effects of rare variants, which may be observed in a pairwise comparison but may not exert a detectable effect within a larger cohort.
Table 1.
SNPs affecting strength of cassette exon splicing sites
Polymorphic SNPs located in the vicinity of exon boundaries for cassette exons differentially spliced between the two samples analyzed. SNPs displaying a difference in maximum entropy score >1 and associated with significant changes in splicing (0.05 FDR correction, see text) are shown.
aLog2 transformation of the fold change (FC) of exon skipping between the two samples.
bStatistical significance of the differential splicing (ratio of exon inclusion) between the two samples.
cΔMES: difference in maximum entropy score for the two samples.
RNA sequencing and microarray eQTL target expression differences are concordant
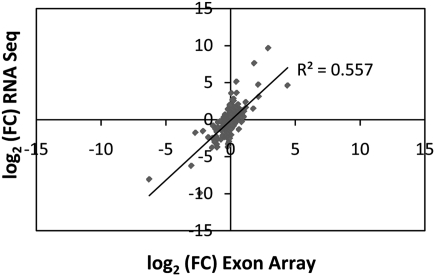
In order to establish the connection between sQTLs and eQTLs, we decided to focus the subsequent analysis on genes that previously have been shown to be genetically controlled. We first set out to demonstrate that our RNA sequencing data are concordant with previous microarray results and do, indeed, support previous eQTL findings. We examined the top 500 eQTLs, based on statistical significance of the correlation between marker genotype and expression levels, from two previous analyses (Stranger et al. 2005; Kwan et al. 2008). We further selected only those genes for which the SNPs associated with the eQTLs were polymorphic in the two individuals used in our study (NA12891 and NA12892) and were expressed above a nominal background level (defined as more than 10 reads targeting the gene in at least one of the samples). Hence, we compared expression level differences predicted from published microarray results to those obtained from RNA sequencing experiments. To estimate gene expression levels, we counted all the sequencing reads mapping to the mRNA of each gene (exons and exon–exon junctions). The results were quantile-normalized and log2-transformed, and the log-ratios (fold change [FC]) were used to measure the difference in gene expression levels between the two individual samples. Previous experiments found general agreement between microarray and RNA-seq results (Marioni et al. 2008; Mortazavi et al. 2008; Wang et al. 2009), particularly for genes with medium expression levels. However, we wanted to verify that this is true for our selected eQTL targets. We found a high level of correlation (Pearson R2 = 0.56) between expression differences from RNA sequencing experiments and those previously obtained from exon array experiments (Fig. 2; Kwan et al. 2008). The correlation was further improved by considering only genes with the highest microarray FCs: R2 = 0.69 for the top 100 genes, R2 = 0.73 for the top 50 genes. This level of correlation is remarkable since it has been obtained using independent biological replicates, distinct cDNA library preparation protocols, and compares hybridization-based (microarray) and direct sequencing approaches. It should be noted that a high level of correlation between microarray and RNA-seq eQTL results has also been observed by others (Montgomery et al. 2010; Pickrell et al. 2010), but our two-sample analysis gives a clear and concise answer as to how the two technologies compare.
Figure 2.
Correlation between expression level differences of the top 500 eQTL targets predicted by an exon array study (Kwan et al. 2008) and our RNA sequencing study. Gene expression levels from RNA sequencing data were obtained by counting all the short sequencing reads mapping to the mRNA of each gene (exons and exon–exon junctions). Results were normalized and log2-transformed, and the log-ratios (fold changes [FC]) between NA12891 and NA12892 were used to measure the difference in gene expression between the two individuals. The Pearson correlation is shown as the R2 value.
The correlation was notably lower for the results obtained using the 3′-targeted Illumina microarray platform: R2 = 0.43 for the top 50 eQTLs. This discrepancy can be explained by the differences in platform design; the exon array tiles the probes roughly evenly across each transcript, a strategy that is similar to mapping sequencing reads to a gene, whereas the Illumina platform relies on a single probe generally targeted to the 3′ region or individual exons of each gene. This lower correlation level suggests that a considerable proportion of eQTLs detected using 3′ microarrays may be isoform eQTLs, or sQTLs. Here, we define isoform eQTLs as those affecting only expression of a particular isoform, as opposed to all transcripts originating from a given locus, and where the overall total amount of mRNA expressed by the locus is not significantly affected. Such isoform eQTLs may affect alternative promoter usage, alternative splicing, or premature termination of the transcript.
The majority of eQTLs act at the level of transcription
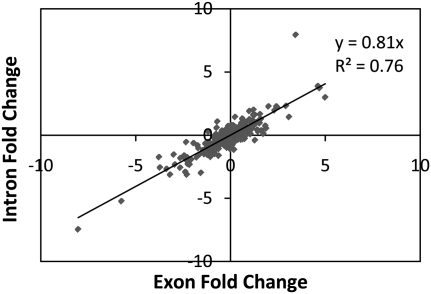
Our method of cDNA preparation for sequencing retains a detectable fraction of unprocessed heteronuclear RNA (Ge et al. 2009). This is the primary, unspliced RNA fraction, or pre-mRNA. Based on the comparison of the number of reads mapping to intronic and exonic reads, respectively, we estimate that ∼3% of all RNA molecules in our data set originate from the heteronuclear fraction. Heteronuclear RNAs contain introns, and since intronic sequences are, on average, an order of magnitude longer than exonic sequences, more than one-third of our reads map to introns (see Methods). Thus, we are able to obtain a considerable amount of information about the levels and types of pre-mRNAs present in our sample. Specifically, comparing intronic and exonic RNA levels provides valuable information regarding the mechanisms controlling gene expression levels. If the expression of a gene is regulated at the level of transcription, the amounts of mature mRNA and unprocessed pre-mRNA levels will be affected equally. However, if a gene is regulated post-transcriptionally, the levels of intronic RNA should remain relatively unchanged, while the level of mature exonic mRNA will vary. We investigated the two hypotheses by comparing the fold-changes in the levels of intronic and exonic sequences within each of the top 500 eQTLs compiled from exon array experiments (Kwan et al. 2008). We find a very high level of correlation (Pearson test, R2 = 0.76) between the expression levels measured in exons and introns (Fig. 3), implying that 76% of the variation in the expression of these genes can be explained by regulation at the level of transcription. Hence, we conclude that transcriptional regulation is responsible for the control of the majority of eQTLs observed in human lymphoblasts.
Figure 3.
Correlation between expression levels of exons and introns within the eQTL targets used in the study (see text). This correlation can be used to infer the number of eQTLs acting at the level of transcription versus the number of eQTLs acting co- or post-transcriptionally. The R2-value represents the Pearson correlation, and the y-value represents the slope of the regression line.
We hypothesize that the remaining eQTL targets may be regulated co- or post-transcriptionally. In accordance with this hypothesis, we observe that the slope of the regression line of intronic versus exonic data is below the diagonal (i.e., slope = 0.81), demonstrating that exonic variation is more pronounced than intronic variation (Fig. 3). The expectation under the hypothesis of purely transcriptional regulation would be a slope of 1, representing effects of equal magnitude observed in exons and introns. The regression analysis again demonstrates that variation within exons alone is responsible for 20%–25% of the observed change in expression. This co- or post-transcriptional regulation may take place at the level of mRNA stability or splicing.
Alternative splicing is involved in regulation of eQTL targets
We have previously described a large number of genes that are affected by common splicing polymorphisms, including variants that alter the strength of the consensus splice sites (Table 1) or regulatory RNA sequences located in the vicinity of the splice sites (Kwan et al. 2007, 2008; Coulombe-Huntington et al. 2009). A number of differentially expressed genes (eQTL targets) exhibit concurrent differences at the levels of total mRNA produced and the type of isoform produced (sQTL targets). We set out to investigate the connection between these two phenomena. Specifically, are the total RNA levels of genes commonly regulated by splicing polymorphisms? If this is the case, eQTL targets should contain detectable alternatively spliced variants more often than expected by chance, or in other words, at a higher frequency than a non-eQTL set of genes. To test this hypothesis, we identified the splicing events (cassette exons, alternative 5′ and 3′ splice site usage) that were differentially represented between the two individuals (at P < 0.001, one-tailed χ2 test). We find a significant, roughly twofold enrichment of these AS events in eQTL targets as compared to non-eQTL genes expressed at comparable levels (Table 2). It should be noted that the overall proportion of AS events within the eQTL set is lower than 10%, meaning that for genes where the overall expression of the transcript is under genetic control, regulation at the level of splicing has a significant effect but is not likely to account for more than 10% of the total variance.
Table 2.
Alternative splicing events within eQTLs
aThe number of alternative splicing events were counted in a set of control non-eQTL genes, in the eQTLs identified in an Illumina microarray study by Stranger et al. (2005), and in an exon microarray (HuEx) study by Kwan et al. (2008). A one-tailed χ2 test of homogeneity was used to determine the significance of the differences between the two eQTL gene sets and the control genes.
We examined our list of AS events within eQTL targets to identify the clearest examples of regulation of mRNA levels by differential splicing. Three types of mechanisms were observed: (1) SNPs altering the efficiency of splice sites leading to alternative splice site usage; (2) SNPs activating pseudoexons resulting in premature termination codons and likely nonsense-mediated decay; and (3) SNPs altering splicing within 5′ UTRs, thereby affecting the stability of the mRNA. We describe in detail the regulatory mechanisms of the SNPs most likely to be responsible for the observed variation in the genes USMG5, MMAB, OAS1, and MRPL43. Furthermore, these four genes show significant inconsistencies in mRNA and pre-mRNA expression differences between the two individuals, indicating a post-transcriptional mode of regulation.
USMG5
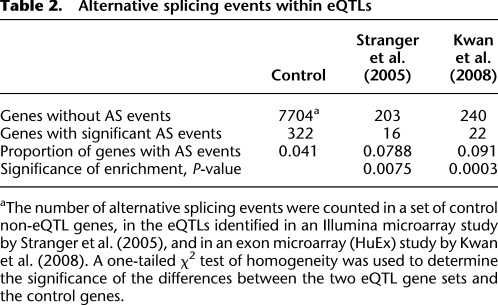
Illumina microarray results predicted a threefold difference in expression of this gene between NA12891 and NA12892, and RNA-seq estimates are in agreement. However, the total number of intronic reads is unchanged between the two samples (FC = 1.02), suggesting that the difference in expression of USMG5 does not take place at the level of transcription. Further examination of the splicing patterns of USMG5 reveals that exon 2, a known cassette exon, is more frequently retained in individual NA12892 (Fig. 4A). On the other hand, individual NA12891 mostly skips this exon and instead exhibits increased read coverage in a region slightly upstream, which was subsequently identified as an alternative exon (1a). These two alternative exons are almost always mutually exclusive in our two samples and have opposite effects on mRNA expression: The increased retention of exon 2 correlates with increased overall transcript levels, whereas inclusion of exon 1a occurs concomitantly with reduced overall expression (Fig. 4).
Figure 4.
In USMG5, inclusion of exon 2 is associated with increased transcript expression, and inclusion of exon 1a is associated with decreased transcript expression. The reference T allele of rs791148 is associated with inclusion of exon 1a, and the alternate G allele is associated with inclusion of exon 2. (A) Screenshots taken from Integrated Genome Viewer (IGV) illustrating the read coverage of the two samples in USMG5. The Y-axis for both tracks is scaled to a maximum read count of 1150 (measured as number of reads mapping to a given nucleotide). (B) The different USMG5 isoforms observed. (C) The percentage of each isoform (color-coded as in B) observed in the allele-specific alignment for both individuals. The height of each pie chart is representative of the gene expression for USMG5 as measured by the number of reads mapping to the exonic and intronic regions of the gene. The number of isoforms present in each individual was deduced by counting the number of splice junction reads spanning a unique splice junction for each of the isoforms (junction 1a-2 for isoform 1; junction1-3 for isoform 2; junction 1a-3 for isoform 3; and junction 1-2 for isoform 4).
We propose that SNP rs7911488, located in exon 2, is responsible for this expression pattern based on Figure 4 and our earlier exon microarray study (Kwan et al. 2008). The alternate G allele, associated with elevated inclusion levels of exon 2, is predicted to introduce the binding sites for two exonic splicing enhancers: SF2/ASF (also known as SRSF1) and SRp55 (also known as SRSF6) (Cartegni et al. 2003). In contrast, no binding sites for any known SR protein splicing factors are present within the sequence containing the reference T allele, associated with skipping of exon 2 and lower transcript coverage (Fig. 4C). The association between rs7911488, USMG5 isoforms, and USMG5 expression level is an example of how an SNP can alter splicing efficiency, in this case by introducing splicing factor binding sites, leading to differential isoform expression and consequently to differences in gene expression in a genotype-dependent manner (mechanism 1; see above). This is further compounded by the fact that the SNP is in the 5′ UTR and thereby may affect mRNA stability (mechanism 3).
MMAB
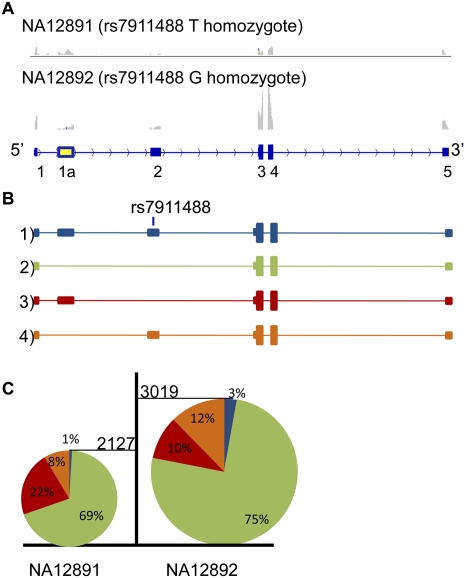
We have previously noted that the MMAB gene contains an unannotated exon (6a) (Fig. 5), whose inclusion is regulated by a trio of SNPs in close proximity to its donor splice site (Coulombe-Huntington et al. 2009). Figure 5A clearly shows the correct splicing and increased inclusion levels of this exon in NA12892. This inclusion is associated with decreased expression levels of the entire gene: Individual NA12892 has a 5.7-fold increase in expression of exon 6a, yet a 1.5-fold decrease in total MMAB expression compared to NA12891 (Fig. 5C). The alternative exon of MMAB exhibits a high level of allelic expression with SNP rs2287180 in individual NA12892 (Fig. 5D). This imbalance is not seen in heterozygous intronic SNPs throughout the gene, suggesting that the regulatory effect on gene expression takes place at the post-transcriptional level, and is most likely due to the inclusion of the alternative exon. This exon is in-frame but introduces a stop codon immediately downstream from exon 6, likely resulting in nonsense-mediated decay and the corresponding decrease in mRNA levels of the gene. This is an example of mechanism 2: The inclusion of exon 6a is increased with the presence of the alternate bases of the three SNPs affecting the donor splice site and results in a premature termination codon and nonsense-mediated decay.
Figure 5.
Correlation of SNP rs2287180 to overall MMAB transcript expression and to expression of MMAB novel exon 6a. (A) Screenshots taken from IGV illustrating the read coverage of MMAB. The maximum height for both tracks is set to 700. (B) The different MMAB isoforms seen in the alignment. (C) The percentage of transcripts (color-coded as in B) including and excluding exon 6a in NA12891 and NA12892. To measure the transcripts including exon 6a, the number of reads mapping to the junctions 6-6a and 6a-7 was averaged, and to measure the number of transcripts excluding exon 6a, the number of reads mapping to junction 6-7 were counted. The height of each pie chart is representative of the gene expression for MMAB. (D) The number of reads supporting the reference and alternate alleles for rs2287180, respectively, in individual NA12892.
MMAB is of particular interest because of its involvement in the cobalamine pathway. Mutations in MMAB result in the metabolic disorder methylmalonic aciduria, type cblB, and polymorphisms near the gene have recently been associated with high-density lipoprotein (HDL) cholesterol levels, the “good cholesterol.” Fogarty et al. (2010) found that the alternate alleles of two MMAB SNPs lead to higher expression of the full-length MMAB transcript resulting in lower HDL levels. The SNP rs2287180 expressed in NA12892 appears to have the opposite effect in terms of MMAB expression. As noted above, the T allele of rs2287180 leads to the inclusion of exon 6a and decreased expression of MMAB. Thus, it is plausible that rs2287180 also plays a role in HDL levels by decreasing MMAB expression.
OAS1
Although the variation in this gene has been studied experimentally (Bonnevie-Nielsen et al. 2005), with microarrays (Kwan et al. 2007, 2008) and, more recently, with RNA-seq (Pickrell et al. 2010), we present it as an example in view of additional isoforms and allelic expression evidence uncovered in our analysis, which further clarifies the genetic control of OAS1 expression. Furthermore, the OAS1 locus has high disease relevance, having been associated with susceptibility to type 1 diabetes (Field et al. 2005), West Nile virus (Lim et al. 2009), dengue virus (Lin et al. 2009), and multiple sclerosis (Fedetz et al. 2006). OAS1 has previously been identified as an eQTL in microarray analyses (Stranger et al. 2005; Kwan et al. 2008), and we and others (Bonnevie-Nielsen et al. 2005; Kwan et al. 2007; Pickrell et al. 2010) have further described a splice eQTL involving the SNP rs10774671.
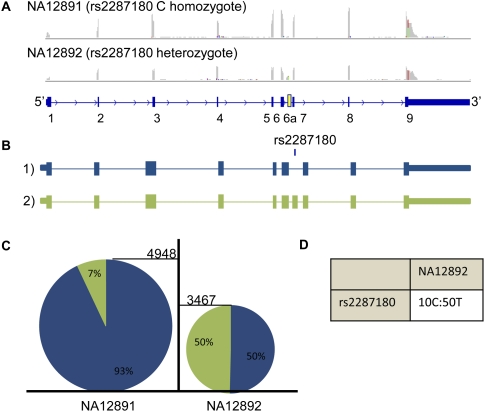
It is previously understood that the alternate A allele of rs10774671 disrupts the splice acceptor site in the terminal exon creating two alternative OAS1 isoforms: p48, which uses a downstream splice site within the last exon; and p52, which shifts the splice site by 1 bp (Bonnevie-Nielsen et al. 2005). Remarkably, from our RNA-seq data, we can additionally show that the decreased level of exonic expression is associated with an increased retention of the last intron in NA12892 (Fig. 6A). This individual expresses two additional isoforms associated with rs10774671 that have been previously identified (Fig. 6B; Pickrell et al. 2010). In the first isoform, p42, the A allele causes preferential use of an alternative, proximal polyadenylation site within the penultimate exon. Until the advent of RNA-seq, it was not known that p42 is preferentially expressed in conjunction with the A allele. The A allele is also responsible for the second isoform (isoform 4 or p44), which uses an alternative, cryptic splice site within the last intron, upstream of the affected splice site, and constitutes ∼25% of the transcripts for this genotype (Fig. 6C). Additional evidence portraying the allele-specific expression of OAS1 is provided by SNPs throughout the transcript (Fig. 6D). Finally, we identify a completely novel isoform (isoform 6), resulting from alternative splice acceptor site usage within exon 3. This isoform appears not to be under genetic control and is present in ∼5% of transcripts produced by each genotype, further increasing the potential proteomic diversity introduced by this locus. In summary, the alternate allele of rs10774671 disrupts a splice acceptor site and instead is preferentially included in alternatively spliced isoforms that use upstream splice sites or retain the last intron, illustrating that this eQTL functions by mechanism 1 (Fig. 6C,D).
Figure 6.
OAS1 isoforms. (A) Screenshots from IGV displaying the read coverage within the OAS1 gene. The maximum height of both tracks is 2000. (B) All possible isoforms seen in OAS1 and their association with SNPs rs1131454, rs60623134, rs10774671, and rs1051042. Larger font indicates stronger allelic imbalance (not to scale), and if both alleles are shown, then there is no evidence for genetic control on isoform production by this SNP. Isoforms 1, 2, 3, and 4 differ only by exon 6: in isoform 1 it is the normal exon 6; in isoform 2 it is shifted by 1 bp (exon 6′); in isoform 3 it is shifted by 98 bp (exon 6″); and in isoform 4 it begins within the intron (exon 6″′). (C) Relative percentages of isoforms seen within individuals NA12891 and NA12892. The number of isoforms is inferred by the number of reads mapping to unique splice junctions for most isoforms (junction 5-6 for isoform 1; junction 5-6′ for isoform2; junction 5-6″ for isoform 3; junction 5 to 6″′ for isoform 4; junction 2-3′ for isoform 6). For isoform 5, expression was measured by the number of reads mapping 25 bp past the end of exon 5 (still within the extended exon). The height of each pie chart is representative of the gene expression for OAS1. There is a 1.9-fold increase in expression in favor of individual NA12892. (D) The number of reads supporting the reference and alternate alleles seen in individual NA12892.
MRPL43
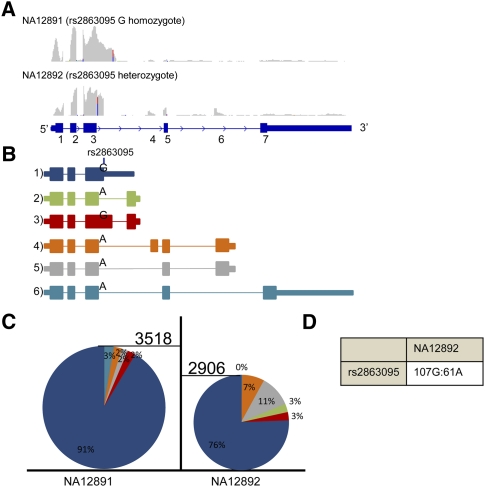
The gene MRPL43 has been detected as an eQTL target in several previous studies (Stranger et al. 2005; Kwan et al. 2008). Exon array analysis indicated that MRPL43 may be controlled by an sQTL with expression of several isoforms of the gene varying in genotype-dependent ratios; however, the precise nature of those isoforms could not be identified. Here, we identify the probable causative SNP, rs2863095, affecting the splicing and overall mRNA isoform levels within this gene. rs2863095 is immediately downstream (3 bp) from the splice donor site of exon 3, and the alternate A allele is predicted computationally to significantly strengthen this splice site (maximum entropy increase of 4.26) (Yeo and Burge 2004). As a result, the vast majority of G-containing transcripts fail to splice this exon and instead use a downstream polyadenylation site to terminate the transcripts, as demonstrated in individual NA12891 (Fig. 7B,C). In contrast, transcripts containing the A allele preferentially use this splice site to produce several longer alternative isoforms. Indeed, individual NA12892, who is heterozygous for rs2863095, exhibits both a much larger proportion of MRPL43 isoforms and higher expression of these isoforms (Fig. 7C). The responsible SNP also demonstrates a considerable expression imbalance within the short isoform (Fig. 7D), providing further evidence that the G allele is associated with production of this transcript. This regulatory effect is an example of how an SNP can alter the efficiency of an existing splice site and introduce alternative isoforms (mechanism 2).
Figure 7.
Isoform eQTLs found in MRPL43. (A) Screenshots from IGV displaying the read coverage within MRPL43. The maximum height of both tracks is 370. (B) Various isoforms detected in MRPL43 and their allele specificity (if any) to SNP rs2863095. (C) Relative percentages of isoforms (color-coded as in B) seen within individuals NA12891 and NA12892. Expression of each isoform was measured by counting the number of reads mapping to a junction specific for most isoforms (junction 3-3a for isoform 2; junction 3′-3a for isoform 3; junction 3-4 for isoform 4; junction 5-7 for isoform 6). For isoform 1, the expression was estimated as the number of reads mapping 110 bp after exon 3, since it is the only isoform that should be expressed at this point. The expression of isoform 5 was deduced by subtracting the number of reads supporting isoform 6 from the number of reads spanning junction 3-4, since they are the two isoforms using this splice junction. The height of each pie chart is representative of the gene expression for MRPL43. (D) Ratio of reference to alternate alleles seen in individual NA12892.
MRPL43 was recently found within a disease locus for spastic paraplegia with mental retardation (Dursun et al. 2009). Resequencing of MRPL43, which was the top candidate out of 87 genes within the locus, identified no mutations; however, a 20-fold increase in mRNA levels was seen between two patients and five controls, all of whom are from the same family. It is possible that the more subtle regulatory effects in the MRPL43 eQTL exert less visible phenotypic effects across human populations.
Discussion
The preponderance of expression quantitative trait loci has now been extensively documented (Morley et al. 2004; Cheung et al. 2005; Stranger et al. 2005; Pastinen et al. 2006; Dixon et al. 2007; Göring et al. 2007; Kwan et al. 2008; Montgomery et al. 2010; Pickrell et al. 2010). Many human genes are variably expressed in a genotype-dependent manner. The estimated number of such genes has reached thousands, as the power of studies and technologies have both improved (Göring et al. 2007; Ge et al. 2009). Many eQTLs have been validated across studies, and many have been found to act in multiple tissues (Heinzen et al. 2008; Bullaughey et al. 2009; Kwan et al. 2009).
More recently, our group (Kwan et al. 2007, 2008; Coulombe-Huntington et al. 2009), followed by several others (ElSharawy et al. 2009; Montgomery et al. 2010; Pickrell et al. 2010), has demonstrated the parallel existence of isoform eQTLs, loci that may not affect the overall gene expression levels but alter their transcript structure in a genotype-dependant manner. These isoform eQTLs can function at the level of alternative splicing, alternative polyadenylation sites, or alternative promoter usage. Subsequently, we have been able to identify and validate a large number of genetically controlled splicing variants, along with their regulatory SNPs whose function can often be predicted in silico (Coulombe-Huntington et al. 2009).
RNA-seq provides an unprecedented opportunity to investigate splicing-level variation. It has been shown to be highly accurate and replicable, resulting in 85% validation rates of detected splicing events by RT-PCR (Griffith et al. 2010). In this study, we demonstrate that we can reliably predict the effect of SNPs on the splicing of cassette exons, even when comparing only two samples. Although an analysis of two samples, carried out with no technical replication, will typically identify a large number of stochastic variation between samples, we show that by focusing on variable cassette exons with SNPs in close proximity to splice sites, we achieve 100% accuracy in predicting the effect of those SNPs on exon inclusion. Only three of the cassette exons reported here have been identified as targets of sQTLs in a previous study (Coulombe-Huntington et al. 2009), while the remainder are novel, further demonstrating the utility of this approach.
On a technical level, we find that current RNA-seq protocols provide, on average, consistent coverage of transcripts with no significant dependence on gene length and little variation in coverage of the interior portions of genes (Fig. 1). This renders the method highly suited for profiling overall transcript expression levels and detecting alternative splicing of interior, cassette exons. However, there are significant biases against both the 3′ and 5′ transcript termini, resulting in loss of coverage and relatively poorer detection of alternative initiation and termination sites of genes. Since these effects are most likely related to RNA-level fragmentation (Wang et al. 2009) and reduced efficiency of random priming in the vicinity of fragmented template (Bemmo et al. 2008), future adjustment of protocols for isoform-level analysis may include the use of poly(T) primers to better capture the 3′ fragments, and cDNA template fragmentation to prevent the loss of signal at the 3′ ends. It should be noted that many of the biases observed in current studies may be overcome entirely by third-generation sequencing technologies (Schadt et al. 2010), which minimize sample preparation requirements, require no template amplification, provide sequence data originating from single RNA molecules, and promise to provide information not only on individual splicing events, but also reveal the exact combinatorial RNA structure of individual transcripts.
Our method of template preparation employed in this study is essentially similar to other current protocols (Marioni et al. 2008; Sandberg et al. 2008; Griffith et al. 2010; Montgomery et al. 2010; Pickrell et al. 2010), with the distinction of using only a single (as opposed to repeated) poly(A) selection step. As a result, our template retains significant amounts of unspliced, pre-mRNA, allowing us to profile the levels of both exonic and intronic sequences. We use this information to investigate the relationship between sQTLs and eQTLs and draw some general conclusions regarding the mechanisms of eQTL action.
It is likely that eQTLs act through a combination of transcriptional, cotranscriptional (e.g., splicing), and post-transcriptional (e.g., RNA stability) mechanisms. However, in order to identify regulatory SNPs, it is of high interest to determine which of the mechanisms are predominant. In our present study, by comparing results based on intronic RNA (unprocessed, heteronuclear) with those based on exonic RNA (predominantly spliced, polyadenylated), we conclude that a large majority (∼75%) of eQTLs control their targets at the level of transcription. The remaining 25% are likely to act co- or post-transcriptionally, possibly via splicing or mRNA stability-related mechanisms. This constitutes the first empirical evidence quantifying the relative importance of transcriptional and post-transcriptional mechanisms on regulating eQTL targets. Furthermore, by focusing on the genes for which the intronic and exonic analyses produce discordant results, we were able to identify and dissect in fine detail four examples of genes regulated by SNPs affecting alternative splicing. This analysis further establishes the connections between eQTLs and sQTLs and points out approaches for pinpointing causative regulatory polymorphisms.
Both eQTLs and sQTLs are hypothesized to be involved in human phenotypic diversity and susceptibility to disease (Faustino and Cooper 2003; Dewan et al. 2006; Emilsson et al. 2008; Lim et al. 2009; Andrés et al. 2010; Cooper 2010; Musunuru et al. 2010; Nica et al. 2010). In order to be able to use eQTLs as diagnostic markers, as well as for any future therapeutic possibilities, it is essential to identify the causative SNPs, along with their mechanisms of action. To date, very few eQTLs have been dissected at the molecular level, and little is known about the predominant mechanisms linking genetic variation and gene expression levels. Our study makes new inroads toward understanding the mechanisms underlying the effects of genetic variation on human gene expression.
Methods
RNA extraction and cDNA synthesis
The two samples analyzed here are the unrelated parents from a HapMap CEU trio: NA12892 and NA12891. Lymphoblastoid cell lines (LCLs) were obtained from Coriell, and cell culture was carried out under standard conditions (Kwan et al. 2008). We extracted total RNA and applied a cDNA synthesis protocol on heteronuclear RNA, which allowed us to measure unspliced primary transcripts. Approximately 150 μg of total RNA was isolated, treated with 6 U of DNase I, and poly(A)-enriched using the MicroPoly(A)Purist protocol (Ambion). The first- and second-strand cDNA synthesis was carried out on 1 μg of poly(A)-enriched RNA using random hexamers, and second-strand cDNA synthesis was performed using the Superscript Double-Stranded cDNA Synthesis Kit (Invitrogen). The protocol was based on that described by Ge et al. (2009) in the study of allelic expression using Illumina bead arrays.
High-throughput sequencing
cDNA from each sample was subjected to sequencing using the Illumina GAIIx technology. We obtained seven lanes of paired-end, 76-bp reads per sample. Reads were processed using ELAND (Cox 2007) to those that passed quality control in FASTQ format. This provided an average of 143 million post-quality-control reads, for a total of 10.1 Gb of sequence per sample. In total, 95% and 94% of reads were mappable to the genome for NA12891 and NA12892, respectively. The majority of the remaining reads likely aligned to multiple positions in the genome and, as a consequence, were filtered and excluded from downstream analysis. These repetitive locations likely represent gene families, such as paralogs and pseudogenes.
In order to estimate the relative proportions of unspliced, heteronuclear pre-mRNA molecules and the processed, mature mRNA molecules, we compared the number of reads mapping to exons and introns in all RefSeq-annotated genes. The total number of intronic reads was then normalized by the total length of introns (in kilobases) contained within all genes expressed at detectable levels, while the number of exonic reads was normalized by the length of exons (in kilobases) for the same set of genes. Genes with an average of at least 1 read per base pair were considered expressed for these calculations. For NA12891 we obtained the following results: normalized mean intronic read count = 0.05742; normalized mean exonic read count = 1.928. For NA12892, the corresponding numbers were very similar: normalized mean intronic read count = 0.05654; normalized mean exonic read count = 1.710. Together, these numbers suggest that exons are targeted on average at a rate 32 times higher than introns, when adjusted for size. This, in turn, implies an ∼3% fraction of unspliced hRNA molecules in the sample.
Sequence alignment
In order to quantify gene and isoform expression levels, the sequencing reads were aligned to each annotated gene in the human genome. We used the BWA aligner due to its proven ability to efficiently call indels (Li and Durbin 2009). Our pipeline involved first aligning the reads to the reference genomic sequence. Since, similarly to SNPs within microarray probe sequences (Benovoy et al. 2008), individual-specific genotype differences are known to introduce biases during the alignment process (Degner et al. 2009), we constructed four individual-specific reference sequences. Each sequence corresponded to a phased haplotype of one set of autosomes. Phased haplotypes were obtained from the 1000 Genomes website (http://www.1000genomes.org/). Since these two individuals had been sequenced at a high coverage as part of the pilot project, we were confident that the resulting sequences were highly accurate and did, indeed, constitute nearly perfect references for the purpose of quantitative mapping. For each individual, we mapped the reads to the two distinct haplotypes, combined the results, and filtered out duplicates to make sure that each read was only counted once.
The reads that were not confidently mappable to the genome were presumed to originate from exon–exon junctions. Thus, the remainder of the reads were aligned to a library of junction sequences constructed from 75 bases (or the length of the exon if less than 75) of each upstream exon, fused to the 75 bases (or the length of the exon) of the downstream exon. In order to account for novel splice isoforms, we created a library of all possible consecutive exon–exon junctions, based on the UCSC gene annotation (Rhead et al. 2010).
Alternative splicing analysis
AS events were detected by identifying mutually exclusive splice junctions. Any two splice junctions with one matching coordinate and one distinct coordinate indicate an AS event. In the case of cassette exons (exon skipping), the read counts for two junctions supporting the inclusion were combined and compared with the read counts for the junction supporting exon skipping. Only sets of adjacent exons were considered. The difference and significance of differential splicing between samples were tested using Fisher's exact test. Supplemental Files with read counts and statistics for gene expression and alternative splicing for the eQTLs used in this study, after filtering out genes with low expression levels, are available online.
From a list of all SNPs present in our two samples obtained from the 1000 Genomes website (http://www.1000genomes.org/), we selected those SNPs that are polymorphic in the two individuals of our study and lying within 200 bp of alternative splice junctions. Splice site strength was scored using a maximum entropy-based method (Yeo and Burge 2004). Exonic splicing enhancer (ESE) motifs were scored as follows: For each polymorphic SNP in the vicinity of a splicing junction, the surrounding 15-bp sequence was analyzed with ESEfinder 3.0 (Cartegni et al. 2003), using sliding windows of 7 bp. For each SNP, the highest score for these sliding windows is reported (Supplemental Table 2).
Expression analysis
We used SAMTools (Li et al. 2009) to extract SNPs with a phred-like score >20 and indels with a phred-like score >50 from the alignment data. Integrated Genome Viewer (http://broadinstitute.org/igv) was used to visualize the data. In order to estimate gene expression levels, we used all reads mapping within the maximal genomic locus containing each gene and its known isoforms, along with all the exon–exon junction reads within the same interval. To estimate expression at the mRNA level, we used only the reads mapping to exonic regions and to junctions for genes with at least 10 mapped reads. To estimate expression at the heteronuclear, pre-mRNA level, we used only reads mapping within introns in the same set of genes. Analysis of alternative splicing was carried out by comparing the numbers of reads mapping to alternative, mutually exclusive junctions. Expression-level counts were quantile-normalized in order to account for systematic differences between the two samples.
Comparison to microarray studies
From the top (statistically most significant) 500 eQTLs of two previous microarray analyses (Stranger et al. 2005; Kwan et al. 2008), we selected a subset of genes for which the SNPs associated with the eQTLs were polymorphic and had at least 10 reads mapping to it in at least one of the two individuals. To estimate gene expression levels, we counted all the sequencing reads mapping to mRNA of each gene (exons and exon–exon junctions). The results were quantile-normalized and log2-transformed, and the log-ratios were used to measure the difference in gene expression levels between the two individual samples. Pearson correlations were used to assess the level of similarity between the two technologies.
Transcriptional regulation
To determine whether a given gene is regulated transcriptionally or post-transcriptionally, high-confidence eQTLs associated with SNPs for which our samples are polymorphic were selected. The number of reads mapping to the gene's exons and introns were counted, and the fold-changes were compared between the sample-specific alignments for each individual.
Acknowledgments
This work was supported by grants from Genome Quebec/Genome Canada awarded to T.P. and J.M., and the Canadian Institutes of Health Research (to J.M.). Both T.P. and J.M. hold Canada Research Chair awards. The RNA sequencing work was performed by the Genome Quebec Expression and Sequencing Platforms at the McGill University and Genome Quebec Innovation Centre.
Footnotes
[Supplemental material is available for this article. The sequencing data from this study have been submitted to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession no. SRA026779.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.111211.110.
References
- Andrés AM, Dennis MY, Kretzschmar WW, Cannons JL, Lee-Lin SQ, Hurle B, NISC Comparative Sequencing Program, Schwartzberg PL, Williamson SH, Bustamante CD, et al. 2010. Balancing selection maintains a form of ERAP2 that undergoes nonsense-mediated decay and affects antigen presentation. PLoS Genet 6: e1001157 doi: 10.1371/journal.pgen.1001157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemmo A, Benovoy D, Kwan T, Gaffney DJ, Jensen RV, Majewski J 2008. Gene expression and isoform variation analysis using Affymetrix Exon Arrays. BMC Genomics 9: 529 doi: 10.1186/1471-2164-9-529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benovoy D, Kwan T, Majewski J 2008. Effect of polymorphisms within probe-target sequences on olignonucleotide microarray experiments. Nucleic Acids Res 36: 4417–4423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnevie-Nielsen V, Field LL, Lu S, Zheng DJ, Li M, Martensen PM, Nielsen TB, Beck-Nielsen H, Lau YL, Pociot F 2005. Variation in antiviral 2′,5′-oligoadenylate synthetase (2′5′AS) enzyme activity is controlled by a single-nucleotide polymorphism at a splice-acceptor site in the OAS1 gene. Am J Hum Genet 76: 623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullaughey K, Chavarria CI, Coop G, Gilad Y 2009. Expression quantitative trait loci detected in cell lines are often present in primary tissues. Hum Mol Genet 18: 4296–4303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR 2003. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res 31: 3568–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung VG, Spielman RS, Ewens KG, Weber TM, Morley M, Burdick JT 2005. Mapping determinants of human gene expression by regional and genome-wide association. Nature 437: 1365–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DN 2010. Functional intronic polymorphisms: Buried treasure awaiting discovery within our genes. Hum Genomics 4: 284–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe-Huntington J, Lam KC, Dias C, Majewski J 2009. Fine-scale variation and genetic determinants of alternative splicing across individuals. PLoS Genet 5: e1000766 doi: 10.1371/journal.pgen.1000766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AJ 2007. ELAND: Efficient large-scale alignment of nucleotide databases. lllumina, San Diego, CA [Google Scholar]
- Degner JF, Marioni JC, Pai AA, Pickrell JK, Nkadori E, Gilad Y, Pritchard JK 2009. Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics 25: 3207–3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewan A, Liu M, Hartman S, Zhang SS, Liu DT, Zhao C, Tam PO, Chan WM, Lam DS, Snyder M, et al. 2006. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science 314: 989–992 [DOI] [PubMed] [Google Scholar]
- Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, Taylor J, Burnett E, Gut I, Farrall M, et al. 2007. A genome-wide association study of global gene expression. Nat Genet 39: 1202–1207 [DOI] [PubMed] [Google Scholar]
- Dursun U, Koroglu C, Kocasoy Orhan E, Ugur SA, Tolun A 2009. Autosomal recessive spastic paraplegia (SPG45) with mental retardation maps to 10q24.3–q25.1. Neurogenetics 10: 325–331 [DOI] [PubMed] [Google Scholar]
- ElSharawy A, Hundrieser B, Brosch M, Wittig M, Huse K, Platzer M, Becker A, Simon M, Rosenstiel P, Schreiber S, et al. 2009. Systematic evaluation of the effect of common SNPs on pre-mRNA splicing. Hum Mutat 30: 625–632 [DOI] [PubMed] [Google Scholar]
- Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, Carlson S, Helgason A, Walters GB, Gunnarsdottir S, et al. 2008. Genetics of gene expression and its effect on disease. Nature 452: 423–428 [DOI] [PubMed] [Google Scholar]
- Faustino NA, Cooper TA 2003. Pre-mRNA splicing and human disease. Genes Dev 17: 419–437 [DOI] [PubMed] [Google Scholar]
- Fedetz M, Matesanz F, Caro-Maldonado A, Fernandez O, Tamayo JA, Guerrero M, Delgado C, López-Guerrero JA, Alcina A 2006. OAS1 gene haplotype confers susceptibility to multiple sclerosis. Tissue Antigens 68: 446–449 [DOI] [PubMed] [Google Scholar]
- Field LL, Bonnevie-Nielsen V, Pociot F, Lu S, Nielsen TB, Beck-Nielsen H 2005. OAS1 splice site polymorphism controlling antiviral enzyme activity influences susceptibility to type 1 diabetes. Diabetes 54: 1588–1591 [DOI] [PubMed] [Google Scholar]
- Fogarty MP, Xiao R, Prokunina-Olsson L, Scott LJ, Mohlke KL 2010. Allelic expression imbalance at high-density lipoprotein cholesterol locus MMAB-MVK. Hum Mol Genet 19: 1921–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge B, Pokholok DK, Kwan T, Grundberg E, Morcos L, Verlaan DJ, Le J, Koka V, Lam KC, Gagné V, et al. 2009. Global patterns of cis variation in human cells revealed by high-density allelic expression analysis. Nat Genet 41: 1216–1222 [DOI] [PubMed] [Google Scholar]
- Gilad Y, Rifkin SA, Pritchard JK 2008. Revealing the architecture of gene regulation: The promise of eQTL studies. Trends Genet 24: 408–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göring HH, Curran JE, Johnson MP, Dyer TD, Charlesworth J, Cole SA, Jowett JB, Abraham LJ, Rainwater DL, Comuzzie AG, et al. 2007. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet 39: 1208–1216 [DOI] [PubMed] [Google Scholar]
- Graham RR, Kyogoku C, Sigurdsson S, Vlasova IA, Davies LR, Baechler EC, Plenge RM, Koeuth T, Ortmann WA, Hom G, et al. 2007. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc Natl Acad Sci 104: 6758–6763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith M, Griffith OL, Mwenifumbo J, Goya R, Morrissy AS, Morin RD, Corbett R, Tang MJ, Hou YC, Pugh TJ, et al. 2010. Alternative expression analysis by RNA sequencing. Nat Methods 7: 843–847 [DOI] [PubMed] [Google Scholar]
- Heinzen EL, Ge D, Cronin KD, Maia JM, Shianna KV, Gabriel WN, Welsh-Bohmer KA, Hulette CM, Denny TN, Goldstein DB 2008. Tissue-specific genetic control of splicing: implications for the study of complex traits. PLoS Biol 6: e1 doi: 10.1371/journal.pbio.1000001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan T, Benovoy D, Dias C, Gurd S, Serre D, Zuzan H, Clark TA, Schweitzer A, Staples MK, Wang H, et al. 2007. Heritability of alternative splicing in the human genome. Genome Res 17: 1210–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan T, Benovoy D, Dias C, Gurd S, Provencher C, Beaulieu P, Hudson TJ, Sladek R, Majewski J 2008. Genome-wide analysis of transcript isoform variation in humans. Nat Genet 40: 225–231 [DOI] [PubMed] [Google Scholar]
- Kwan T, Grundberg E, Koka V, Ge B, Lam KC, Dias C, Kindmark A, Mallmin H, Ljunggren O, Rivadeneira F, et al. 2009. Tissue effect on genetic control of transcript isoform variation. PLoS Genet 5: e1000608 doi: 10.1371/journal.pgen.1000608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup 2009. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JK, Lisco A, McDermott DH, Huynh L, Ward JM, Johnson B, Johnson H, Pape J, Foster GA, Krysztof D, et al. 2009. Genetic variation in OAS1 is a risk factor for initial infection with West Nile virus in man. PLoS Pathog 5: e1000321 doi: 10.1371/journal.ppat.1000321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin RJ, Yu HP, Chang BL, Tang WC, Liao CL, Lin YL 2009. Distinct antiviral roles for human 2′,5′-oligoadenylate synthetase family members against dengue virus infection. J Immunol 183: 8035–8043 [DOI] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y 2008. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18: 1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery SB, Sammeth M, Gutierrez-Arcelus M, Lach RP, Ingle C, Nisbett J, Guigo R, Dermitzakis ET 2010. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature 464: 773–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, Cheung VG 2004. Genetic analysis of genome-wide variation in human gene expression. Nature 430: 743–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Musunuru K, Strong A, Frank-Kamenetsky M, Lee NE, Ahfeldt T, Sachs KV, Li X, Li H, Kuperwasser N, Ruda VM, et al. 2010. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature 466: 714–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nica AC, Montgomery SB, Dimas AS, Stranger BE, Beazley C, Barroso I, Dermitzakis ET 2010. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet 6: e1000895 doi: 10.1371/journal.pgen.1000895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastinen T, Ge B, Hudson TJ 2006. Influence of human genome polymorphism on gene expression. Hum Mol Genet 15: R9–R16 [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE, Nkadori E, Veyrieras JB, Stephens M, Gilad Y, Pritchard JK 2010. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 464: 768–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinyol M, Bea S, Plà L, Ribrag V, Bosq J, Rosenwald A, Campo E, Jares P 2007. Inactivation of RB1 in mantle-cell lymphoma detected by nonsense-mediated mRNA decay pathway inhibition and microarray analysis. Blood 109: 5422–5429 [DOI] [PubMed] [Google Scholar]
- Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. 2010. The UCSC Genome Browser database: update 2010. Nucleic Acids Res 38: D613–D619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB 2008. Proliferating cells express mRNAs with shortened 3' untranslated regions and fewer microRNA target sites. Science 320: 1643–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Turner S, Kasarskis A 2010. A window into third generation sequencing. Hum Mol Genet 19: R227–R240 [DOI] [PubMed] [Google Scholar]
- Stranger BE, Forrest MS, Clark AG, Minichiello MJ, Deutsch S, Lyle R, Hunt S, Kahl B, Antonarakis SE, Tavaré S, et al. 2005. Genome-wide associations of gene expression variation in humans. PLoS Genet 1: e78 doi: 10.1371/journal.pgen.0010078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyrieras JB, Kudaravalli S, Kim SY, Dermitzakis ET, Gilad Y, Stephens M, Pritchard JK 2008. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet 4: e1000214 doi: 10.1371/journal.pgen.1000214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Burge CB 2004. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 11: 377–394 [DOI] [PubMed] [Google Scholar]