Abstract
In mature human sperm, genes of importance for embryo development (i.e., transcription factors) lack DNA methylation and bear nucleosomes with distinctive histone modifications, suggesting the specialized packaging of these developmental genes in the germline. Here, we explored the tractable zebrafish model and found conceptual conservation as well as several new features. Biochemical and mass spectrometric approaches reveal the zebrafish sperm genome packaged in nucleosomes and histone variants (and not protamine), and we find linker histones high and H4K16ac absent, key factors that may contribute to genome condensation. We examined several activating (H3K4me2/3, H3K14ac, H2AFV) and repressing (H3K27me3, H3K36me3, H3K9me3, hypoacetylation) modifications/compositions genome-wide and find developmental genes packaged in large blocks of chromatin with coincident activating and repressing marks and DNA hypomethylation, revealing complex “multivalent” chromatin. Notably, genes that acquire DNA methylation in the soma (muscle) are enriched in transcription factors for alternative cell fates. Remarkably, whereas H3K36me3 is located in the 3′ coding region of heavily transcribed genes in somatic cells, H3K36me3 is present in the promoters of “silent” developmental regulators in sperm, suggesting different rules for H3K36me3 in the germline and soma. We also reveal the chromatin patterns of transposons, rDNA, and tDNAs. Finally, high levels of H3K4me3 and H3K14ac in sperm are correlated with genes activated in embryos prior to the mid-blastula transition (MBT), whereas multivalent genes are correlated with activation at or after MBT. Taken together, gene sets with particular functions in the embryo are packaged by distinctive types of complex and often atypical chromatin in sperm.
A central question in early development is how totipotency and pluripotency are established in germline and embryonic stem (ES) cells, respectively. Studies in ES cells cultured in vitro have provided many interesting concepts for pluripotency, such as the use of special transcription factor combinations (POU5F1 [also known as OCT4], SOX2, others) that operate as a network to promote pluripotency and self renewal, and the presence of specialized chromatin at developmental regulators in ES cells to ensure their silencing and poising until later in development (Rao and Orkin 2006; Wang et al. 2006; Jaenisch and Young 2008; Kim et al. 2008). Due to technical limitations, little is known about the mechanisms underlying the totipotency of vertebrate eggs (Seydoux and Braun 2006), but multiple contributing mechanisms can be envisioned, including maternal RNAs (coding and noncoding) that promote totipotency, the loading and function of key transcription factor proteins (including pluripotency/self-renewal factors), and chromatin structures that enable (or prevent) the expression of particular embryonic developmental regulators.
Until recently, options (beyond DNA methylation) for the sperm genome to provide epigenetic contribution to totipotency appeared limited (Seydoux and Braun 2006), as spermatogenesis involves the replacement of the vast majority of the histone-based chromatin by protamine (Ward and Coffey 1991; Wykes and Krawetz 2003), which is not known to propagate information via modifications. One view is that the male genome need not carry any gene packaging information (beyond methylated imprinted genes), as it can simply be “reprogrammed” and repackaged by the egg following fertilization to achieve totipotency. One clear event in mice and humans involves active DNA demethylation of the paternal genome upon fertilization, prior to the fusion of the two nuclei and the onset of replication (Mayer et al. 2000; Oswald et al. 2000; Hajkova et al. 2008, 2010; Abdalla et al. 2009; Okada et al. 2010). Although this clearly occurs, we currently lack an understanding of which genes in the genome are susceptible or resistant to DNA demethylation, beyond the resistant imprinted genes. Indeed, a full understanding of the methylation state of the gametes before and after fertilization will be an important step forward.
Another view is that specialized gene packaging may indeed occur in sperm. Recently, a small percentage of the human and mouse genomes have been shown to remain enriched with modified nucleosomes in mature sperm (Arpanahi et al. 2009; Hammoud et al. 2009; Brykczynska et al. 2010). These studies revealed the packaging of the genes encoding most key embryonic developmental and morphogenesis regulators by nucleosomes with histone modifications that correlate both with activation and silencing. The observed silencing mark was trimethylation of lysine 27 on histone H3 (H3K27me3), a modification catalyzed by the PRC2 (Polycomb Repressive Complex 2) (Cao et al. 2002; Czermin et al. 2002; Kuzmichev et al. 2002; Muller et al. 2002). Resident activating marks included di- and trimethylation of lysine 4 on histone H3 (H3K4me2/3), a modification catalyzed in the soma by MLL, Set1, and Set7/9 histone methyltransferases (Byrd and Shearn 2003; Dou et al. 2005; Wysocka et al. 2005). The coincidence of these two marks is termed bivalency and has been described previously at the promoters of developmental regulators in ES cells (Bernstein et al. 2006). These antagonistic marks are believed to help poise these genes in a repressed state. Furthermore, developmental regulators in human sperm are profoundly DNA demethylated (Down et al. 2008; Farthing et al. 2008; Hammoud et al. 2009), a result that makes the genome-wide demethylation of the paternal genome all the more curious, if needed (in part) for totipotency. Finally, genes that are involved in spermatogenesis and metabolic processes are solely coated with the active mark H3K4me3 in both human and mouse sperm (Hammoud et al. 2009; Brykczynska et al. 2010). Thus, human and mouse sperm display two very distinct chromatin patterns for two different classes of genes: active genes (in sperm) and embryonic transcription factors.
These studies prompt several key questions. First, why place embryonic developmental regulators in bivalent chromatin in mature sperm cells? One possibility is to poise these genes for expression in the embryo. Another possibility is to protect them from DNA methylation through the use of “activating” chromatin, while simultaneously placing repressive chromatin on them to prevent their expression in the germline (Bernstein et al. 2006; Okitsu and Hsieh 2007; Ooi et al. 2007; Ciccone et al. 2009). Another key question is the complexity of the packaging and marking of developmental genes. Also, one might expect this packaging/marking system, if indeed instructive, to require additional modifications or histone variants to maintain robustness in the embryo. Furthermore, is this system for gene packaging in sperm identical to the system already established in ES cells, or are there unique aspects to packaging developmental regulators in the germline? Finally, it is of high interest to understand how this packaging is established and maintained in the germline and whether a similar system exists in the egg.
To begin to investigate these questions, we turned to the zebrafish animal model, due to the ease of obtaining both sperm and eggs and to the possibilities for future genetic manipulation. One apparent difference between humans and zebrafish is the use (revealed below) of nucleosomes rather than protamine to package the sperm genome. In spite of this difference, our studies reveal remarkable conservation between humans and zebrafish in the use of distinctive chromatin states to package embryonic developmental regulators in sperm, including the use of coincident activating and silencing modifications, and DNA demethylation. In addition, we greatly extended the analysis of chromatin composition to several other histone modifications and a histone variant, showing contiguous blocks of “multivalent” programmatic chromatin built over the promoters (or full genes) for developmental regulators, suggesting a complex and robust packaging network. Furthermore, we find features in this germline chromatin not observed in ES cells or in the soma, including the presence of H3K36me3 in these large blocks at developmental regulators.
Results
Biochemical composition of the zebrafish sperm genome
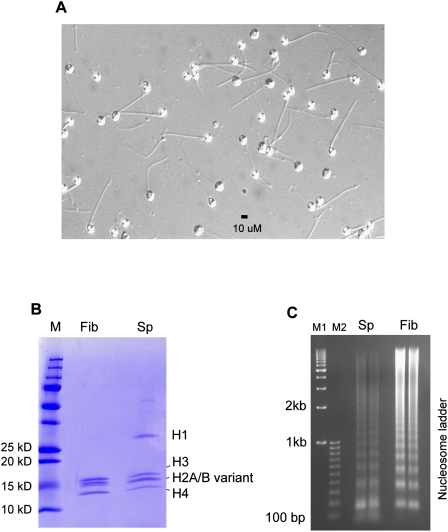
We began by characterizing the proteins that package the genome of mature zebrafish sperm. Sperm cells were collected from sexually mature males, which exhibited a sperm head morphology more rounded than those derived from mammals, suggesting an alternative compaction strategy (Fig. 1A). Chromatin preparations from sperm and a zebrafish fibroblast cell line were analyzed by SDS-PAGE, revealing prominent bands migrating at the exact size and distribution of histones. Mass spectrometric analysis revealed the presence of the canonical histones H2A, H2B, H3, and H4, as well as certain canonical histone variants (H2AFX, and H2AFV), and linker H1 histones and variants (related to human HISTH1C, -D, and -E; and H1FX; H1F0) (Fig. 1B; Supplemental Table 1), known to package the somatic genome. Here, we note that Western analysis reveals the clear presence of H1 linker histone in ZF4 fibroblasts, but at levels that are about 25% of those in sperm, normalized for ploidy (Supplemental Fig. 1A,B). Somewhat surprisingly, protamine, transition proteins, and testis-specific histone variants—proteins that constitute the vast majority of proteins in mammalian sperm—were not identified in zebrafish sperm, either through acid-urea gel analysis (data not shown) or mass spectrometry. Furthermore, the current zebrafish genome (Zv8) lacks the structural genes encoding orthologs of mammalian testis-specific histones, transition proteins, or protamine; though we note the presence of a gene distantly-related to protamine (zgc:114104, 492 amino acids), of unknown function and not detected in our sperm chromatin preparations. Consistent with the lack of protamine, the round head morphology of mature zebrafish sperm is similar to the “round spermatid” stage of human sperm development, the stage preceding protamine replacement of histones. Taken together, the mature zebrafish sperm genome is packaged by histones but contains a higher relative level of linker histone than does a somatic cell (Fig. 1B).
Figure 1.
Sperm morphology and biochemical composition of zebrafish sperm chromatin. (A) Zebrafish sperm have a round head morphology (∼10-μM diameter). (B) Whole-cell extracts were prepared from sperm and fibroblasts (ZF4) by lysing cells in 2× SDS-PAGE sample buffer. SDS-PAGE analysis of the cell extracts reveals a shared canonical histone pattern, with sperm displaying a higher level of linker histones. Histone components in sperm were verified by mass spectrometry (Supplemental Table 1). (C) Internucleosomal distances are longer in zebrafish sperm. Nucleosomes in sperm and fibroblasts (ZF4) were released by MNase digestion, and the DNA was analyzed by agarose gel electrophoresis and staining with EtBr (duplicate loading for each sample). Sperm displayed a longer average internucleosomal distance, 50 bp, than did the fibroblast, 40 bp. M indicates protein marker; Sp, sperm; Fib, fibroblast; and M1 and M2, DNA markers start from 1 kb with 1-kb increments and from 100 bp in 100-bp increments, respectively.
Sperm genome packaging and condensation
As zebrafish sperm lack protamine, one interesting question is how the genome is condensed to a moderately-high level in zebrafish sperm. As previewed above, sperm chromatin has abundant linker histone H1 variants, appearing nearly stoichiometric with histone octamers (Fig. 1B). Linker histones promote higher-order chromatin compaction, bind nucleosomes in a 1:1 ratio to form “chromatosomes,” and increase the average nucleosome repeat length (Huynh et al. 2005; Robinson et al. 2006; Zhou et al. 2007). To determine repeat length, nucleosomes from sperm or fibroblast cells were released by limited micrococcal nuclease (MNase) digestion (Fig. 1C), revealing a longer average internucleosomal distance in sperm (50 bp) than fibroblasts (40 bp), consistent with global H1 packaging in sperm (Godde and Ura 2009). Our interpretation is that zebrafish sperm, in comparison to fibroblasts, have a higher proportion of their genome in chromatosomes as opposed to nucleosomes.
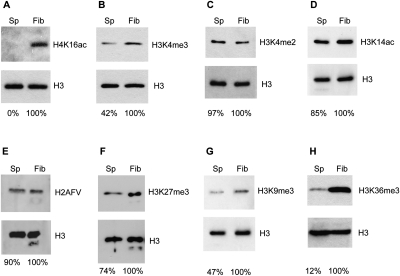
A second mode of promoting compaction was suggested through our analysis of histone modifications. The histone modification most closely associated with regulating compaction is the acetylation of lysine 16 on histone H4 (H4K16ac), which alone deters the transition of 10-nm nucleosome arrays to more compact 30-nm fibers (Shogren-Knaak et al. 2006; Zhou et al. 2007; Robinson et al. 2008; Kan et al. 2009). Furthermore, H4K16ac uniquely antagonizes the chromatin remodeling ATPases (ISWI- and CHD-family) responsible for organizing nucleosomes in ordered arrays of consistent spacing (Clapier et al. 2001; Corona et al. 2002). Remarkably, immunoblot analyses demonstrate that H4K16ac is virtually absent in zebrafish sperm chromatin (Fig. 2A; Supplemental Fig. 1C), a result of distinction given the relative abundance of other histone modifications in zebrafish sperm (detailed below). We note that H4R3me2 is robustly detected (Supplemental Fig. 1D), ensuring that the H4 tail is present and intact in zebrafish sperm chromatin. Furthermore, we detect in the zebrafish male gonad (data not shown) robust expression of the two zebrafish orthologs of the human ISWI family members SMARCA1 and SMARCA5. These results raise the possibility that zebrafish sperm promotes genome condensation by utilizing linker histones and the lack of H4K16ac, which coordinate with assembly remodelers to promote compaction in somatic cells.
Figure 2.
Bulk levels of histone modifications and histone variants in sperm. Immunoblotting was applied to quantify the level of modifications in sperm by comparing to ZF4 fibroblast cells and normalizing with H3 levels. (A) H4K16ac is not detected in sperm. (B–H) The levels of active and repressive marks and a histone variant (H2AFV/H2A.Z) in sperm compared with ZF4 fibroblast cells.
Active and repressive histone modifications in zebrafish sperm chromatin
Histone modifications in sperm chromatin were assessed by comparison to levels in fibroblasts by immunoblotting (normalized to histone H3 levels) (Supplemental Fig. 1B). We find the common “repressive/silencing” modifications (those correlated with gene silencing) such as H3K9me3 and H3K27me3 present and at levels comparable to or slightly lower than that of fibroblasts (Fig. 2F,G). We note that mature sperm of humans and mice are transcriptionally inactive and lack RNA polymerase (Pol) II (Miteva et al. 1995; Naz 1998; Grootegoed et al. 2000; data not shown). In keeping, we find RNA Pol II barely detectable in zebrafish sperm (<0.5% the level in fibroblasts). Nevertheless, sperm chromatin retains “active/positive” histone modifications (those correlated with gene activity/competency) at levels within twofold of fibroblasts, including H3K4me3, H3K4me2, and H3K14ac (Fig. 2B–D)—though as discussed above, H4K16ac is absent. Furthermore, levels of the histone variant H2AFV (ortholog of H2A.Z in mammals), which has been linked with gene poising and chromatin boundaries (Meneghini et al. 2003; Zhang et al. 2005), are comparable in sperm and fibroblasts (Fig. 2E). Thus, sperm chromatin resembles somatic chromatin in bulk levels of the many active and repressive histone modifications tested, as well as a key histone variant.
Approaches for localizing histone modifications and DNA methylation
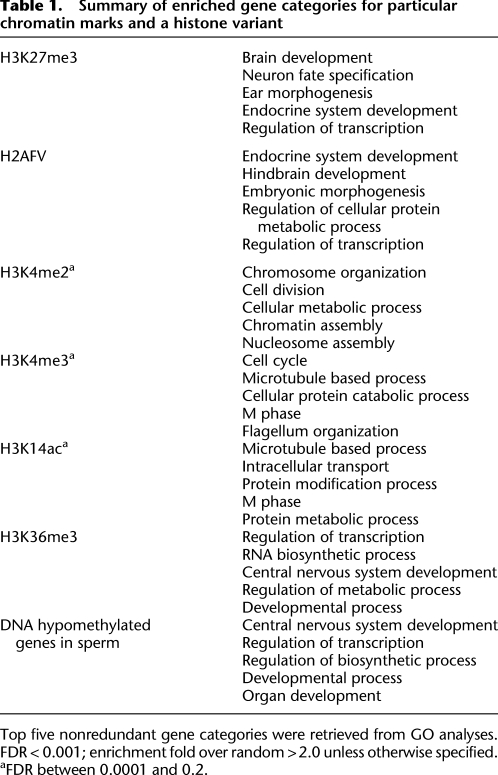
Chromatin immunoprecipitation (ChIP) and methylated DNA immunoprecipitation (MeDIP) were performed in mature sperm to localize histone modifications and DNA methylation, respectively. From native sperm chromatin, nucleosomes were released by MNase digestion to mainly di- and mononucleosomes and utilized for ChIP, involving multiple modification-specific antibodies (Supplemental Fig. 2). For MeDIP, sperm DNA was purified and sheared into fragments (∼400 bp) and then immunoprecipitated with anti-5-methylcytosine antibodies (Weber et al. 2007). Following amplification and labeling (see Methods), the ChIP eluate and input were then hybridized to customized zebrafish promoter arrays that tiled (at 250-bp resolution) 12 kb of the promoter region of about 13,000 genes, as well as many other chromosomal elements and loci (miRNAs, repetitive sequences, tRNAs, etc.) to provide a genome-wide perspective of chromatin packaging. Enriched loci were determined and ranked, and enriched gene promoters were subjected to Gene Ontology (GO) term analyses, summarized in Table 1 (for full lists, see Supplemental Materials; for GO analyses, see Methods).
Table 1.
Summary of enriched gene categories for particular chromatin marks and a histone variant
Top five nonredundant gene categories were retrieved from GO analyses. FDR < 0.001; enrichment fold over random >2.0 unless otherwise specified.
aFDR between 0.0001 and 0.2.
Multivalent and regional chromatin packages developmental loci
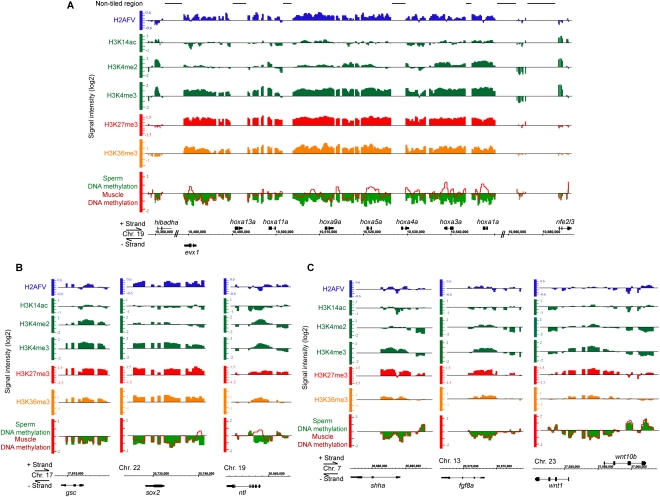
One clear and striking feature of our chromatin maps is regional blocks of coincident “multivalent” chromatin at developmental transcription factors utilized in embryonic development. First, blocks of H3K27me3 are strikingly enriched at developmental loci, a feature most clearly evident at hox loci (but not at flanking genes) and also evident at stand-alone transcription factors, with varying degrees of spreading (Fig. 3A–C; Table 1; Supplemental Table 2). Remarkably, of the 250 loci most enriched with H3K27me3, 90% are the promoters of embryonic transcription factors, including the vast majority of homeobox, T-box, GATA, and ETS family members (Fig. 3A,B; Supplemental Table 3). In addition, the promoters of many embryonic signaling molecules also displayed H3K27me3 enrichment, including shha, fgf8a, wnt1, and wnt10b genes (Fig. 3C). Indeed, GO analysis of the top 800 enriched loci generates almost exclusively developmental gene classes with high significance (Supplemental Table 2).
Figure 3.
Developmental loci contain multivalent and regional chromatin features. ChIP-chip and MeDIP-chip profiles of developmental loci, with fold enrichment over input in log2 scale (y-axis). (A) hoxa cluster. Lines above the tracks indicate nontiled regions on the array. (B) Stand-alone transcription factors: gsc, sox2, and ntl. (C) Signaling factors: shha, fgf8a, wnt1, and wnt10b. Distance between minor scales is 500 bp. Chr indicates chromosome.
Interestingly, we observe general coincidence of H3K27me3 with the histone variant H2AFV (the zebrafish ortholog of mammalian H2A.Z) at developmental loci (Fig. 3A–C; top 250 genes, Supplemental Table 4); intersecting of the respective top 800 genes displays a 40% overlap (correlation P-value < 0.001). GO term analysis showed H2AFV is highly enriched at the promoters of genes involved in transcription and developmental processes (Table 1; Supplemental Table 5), in keeping with previous observations in stem cells (Creyghton et al. 2008).
Notably, H3K4me2 and H3K4me3 are moderately enriched at developmental loci bearing H3K27me3 (Supplemental Table 3). H3K4me2/3 levels at developmental loci are higher than at average promoters (2.3- or 2.0-fold higher for H3K4me3 and H3K4me2, respectively) and coincident with H3K27me3 (Fig. 3A–C). This aligns with the observations in human sperm and ES cells of the presence of “bivalent domains”—coincident H3K4me3 and H3K27me3—on the orthologous developmental regulators (Bernstein et al. 2006; Mikkelsen et al. 2007). To address whether H3K27me3 and H3K4me3 are present on the same nucleosome and at high density, we performed a sequential ChIP experiment using mononucleosomes from sperm. However, this procedure failed to provide enrichment (data not shown), suggesting that most nucleosomes do not contain both marks, a result supported by experiments described later suggesting that H3K4me3 is only at moderate density at developmental genes, while H3K27me3 is at high density. Curiously, certain gene clusters are enriched in H3K4me3 while also deficient in H3K4me2 (i.e., hoxb and hoxd clusters, but not hoxa) (Fig. 3A; Supplemental Fig. 3A,B), suggesting precise and regional H3K4me states at certain loci. Finally, developmental loci lack significant H3K14ac enrichment, a mark correlated with transcription in somatic cells (Supplemental Table 3), and also generally lack significant enrichment of the repressive mark H3K9me3 (data not shown), a mark present at other types of repressed loci, described later.
Taken together, the vast majority of developmental transcription factors, as well as considerable number of key signaling proteins, is bound by coincident blocks of H3K27me3, H3K4me2/3, and H2AFV but lacks significant H3K14ac enrichment. These blocks can extend for tens of kilobases at clustered gene loci, whereas at stand-alone transcription factors genes, the blocks are typically 2–5 kb (with variation), extending from the TSS into the proximal promoter and sometimes encompassing the entire gene.
Pronounced DNA hypomethylation at developmental loci
Although the bulk zebrafish sperm genome is hypermethylated (Mhanni and McGowan 2004), GO analysis reveals three categories of DNA hypomethylated promoters (false discovery rate [FDR] < 0.1%): developmental regulators, transcription factors, and metabolism/biosynthesis (Table 1; Fig. 3A–C; Supplemental Table 6). Indeed, the top 250 hypomethylated genes are dominated by embryonic transcription factors (Supplemental Table 7). One example is the hoxa locus, where hypomethylation covers almost the entire locus (Fig. 3A). This observation extends beyond the clustered developmental transcription factors to many stand-alone developmental regulators (Fig. 3B). DNA hypomethylation at developmental loci in sperm was further confirmed by bisulfite sequencing (Supplemental Fig. 4), which revealed a nearly quantitative absence of methylation. To identify loci that acquire methylation in development, we compared DNA methylation profiles in sperm to a differentiated cell type, adult zebrafish muscle. These examinations revealed extensive methylation of developmental transcription factor promoters not expressed in muscle cells (almost all hox genes), but the lack of methylation at the mef2 and hoxa13 promoters, which are expressed in muscle (Fig. 3; data not shown). Indeed, 35% of the top 250 genes that acquire DNA methylation in their proximal promoter region in muscle cells are developmental regulators such as transcription factors (Supplemental Table 8). In contrast, along the majority of the genome, DNA methylation profiles are virtually superimposable in sperm and muscle (Supplemental Fig. 3C). The acquisition of methylation at developmental promoters in muscle was further confirmed by bisulfite sequencing (Supplemental Fig. 4). Together, these results suggest that development in zebrafish is accompanied by the methylation of developmental loci for alternative transcription programs.
We find multivalent chromatin and DNA hypomethylation residing at the promoters of the vast majority of developmental transcription factors and a large number of signaling factors in zebrafish sperm. Exceptions to this include the promoters for the transcription factors atoh7, atoh2a, pou1f1, foxn1, tal2, and gata1a, among others (data not shown). Notably, these genes are not generally expressed during early patterning or organ specification but rather are expressed during or after segmentation—with many of these types of factors being expressed during the later stages of eye, thymus, blood, or neuronal development. Thus, the multivalent chromatin may be more important for helping to regulate gene expression in early embryos than during terminal differentiation. Furthermore, the promoters of a subset of signaling factors such as wnt5b, fgf10, egf, and bmp7 are also not marked with H3K27me3 (or H3K4me2/3) and are DNA methylated (data not shown). However, we have not identified a clear biological or temporal attribute of this group of signaling factors that might underlie their alternative marking, but these loci are of interest to examine for DNA demethylation in embryos.
Loci bearing high levels of multiple active marks in zebrafish sperm
We find the general coincidence of the three active marks tested (H3K4me3, H3K4me2, and H3K14ac), as pairwise intersection analyses of their respective top 250 enriched genes show significant overlap (∼25%–35%; P < 0.001) (Supplemental Tables 9–11). GO analysis reveals H3K4me3 and H3K14ac significantly (FDR <10%) enriched at the promoters of housekeeping genes and genes for sperm-specific processes, including flagella/microtubules, catabolic processes, regulation of M-phase, DNA repair, and nucleotide metabolism (Table 1; Supplemental Tables 12, 13). Intuitively, their presence at genes for sperm-related functions likely represents retention of chromatin modifications from spermatogenesis. Thus, to provide proper context, developmental loci show only low-moderate enrichment with H3K4me2/3 (2.0- to 2.5-fold) in comparison to the gene categories for sperm-specific processes (H3K4me2/3, 4.0- to 5.0-fold; H3K14ac, 2.3-fold). Finally, we observed one large and curious region on chromosome 19 with a continuous block of very high H3K14ac extending through genes and intergenics, including the genes daxx and sdha and certain protease subunits (data not shown).
Atypical placement of H3K36me3 in zebrafish sperm
Methylation on H3K36 is catalyzed by Set2-family methyltransferases, and deposition is generally coupled with RNA Pol II elongation (Krogan et al. 2003; Carrozza et al. 2005; Joshi and Struhl 2005; Keogh et al. 2005), leading to H3K36me3 placement along gene bodies. In yeast, H3K36me3 recruits the Rpd3S complex bearing histone deacetylase (HDAC) activity to prevent spurious transcription in the coding region.
Remarkably, we do not observe H3K36me3 on the coding regions of genes known to be expressed during spermatogenesis and genes displaying robust levels of H3K4me3 and H3K14ac at their TSS, known marks of transcriptional activity (Fig. 4A). Rather, we find H3K36me3 at two types of loci. The type with the highest levels of H3K36me3 is found at certain repetitive genes (U6 genes) and elements (Fig. 4B). The second enriched type has clear H3K36me3 in regional blocks and is coincident with developmental loci (Fig. 3A-C). This is most strikingly observed at the clustered hox gene loci and also at divergent developmental genes, which display clear enrichment of H3K36me3 in the intergenic, such as the divergent wnt1-wnt10 genes (Fig. 3C). We found 137 unique genes overlapping or adjacent to high levels of H3K36me3 (more than 2.7-fold enriched) (Supplemental Table 14), and of these, over 37% are categorized into transcription and developmental processes (FDR < 0.001) (Supplemental Table 15). Intersection analyses show that H3K36me3 in gene promoters is strongly correlated with H3K27me3, H2AFV, or H3K4me3 (each correlation P < 0.001) (Supplemental Fig. 5), but not with H3K14ac—properties evident at developmental genes (Fig. 3). Here, sequential ChIP was performed to test H3K36me3 and H3K27me3 coincidence, and co-enrichment was observed at the promoter region of several developmental regulators tested (Supplemental Fig. 6). These correlation data and localization of H3K36me3 indicate a potential repressive role for H3K36me3 in zebrafish sperm. Importantly, genes with H3K36me3 lack significant H3K14ac in zebrafish sperm, consistent with observations in somatic cells (Figs. 3A–C, 5C; Carrozza et al. 2005; Bell et al. 2007). This raises the possibility that in spite of its atypical placement, H3K36me3 in the germline might still recruit HDACs to help silence loci. Notably, the atypical placement of H3K36me3 is not restricted to clustered genes, as stand-alone developmental transcription and signaling factors contain blocks of H3K36me3 in their promoter region, coincident with H3K27me3, H3K4me2/3, and H2AFV (Fig. 3B,C).
Figure 4.
Chromatin features of genes expressed during spermatogenesis and of repetitive regions reveal atypical use of H3K36me3. (A) Genes involved in spermatogenesis show strong enrichment of active marks, but not H3K36me3 or other repressive marks. (B) U1 and U6 repetitive genes are enriched with H3K36me3, DNA methylation, and the repressive mark, H3K9me3.
The presence of H3K36me3 enrichment at intergenics and gene promoters/5′ ends has not been reported previously in vertebrates. This anomaly is not simply species specificity, as zebrafish late embryos display typical H3K36me3 profiles involving enrichment at 3′ ends of transcribed genes (Vastenhouw et al. 2010), a result confirmed here (Supplemental Fig. 7A). One question is which methyltransferase is responsible for the placement of atypical H3K36me3. Previous work in mice and humans has revealed additional H3K36 methyltransferases, including nsd1 and nsd2 (Wang et al. 2007). We find the orthologs nsd1a, nsd1b, and nsd2 expressed in zebrafish testis (data not shown), revealing candidates for future investigation.
As H3K36me3 distribution was atypical, we therefore examined H3K36me2 at selected genes. We examined two genes expressed in spermatogenesis (bactin2, ddx5) by ChIP-qPCR and found H3K36me2 higher in their coding regions than their 5′ ends (Supplemental Fig. 7B). Furthermore, the levels of H3K36me2 in coding regions of expressed genes were much higher than were levels detected at two developmental genes (gsc, klf4) that are not expressed in spermatogenesis (Supplemental Fig. 7B). Therefore, the distribution of H3K36me2 in sperm appears similar in concept to somatic cells, whereas the distribution of H3K36me3 is atypical; though for H3K36me2, we have only examined a few genes and have not extended this examination genome-wide.
Chromatin features of repeat regions and noncoding RNAs
Our arrays included various regions of the genome, enabling an analysis of chromatin profiles in repetitive regions, 5S/5.8S rDNA repeats, tRNAs, and tRNA clusters. We emphasize that the majority of the regions discussed in this section are nonunique and that chromatin maps derived from array formats represent a class average of the element described. First, we find that type 1 transposons (LINE) are typically DNA methylated and deficient in the activating marks tested but are (somewhat surprisingly) not enriched in any of the repressive marks tested (including H3K9me3), nor are they deficient in H3K14ac (Supplemental Fig. 8A). Type 2 transposons (SINE) displayed a similar pattern, though with only moderate levels of DNA methylation (Supplemental Fig. 8B). We note that our examination of data sets from human T cells (Barski et al. 2007) likewise does not generally reveal H3K9me3 at LINE or SINE elements (data not shown). In counter-distinction, we find the repeated genes encoding the noncoding RNAs U1 and U6 highly DNA-methylated and associated with particular silencing marks (high H3K36me3 and moderate H3K9me3), while highly deficient in activating marks (H3K4me2/3, H3K14ac, or H2AFV) and lacking H3K27me3 (Fig. 4B). A similar pattern is noted for the zebrafish 5S and 5.8S rRNA repeats, although 5.8S rDNA repeats display much higher levels of H3K9me3 than do the 5S repeats and lack H2AFV deficiency (Supplemental Fig. 8C,D). In contrast, our examination of data sets from human T cells (Barski et al. 2007) reveals a general lack of H3K9me3 at the U1/U6 and 5S/5.8S loci, suggesting some differences in the packaging of repetitive loci in zebrafish and humans. The tRNA clusters all showed profiles similar to that of the U1/U6 repeats, while most stand-alone tRNAs were found in regions of active chromatin—bearing moderate to high levels of H3K14ac and H3K4me2/3 while lacking H3K27me3, H3K9me3, or H3K36me3 (Supplemental Fig. 8E,F). This partitioning of tRNAs into very different chromatin environments (active or repressive) is reminiscent of recent studies in human cells, where those tRNAs in active chromatin environments were occupied by the RNA Pol III machinery and those unoccupied by Pol III were in repressive chromatin (Oler et al. 2010).
Transcripts retained in mature sperm and their chromatin profiles
Mature sperm of vertebrates are transcriptionally inert (Miteva et al. 1995; Naz 1998; Grootegoed et al. 2000). Consistent with this observation, RNA Pol II protein levels in mature sperm are barely detectable (<0.5% compared with fibroblasts) (data not shown). In spite of this, we reasoned that an examination of transcript profiles of mature sperm might reflect, in part, the recent transcriptional program of spermatocytes and spermatogenesis and might also correlate with regions of active chromatin retained following the cessation of transcription. To this end, we performed expression profiling by microarray, and enriched RNAs (more than 5.8-fold over background) were retrieved, yielding 1731 genes, which revealed largely genes predicted to be highly expressed (encoding ribosomal proteins, actin, tubulin, etc.) (Supplemental Table 16). As expected, transcripts in mature sperm were well correlated with active chromatin marks (H3K4me2/3 or H3K14ac) and DNA hypomethylation (all P < 0.01), but not with the repressive mark H3K27me3 or the poising mark H2AFV (Fig. 4A; Supplemental Table 17).
Sperm chromatin features and timing of gene expression in embryos
Previous work suggests that a small number of loci might be transcribed before or during an early phase of the mid-blastula transition (MBT), the time when general zygotic transcription initiates in the embryo (Mathavan et al. 2005). These genes include factors that promote the cell cycle and those that assist with RNA translation and metabolism, whose increased levels might help prepare cells for handling the RNAs generated during zygotic transcription. One possibility is that genes that are transcribed prior to (or early in) the MBT in zebrafish (3 h post-fertilization [hpf]) might have been marked in the sperm with particular chromatin modifications or histone variants that might help facilitate their early activation in the embryo. Although certain marks/proteins are likely lost at fertilization (see Discussion), some might be retained and instructive. Here, we reasoned that if marking in the sperm does occur, then genes that contain high levels of active marks in mature sperm might be correlated with the genes activated prior to general zygotic transcription. We find that genes bearing active histone marks (H3K4me2/3 or H3K14ac) in sperm are indeed correlated with an early zygotic expression profile (genes that others defined as initiating their expression at 2 hpf) (Fig. 5A; Mathavan et al. 2005). We note that genes that drive the cell cycle and promote metabolism in very early embryos bear very high levels of active histone marks in sperm (Table 1; Fig. 5B). In contrast, genes in sperm bearing high levels of the repressive mark H3K27me3 and also the histone variant H2AFV are better correlated with a post-MBT expression profile, when developmental regulators begin to be expressed (Fig. 5A; exemplified in Fig. 3A–C). Genes that lack DNA methylation in sperm are correlated with embryonic transcription, both pre- and post-MBT, but better with post-MBT. In conclusion, although a causal relationship has not been established, we do observe correlations between chromatin modification patterns in sperm and gene expression timing in embryos.
Figure 5.
Certain chromatin modification patterns in sperm correlate with the timing of embryonic gene expression. (A) Heat map showing the correlations between genes that initiate expression at particular times during early development (2-hpf, 4-hpf, and 6-hpf columns from Mathavan et al. 2005) and particular histone modifications (or H2AFV enrichment) present at those genes in sperm (rows). Here, the color gradient shows the level of fold enrichment (over random) of a pairwise intersection between the list of expressed genes (Supplemental Table 18), and the list of genes with the respective histone modification/composition. A lack of significant intersection is depicted with a gray box. Genes expressed early/before MBT tend to have the highest levels of active histone modifications (acetylation and H3K4me2/3) in sperm, and those expressed after MBT tend to have repressive modifications (H3K27me3) in sperm. (B) Genes expressed before/early in MBT (3 hpf in zebrafish) are those that drive the cell cycle and promote metabolism, and are enriched with high levels of active histone modifications.
Discussion
Zebrafish sperm utilize nucleosomes and linker histones for packaging
Our goals are to understand chromatin-transcription relationships and their contributions to totipotency in germ cells and early embryos, and here we explore the zebrafish model. We find that zebrafish packages its sperm genome in nucleosomes rather than protamine, which is widely utilized in mammals. Curiously, sperm genome packaging systems utilized in teleost fish vary widely; some utilize only histone, while others use protamine or protamine-like proteins (Shimizu et al. 2000). At present, it is not clear why certain fish utilize particular packaging strategies, as they cannot be partitioned on simple attributes such as salt versus fresh water. However, our analysis of histone modification states and packaging composition suggest an intuitive strategy for the compaction of the histone genome involving the coordinated use of nucleosomes lacking H4K16ac, various linker histones, and possibly ISWI-type remodelers—based on extensive work on this process in somatic cells. We note that the packaging of the sperm genome by nucleosomes prevents its effectiveness as a model for understanding the histone–protamine transition. However, as discussed below, zebrafish appear very similar to humans in their packaging of genes important for embryonic development, suggesting they may serve as an important model for developmental gene packaging and marking.
Features of gene packaging in germ cells and ES cells
Key issues in germ cells include understanding the mechanisms for totipotency and the relative contributions of the egg and sperm genomes to this process. Here, chromatin structure could play an important role, given its known roles in gene regulation and gene poising. Currently, little is known in vertebrate eggs regarding the chromatin structure that packages the key embryonic self-renewal and developmental transcription factors. However, previous work in mature human and mouse sperm has revealed that genes of importance for embryo development (i.e., transcription factors) lack DNA methylation and bear nucleosomes with distinctive histone modifications, suggesting the specialized packaging and/or poising of developmental genes in the germline (Weber et al. 2007; Down et al. 2008; Farthing et al. 2008; Arpanahi et al. 2009; Hammoud et al. 2009; Brykczynska et al. 2010). This packaging is similar in some respects to their packaging in ES cells, as they share H3K4me3/H3K27me3 “bivalency,” DNA hypomethylation near the TSS, and the presence of H2AFV (Bernstein et al. 2006; Creyghton et al. 2008; Fouse et al. 2008). These similarities strongly suggest some degree of overlap in the chromatin mechanisms that contribute to the pluripotency of ES cells and totipotency in germ cells. This also raises the interesting possibility that the chromatin signatures of totipotency arose initially in the germline—and were then largely maintained (or re-established) to achieve pluripotency in somatic cells.
Furthermore, our studies extend on those before to reveal considerable additional complexity in gene packaging in sperm, as well as features thus far unique to germ cells. First, our examination of multiple modifications argues for complex “multivalent” chromatin involving multiple positive and negative marks. Likewise, we speculate that additional histone modifications and packaging proteins might be present at developmental genes in ES cells and contribute to the robustness of their transcription states.
Also, whereas developmental genes in human or mouse ES cells display bivalency in a relatively narrow range near the TSS of genes (Bernstein et al. 2006; Mikkelsen et al. 2007; Hammoud et al. 2009; Brykczynska et al. 2010), hox loci in zebrafish sperm display a contiguous block of modifications that typically (though not uniformly) extend throughout the locus. For stand-alone transcription factors in sperm, blocks of multivalent chromatin are also present and typically extend over many kilobases. The size and typical uniformity of these chromatin blocks strongly suggests a program for their establishment in the germline. Although the mechanistic basis for this program (including the targeting, establishment, and maintenance) has not been determined, we speculate that the self-renewal factors may have roles in targeting chromatin, based on a precedent in ES cells for the targeting and establishment of bivalent chromatin (Boyer et al. 2005, 2006a,b; Orkin 2005; Welstead et al. 2008). Another outstanding question is how these regions are bounded, as the boundaries for this chromatin are often quite sharp, raising the possibility of a boundary factor such as CTCF. Further work in this area will involve an examination of the packaging of these genes in spermatogonial stem cells.
H3K36me3 at developmental genes and relationships to transcription
Remarkably, we find H3K36me3 located in developmental gene promoters in sperm and not present at the 3′ ends of coding regions of genes, even when examining genes heavily transcribed in spermatogenesis and spermiogenesis. Our results raise multiple possibilities, which are not mutually exclusive. One possibility is that the rules for H3K36me3 deposition are the same in the germline and soma—with H3K36me laid down during Pol II elongation (Krogan et al. 2003; Keogh et al. 2005)—but that histone demethylases remove the mark from highly transcribed genes before sperm cell maturation. Another possibility is that the Set2d histone H3K36 methyltransferase, or a Set2 paralog such as nsd1/2, does not methylate highly transcribed genes but instead methylates genes inside the chromatin domains of developmental genes (such as hox clusters) in a manner independent of transcription. Finally, one attractive but speculative model is that the promoters of developmental loci (and perhaps more extensive regions at clustered hox loci) are briefly and promiscuously transcribed in the germline, leading to blocks of overlapping H3K4me2/3, H3K27me3, and H3K36me3 patterns. By this model, the observed chromatin patterns are dependent on initial promiscuous transcription (generating H3K4me3 in initiation regions and H3K36me in elongation regions), followed by the silencing of the gene by the attraction of PRC2/H3K27 by the nascent RNAs produced (Zhao et al. 2008; Lee 2009; Tsai et al. 2010) and deacetylation through resident H3K36me3 and its recruitment of HDACs (Carrozza et al. 2005; Joshi and Struhl 2005; Keogh et al. 2005).
The purpose of zebrafish sperm chromatin
Loci of developmental importance in the embryo bear complex multivalent chromatin in sperm and resemble several ways the chromatin present at these loci during MBT, when developmental loci must be repressed but able to be activated (Hammoud et al. 2009; Lindeman et al. 2010; Vastenhouw et al. 2010). Indeed, work from several laboratories now suggests that totipotent/pluripotent states of the germline and embryos share at developmental promoters unmethylated and complex “multivalent” chromatin (Hammoud et al. 2009; Lindeman et al. 2010; Vastenhouw et al. 2010). A clear question is whether this complex multivalent chromatin built in the sperm is needed solely for the proper gene regulation in the germline, or whether this multivalent chromatin (or any portion) is maintained and instructive in the embryo. One clear role for complex multivalent chromatin in the germline is to repress the many fate-determining developmental embryonic transcription factors. However, this does not explain the presence at these genes of positive marks such as H3K4me2/3 and H2AFV, which one would expect to be absent if the sole purpose is repression. Alternatively, we speculate that the negative marks are indeed present to repress these genes in the germline but that the positive marks are present to prevent DNA methylation of these loci, as even modest DNA methylation might bias these genes for repression in the embryo. This notion is in keeping with previous results that both H3K4me3 and H2A.Z/H2AFV are strongly anti-correlated with DNA methylation (Okitsu and Hsieh 2007; Weber et al. 2007; Zilberman et al. 2008).
The next issue is whether the epigenetic marks other than DNA methylation (histone modifications and variants) are maintained in the embryo and are instructive. We observe correlations of chromatin marking in sperm with expression in the embryo, which might suggest the use of a chromatin memory to instruct expression timing. However, for two reasons, these data must be interpreted with caution. First, the genes with high H3K4me3 and H3K14ac in sperm that are transcribed prior to MBT are strongly transcribed in virtually every somatic cell type, so early expression might be a result of their exceptionally strong promoters, not prior marking in the sperm. Second, recent experiments in zebrafish have demonstrated that the bulk levels of H3K4me3 and H3K27me3 are greatly diminished between fertilization and MBT, and these reductions also occur at developmental genes (Vastenhouw et al. 2010). Furthermore, these marks are then visible again after MBT, thus re-establishing this signature of pluripotency after zygotic transcription. As they are diminished prior to MBT, these two marks cannot solely be instructive for regulating the timing of gene activation.
However, our description of multivalent chromatin raises the possibility that one or more of the alternative marks, histone variants, or possibly noncoding RNAs may still be maintained and utilized to affect gene expression, though this remains to be tested. However, we note that if a mark/protein does persist, it must have a mechanism for self-renewal as new histones deposited during each cell division must be newly marked. An alternative view proposed previously (Vastenhouw et al. 2010) is of a naïve chromatin genome prior to MBT—with developmental loci completely unmarked prior to MBT and with chromatin modifiers (like H3K4 MTases) targeted to these loci at or just prior to MBT. A blending of these mechanisms is also possible, with the use of transcription factors to gain access to the open chromatin soon after fertilization, which then help facilitate the reestablishment of those chromatin marks and RNA Pol II recruitment at MBT. Future experiments will help distinguish between these and other models.
Methods
Zebrafish stocks and cells
The wild-type zebrafish line (Tübingen) and ZF4 cells (zebrafish fibroblast cells) were maintained as described previously (Rai et al. 2007). Sperm cells were collected from sexually mature zebrafish males by standard procedures (Westerfield 2000).
Western blotting and antibodies
The sperm or fibroblasts were counted using a hemacytometer, washed with PBS, and then lysed in 2× sample buffer. Western blotting was done according to standard procedures. Antibodies were as follows: H3K4me3 (Abcam ab8580 and Active Motif 39159), H3K4me2 (Abcam ab7766), H3K14Ac (Upstate 07-353), H2AZ (Abcam ab4174), H3K27me3 (Upstate 07-449), H3K9me3 (Active Motif 39161), H3K36me3 (Abcam ab9050), H3K36me2 (Abcam ab9049), H4K16Ac (Abcam ab1762), H4tetraAc (Upstate 06-866), and H3 (Abcam ab1791).
MeDIP, ChIP, and sequential ChIP
The procedures of MeDIP and ChIP were described previously (Hammoud et al. 2009). Briefly, for MeDIP, 4 μg of sheared genomic DNA (∼400 bp) from mature sperm cells or dissected muscle tissues was incubated with Dynabeads (Invitrogen) conjugated with antibody against 5-methylcytidine (Eurogentec BI-MECY-1000). For ChIP, 107 of sperm cells was treated with 0.05% lysophosphatidylcholine (Sigma L1381) in PBS on ice for 10 min. Fifteen units of MNase (USB 70196Y) was then applied to release oligonucleosomes for 5 min at 37°C. Two micrograms of specific antibody conjugated to Dynabeads was incubated with the oligonucleosomes. The pulled-down fragments and input from MeDIP and ChIP were amplified by whole-genome amplification kit (Sigma WGA2), labeled with cy3 or cy5, and then hybridized to customized Agilent 244k-feature slides. Two biological replicas were used for each ChIP and labeled with dye swap.
For sequential ChIP, 2 × 107 of sperm cells was crosslinked and lysed in sperm cell lysis buffer (50 mM Tris at pH 8.0, 10 mM EDTA, 1 mM EGTA, 150 mM NaCl, 1% SDS, 3 mM DTT). The lysate was diluted with IP dilution buffer (16.7 mM Tris at pH 8.0, 167 mM NaCl, 1.2 mM EDTA, 1.1% Triton X-100, 0.01% SDS) and sheared by sonication to obtain fragments of mononucleosome size. Antibody was applied to first IP as above. After washing out the nonspecific binding, chromatin-associated beads were incubated in elution buffer (30 mM DTT, 0.1% SDS, 500 mM NaCl) for 15 min at 37°C. The eluate was diluted 30-fold with IP dilution buffer for the second IP and then treated as in standard IP procedures (Bernstein et al. 2006).
Customized zebrafish promoter microarray
The customized zebrafish promoter microarray, Z-array+, was adapted from the Whitehead zebrafish promoter chipsets (Wardle et al. 2006), by adding hundreds of additional genes and genetic elements. Z-array+ constitutes of a pair of 244,000 feature arrays with 60-mer oligos tiled an average 250 bp in the extended promoter region (−9 to +3 kb) of approximately 13,000 mRNA genes, miRNAs, tRNAs, rRNAs, repeat elements, CpG islands, and two centromeric heterochromatin regions.
RNA extraction and expression microarray
As in standard procedures (Westerfield 2000), mature sperm cells were screened under a microscope. RNA from 6 × 107 of mature sperm cells was extracted with a Qiagen RNA isolation kit. The zebrafish gene expression microarray (Agilent) includes 43,803 probes covering approximately 21,500 genes.
Data availability
The data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE26609. The processed data are available for programmatic access using the GenoPub DAS/2 data distribution server (description, http://bioserver.hci.utah.edu/BioInfo/index.php/Software:DAS2; GenoPub web app, http://bioserver.hci.utah.edu:8080/DAS2DB/genopub (type “guest” for both the user name and password); and the DAS/2 Data Access URL, http://bioserver.hci.utah.edu:8080/DAS2DB/genome). DAS/2 compliant genome browsers such as IGB (http://www.bioviz.org/igb/) can be used to view the data sets, found under Danio rerio→D_rerio_Jul_2007→Wu_2011.
Acknowledgments
We thank G. Bell (Whitehead Institute) for the design of a majority of the microarray probes, D. Nix for computational analysis and discussion, B. Dalley for microarray expertise, T. Parnell for technical advice, B. Demarest for imaging help, I. Jafri for initial validation of MeDIP, V. Khoddami for bisulfite sequencing, J. Hansen for H3K36me3 qPCR, and S. Hammoud for comments. Financial support was provided by NIH 5R01HD058506 and the Howard Hughes Medical Institute. We are grateful for grant CA24014 to the Huntsman Cancer Institute for support of core facilities. B.R.C. is an investigator with HHMI.
Footnotes
[Supplemental material is available for this article. The microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE26609.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.113167.110.
References
- Abdalla H, Yoshizawa Y, Hochi S 2009. Active demethylation of paternal genome in mammalian zygotes. J Reprod Dev 55: 356–360 [DOI] [PubMed] [Google Scholar]
- Arpanahi A, Brinkworth M, Iles D, Krawetz SA, Paradowska A, Platts AE, Saida M, Steger K, Tedder P, Miller D 2009. Endonuclease-sensitive regions of human spermatozoal chromatin are highly enriched in promoter and CTCF binding sequences. Genome Res 19: 1338–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Bell O, Wirbelauer C, Hild M, Scharf AN, Schwaiger M, MacAlpine DM, Zilbermann F, van Leeuwen F, Bell SP, Imhof A, et al. 2007. Localized H3K36 methylation states define histone H4K16 acetylation during transcriptional elongation in Drosophila. EMBO J 26: 4974–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Mathur D, Jaenisch R 2006a. Molecular control of pluripotency. Curr Opin Genet Dev 16: 455–462 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006b. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, Beisel C, Schubeler D, Stadler MB, Peters AH 2010. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol 17: 679–687 [DOI] [PubMed] [Google Scholar]
- Byrd KN, Shearn A 2003. ASH1, a Drosophila trithorax group protein, is required for methylation of lysine 4 residues on histone H3. Proc Natl Acad Sci 100: 11535–11540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592 [DOI] [PubMed] [Google Scholar]
- Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J, Xu G, Li E, Chen T 2009. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 461: 415–418 [DOI] [PubMed] [Google Scholar]
- Clapier CR, Langst G, Corona DF, Becker PB, Nightingale KP 2001. Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol Cell Biol 21: 875–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona DF, Clapier CR, Becker PB, Tamkun JW 2002. Modulation of ISWI function by site-specific histone acetylation. EMBO Rep 3: 242–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Markoulaki S, Levine SS, Hanna J, Lodato MA, Sha K, Young RA, Jaenisch R, Boyer LA 2008. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell 135: 649–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V 2002. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111: 185–196 [DOI] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG 2005. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell 121: 873–885 [DOI] [PubMed] [Google Scholar]
- Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, Graf S, Johnson N, Herrero J, Tomazou EM, et al. 2008. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol 26: 779–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farthing CR, Ficz G, Ng RK, Chan CF, Andrews S, Dean W, Hemberger M, Reik W 2008. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet 4: e1000116 doi: 10.1371/journal.pgen.1000116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, Van Neste L, Jaenisch R, Fan G 2008. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell 2: 160–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godde JS, Ura K 2009. Dynamic alterations of linker histone variants during development. Int J Dev Biol 53: 215–224 [DOI] [PubMed] [Google Scholar]
- Grootegoed JA, Siep M, Baarends WM 2000. Molecular and cellular mechanisms in spermatogenesis. Best Pract Res Clin Endocrinol Metab 14: 331–343 [DOI] [PubMed] [Google Scholar]
- Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, Lee C, Almouzni G, Schneider R, Surani MA 2008. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature 452: 877–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA 2010. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science 329: 78–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR 2009. Distinctive chromatin in human sperm packages genes for embryo development. Nature 460: 473–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh VA, Robinson PJ, Rhodes D 2005. A method for the in vitro reconstitution of a defined “30 nm” chromatin fibre containing stoichiometric amounts of the linker histone. J Mol Biol 345: 957–968 [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Young R 2008. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell 132: 567–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AA, Struhl K 2005. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell 20: 971–978 [DOI] [PubMed] [Google Scholar]
- Kan PY, Caterino TL, Hayes JJ 2009. The H4 tail domain participates in intra- and internucleosome interactions with protein and DNA during folding and oligomerization of nucleosome arrays. Mol Cell Biol 29: 538–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. 2005. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123: 593–605 [DOI] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH 2008. An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132: 1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, et al. 2003. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol 23: 4207–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D 2002. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 16: 2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT 2009. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev 23: 1831–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeman LC, Winata CL, Aanes H, Mathavan S, Alestrom P, Collas P 2010. Chromatin states of developmentally-regulated genes revealed by DNA and histone methylation patterns in zebrafish embryos. Int J Dev Biol 54: 803–813 [DOI] [PubMed] [Google Scholar]
- Mathavan S, Lee SG, Mak A, Miller LD, Murthy KR, Govindarajan KR, Tong Y, Wu YL, Lam SH, Yang H, et al. 2005. Transcriptome analysis of zebrafish embryogenesis using microarrays. PLoS Genet 1: 260–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T 2000. Demethylation of the zygotic paternal genome. Nature 403: 501–502 [DOI] [PubMed] [Google Scholar]
- Meneghini MD, Wu M, Madhani HD 2003. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112: 725–736 [DOI] [PubMed] [Google Scholar]
- Mhanni AA, McGowan RA 2004. Global changes in genomic methylation levels during early development of the zebrafish embryo. Dev Genes Evol 214: 412–417 [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miteva K, Valkov N, Goncharova-Peinoval J, Kovachev K, Zlatarev S, Pironcheva G, Russev G 1995. Electron microscopic data for the presence of post-meiotic gene expression in isolated ram sperm chromatin. Cytobios 83: 85–90 [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111: 197–208 [DOI] [PubMed] [Google Scholar]
- Naz RK 1998. Effect of actinomycin D and cycloheximide on human sperm function. Arch Androl 41: 135–142 [DOI] [PubMed] [Google Scholar]
- Okada Y, Yamagata K, Hong K, Wakayama T, Zhang Y 2010. A role for the elongator complex in zygotic paternal genome demethylation. Nature 463: 554–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okitsu CY, Hsieh CL 2007. DNA methylation dictates histone H3K4 methylation. Mol Cell Biol 27: 2746–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oler AJ, Alla RK, Roberts DN, Wong A, Hollenhorst PC, Chandler KJ, Cassiday PA, Nelson CA, Hagedorn CH, Graves BJ, et al. 2010. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol 17: 620–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. 2007. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448: 714–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH 2005. Chipping away at the embryonic stem cell network. Cell 122: 828–830 [DOI] [PubMed] [Google Scholar]
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J 2000. Active demethylation of the paternal genome in the mouse zygote. Curr Biol 10: 475–478 [DOI] [PubMed] [Google Scholar]
- Rai K, Chidester S, Zavala CV, Manos EJ, James SR, Karpf AR, Jones DA, Cairns BR 2007. Dnmt2 functions in the cytoplasm to promote liver, brain, and retina development in zebrafish. Genes Dev 21: 261–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Orkin SH 2006. Unraveling the transcriptional network controlling ES cell pluripotency. Genome Biol 7: 230 doi: 10.1186/gb-2006-7-8-230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PJ, Fairall L, Huynh VA, Rhodes D 2006. EM measurements define the dimensions of the “30-nm” chromatin fiber: evidence for a compact, interdigitated structure. Proc Natl Acad Sci 103: 6506–6511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PJ, An W, Routh A, Martino F, Chapman L, Roeder RG, Rhodes D 2008. 30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction. J Mol Biol 381: 816–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G, Braun RE 2006. Pathway to totipotency: lessons from germ cells. Cell 127: 891–904 [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Mita K, Tamura M, Onitake K, Yamashita M 2000. Requirement of protamine for maintaining nuclear condensation of medaka (Oryzias latipes) spermatozoa shed into water but not for promoting nuclear condensation during spermatogenesis. Int J Dev Biol 44: 195–199 [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL 2006. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311: 844–847 [DOI] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY 2010. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329: 689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, Liu XS, Rinn J, Schier AF 2010. Chromatin signature of embryonic pluripotency is established during genome activation. Nature 464: 922–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH 2006. A protein interaction network for pluripotency of embryonic stem cells. Nature 444: 364–368 [DOI] [PubMed] [Google Scholar]
- Wang GG, Cai L, Pasillas MP, Kamps MP 2007. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol 9: 804–812 [DOI] [PubMed] [Google Scholar]
- Ward WS, Coffey DS 1991. DNA packaging and organization in mammalian spermatozoa: comparison with somatic cells. Biol Reprod 44: 569–574 [DOI] [PubMed] [Google Scholar]
- Wardle FC, Odom DT, Bell GW, Yuan B, Danford TW, Wiellette EL, Herbolsheimer E, Sive HL, Young RA, Smith JC 2006. Zebrafish promoter microarrays identify actively transcribed embryonic genes. Genome Biol 7: R71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D 2007. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 39: 457–466 [DOI] [PubMed] [Google Scholar]
- Welstead GG, Schorderet P, Boyer LA 2008. The reprogramming language of pluripotency. Curr Opin Genet Dev 18: 123–129 [DOI] [PubMed] [Google Scholar]
- Westerfield M 2000. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio), 4th ed University of Oregon Press, Eugene, Oregon [Google Scholar]
- Wykes SM, Krawetz SA 2003. The structural organization of sperm chromatin. J Biol Chem 278: 29471–29477 [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD 2005. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121: 859–872 [DOI] [PubMed] [Google Scholar]
- Zhang H, Roberts DN, Cairns BR 2005. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123: 219–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT 2008. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322: 750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Fan JY, Rangasamy D, Tremethick DJ 2007. The nucleosome surface regulates chromatin compaction and couples it with transcriptional repression. Nat Struct Mol Biol 14: 1070–1076 [DOI] [PubMed] [Google Scholar]
- Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S 2008. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 456: 125–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE26609. The processed data are available for programmatic access using the GenoPub DAS/2 data distribution server (description, http://bioserver.hci.utah.edu/BioInfo/index.php/Software:DAS2; GenoPub web app, http://bioserver.hci.utah.edu:8080/DAS2DB/genopub (type “guest” for both the user name and password); and the DAS/2 Data Access URL, http://bioserver.hci.utah.edu:8080/DAS2DB/genome). DAS/2 compliant genome browsers such as IGB (http://www.bioviz.org/igb/) can be used to view the data sets, found under Danio rerio→D_rerio_Jul_2007→Wu_2011.