Abstract
Histone modifications regulate gene expression in eukaryotes, but their effects on transcriptomes of a multicellular organism and on transcriptomic divergence between species are poorly understood. Here we present the first nucleotide-resolution maps of histone acetylation, methylation, and core histone in Arabidopsis thaliana and a comprehensive analysis of these and all other available maps with gene expression data in A. thaliana, Arabidopsis arenosa, and allotetraploids. H3K9 acetylation (H3K9ac) and H3K4 trimethylation (H3K4me3) are correlated, and their distribution patterns are associated with Gene Ontology (GO) functional classifications. Highly dense and narrow distributions of these modifications near transcriptional start sites are associated with constitutive expression of genes involved in translation, whereas broad distributions toward coding regions correlate with expression variation of the genes involved in photosynthesis, carbohydrate metabolism, and defense responses. Compared to animal stem cells, dispersed distributions of H3K27me3 without bivalent H3K4me3 and H3K9ac marks correlate with developmentally repressed genes in Arabidopsis. Finally, genes affected by A. thaliana histone deacetylase 1 mutation tend to show high levels of expression variation within and between species. The data suggest that genome-wide coordinated modifications of histone acetylation and methylation provide a general mechanism for gene expression changes within and between species and in allopolyploids.
Histone modifications mediate epigenetic regulation of gene expression, growth, and development in plants and animals (Berger 2007; Li et al. 2007; Zhang 2008). Some histone modifications, such as acetylation of histones H3 and H4 and trimethylation of H3 lysine 4 (H3K4me3), are known as euchromatic marks and often associated with active transcription, whereas other modifications, such as methylation of H3K9 and H3K27, are known as heterochromatic marks and related to gene repression (Jenuwein and Allis 2001; Li et al. 2007). In mouse embryonic stem (ES) cells, developmentally regulated genes are predominantly found in bivalent domains that contain large regions of H3K27me3 covering narrow distributions of H3K4me3, and ∼20% of transcription start sites (TSS) containing high CpG have bivalent domains (Mikkelsen et al. 2007). In multipotent human hematopoietic stem cells, histone marks are associated with the fate of bivalent genes during differentiation (Cui et al. 2009). The data suggest that developmentally regulated genes are repressed by H3K27me3 in stem cells but poised for activation following differentiation. In contrast to animals, H3K27me3 in Arabidopsis does not correlate with gene expression (Zhang et al. 2007). In Arabidopsis thaliana, at least one of the H3K4me1, 2, and 3 modifications is found in ∼75% of genes, and H3K4me3 is associated with highly expressed genes (Zhang et al. 2009). H3K4me2 and H3K4me3 are independent of DNA methylation, whereas H3K4me1 is correlated with CG methylation. In addition, H3K9ac is correlated with gene expression levels in A. thaliana (Zhou et al. 2010). However, the relationship between H3K9ac and H3K4me3 euchromatic marks is unknown, and much less is known about the role of these marks in expression changes between species. To address these issues, we used chromatin immunoprecipitation sequencing (ChIP-seq) to obtain nucleotide-resolution genome-wide maps of H3 nucleosome, H3K9ac, and H3K4me3 in A. thaliana. We then analyzed these data, ChIP-chip data of histone modifications and cytosine methylation (MeC) (Zhang et al. 2006, 2007, 2009; Lister et al. 2008; Oh et al. 2008), and gene expression data in response to developmental and environmental changes (Ha et al. 2007), in histone deacetylase mutant and wild-type plants (Tian et al. 2005), and in the allotetraploids and their progenitors, A. thaliana and Arabidopsis arenosa (Ha et al. 2009). We found associations of H3K9ac, H3K4me3, and H3K27me3 with constitutive gene expression, developmental gene regulation, and differential gene expression within diploid A. thaliana, and among A. thaliana, A. arenosa, and their allotetraploids (Allo733 and Allo738).
Results
General relationships among histone modifications
We compared histone modifications and DNA methylation with their biological functions in A. thaliana using relevant data (Supplemental Table 1). H3 nucleosomes were evenly distributed across the genome, whereas H3K9ac and H3K4me3 were predominantly enriched in genic regions (Supplemental Fig. S1), consistent with the report that H3K4 mono-, di-, and tri-methylation are highly enriched in gene-rich euchromatin (Zhang et al. 2009). The overall distribution of H3 and H3K4me3 in the ChIP-seq maps was consistent with that in the ChIP-chip maps (Oh et al. 2008; Zhang et al. 2009), suggesting that ChIP-seq data are highly reproducible and increase the resolution to the nucleotide level (Supplemental Figs. S2, S6, S7).
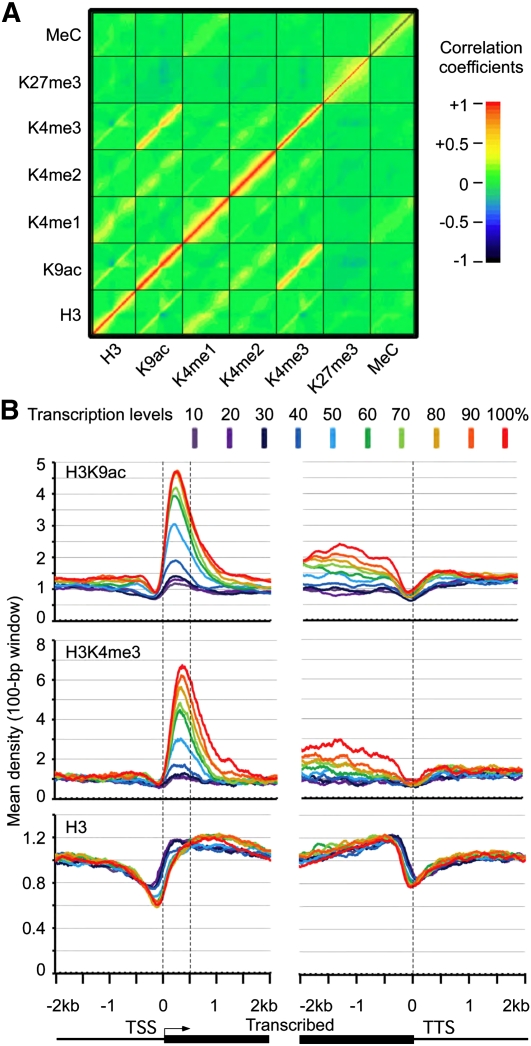
Genetic and biochemical studies suggest that histone methylation controls DNA methylation (Tamaru and Selker 2001), and histone acetylation affects histone methylation (Lawrence et al. 2004). To study the relationships among various modifications, we analyzed H3K4me1, H3K4me2, H3K4me3, H3K9ac, H3K27me3, and DNA methylation patterns within 2-kb upstream and downstream of the transcription start site (TSS) using 100-bp sliding windows. ChIP-seq data of H3, H3K4me3, and H3K9ac (this study) and ChIP-chip data of H3K4me1, H3K4me2 (Zhang et al. 2009), H3K27me3 (Zhang et al. 2007; Oh et al. 2008), and DNA methylation (Zhang et al. 2006; Zilberman et al. 2007) were normalized for corresponding genes (∼18,000) that were detected in gene expression microarrays (Ha et al. 2007, 2009). Here the “density” is used to infer the hybridization intensity of a locus in ChIP-microarray chip (ChIP-chip) data or the frequency of sequencing reads covering the locus normalized by mean coverage of total sequence reads in the whole genome in ChIP-seq data. Pearson's correlation coefficient (R) was used to estimate the relationship between histone modification and DNA methylation densities at all possible pairwise loci from −2 kb to +2 kb around the TSS (Fig. 1A). The correlation between H3K4me3 and H3K9ac at the same loci is highly significant (R = 0.54, P = 0, N = 1,023,640). In addition, positive correlations between H3K4me1 and H3, H3K4me2 and H3K9ac, or H3K4me2 and H3Kme3 at the same loci are observed. Heterochromatic marks, such as DNA methylation and H3K27me3, did not show any significant correlation with euchromatic histone marks (Zhang et al. 2007, 2009). Correlative relationships between euchromatic and heterochromatic marks may lead to enforcing and coordinated effects on transcriptional activity. In particular, H3K4me3 and H3K9ac were coordinately modified at the genome-wide level, as supported by biochemical studies on individual genes (Martin et al. 2006; Berger 2007). In humans, H3K4me3 facilitates histone acetylation (Wang et al. 2009), and bivalent modifications of H3K4me3 and H3K27me3 are associated with developmental gene regulation in human hematopoietic stem cells (Cui et al. 2009) and mouse stem cells (Bernstein et al. 2006; Mikkelsen et al. 2007). Arabidopsis may have developed a similar mechanism of gene regulation through coordination of H3K4 and H3K9 modifications.
Figure 1.
Histone modifications and DNA methylation and their effects on gene expression variation. (A) Density plot of correlation coefficients between H3, H3K9ac, H3K4me1, H3K4me2, H3K4me3, H3K27me3, and cytosine methylation (MeC) around the TSS. The x-axis and y-axis in each square (modification) represent the distribution frequency of mapped reads from −2 kb to +2 kb around the TSS. (B) Mean densities of H3K4me3 and H3K9ac within 500-bp downstream from TSS (y-axis) are correlated with the transcription level of genes from −2 kb to +2 kb (x-axis). A total of 26,000 genes were divided into 10 groups based on their transcription levels in leaves, from the top 10% (red) to the lowest 10% (deep purple). Mean-normalized ChIP-seq densities of 1-bp resolution were plotted within a 4-kb region flanking the TSS or the transcription termination sites (TTS). The densities of H3K4me3 and H3K9ac in the region between dotted lines (TSS and TTS) showed significant positive correlation with the transcription levels of genes, whereas H3 densities in the same region are negatively correlated with the transcription level.
Effects of H3K4me3 and H3K9ac on gene expression variation in leaves
H3K9ac and H3K4me3 densities within a 4000-bp region flanking the TSS in all genes were used to test their roles in gene expression. Using multiple linear regression analysis of histone H3 occupancy and modifications (H3K4me3 and H3K9ac) around the TSS, we found that depletion of H3 nucleosomes and enrichment of H3K4me3 and H3K9ac at 500 bp downstream from the TSS significantly correlated with gene expression levels (Supplemental Fig. S3). The levels of H3K9ac, H3K4me3, and H3 sharply decreased within 100 bp upstream of the TSS, suggesting that nucleosomes are depleted near the TSS to facilitate binding of transcriptional machineries including RNA polymerase II and associated transcription factors (Hassan et al. 2001). H3K9ac and H3K4me3 levels from the +1 position to the +500 position were significantly correlated with transcript abundance (Fig. 1B), when the mean densities of H3K9ac, H3K4me3, and H3 were compared for 10 groups of genes with varied degrees of expression levels in A. thaliana leaves (from the top 10% to the lowest 10%) using public microarray data (Supplemental Table 2). As the transcript levels increased, the H3K9ac and H3K4me3 densities increased, whereas H3 nucleosome occupancy levels decreased. This genome-wide observation is consistent with the notion that H3K9ac and H3K4me3 near the TSS destabilize the interaction between histones and DNA, leading to nucleosome loss (Boeger et al. 2003; Reinke and Horz 2003).
Roles of H3K4me3 and H3K9ac in gene expression variation during development
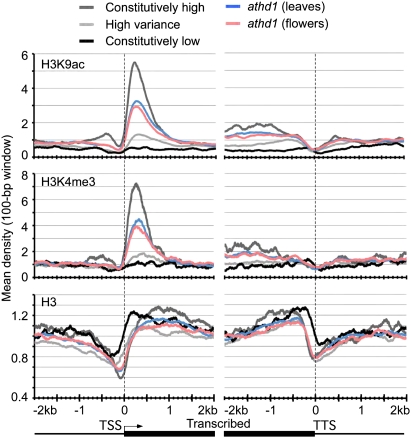
To test if chromatin modifications are associated with gene expression changes during development, H3K9ac, H3K4me3, and H3 densities in the genes were compared with gene expression patterns at various developmental stages (Supplemental Table 2; Ha et al. 2007). The variance of gene expression values obtained from 63 different developmental stages or organs was analyzed and divided into two groups showing the highest (10%) and the lowest (10%) expression variance, respectively. The high-variance genes displayed developmental and tissue-specific expression patterns, whereas the low-variance (var < 0.03) genes were constantly expressed across developmental stages. The low-variance genes with relatively constant expression levels were further divided into genes with constitutively high and low expression abundance, representing the top 10% and the lowest 10% expression levels in all tissues, respectively. The densities of H3K9ac and H3K4me3 were higher in the low-variance genes with constitutive high expression levels across developmental stages than in the genes with top 10% high expression levels in the leaves (Fig. 2). The low-variance genes with constantly low expression levels had barely detectable levels of H3K4me3 or H3K9ac but high levels of nucleosome occupancy. The high-variance genes showed moderate levels of nucleosome, H3K9ac, and H3K4me3 densities. These data suggest that H3K9ac and H3K4me3 levels are associated with gene expression variation during development.
Figure 2.
H3K9ac, H3K4me3, and nucleosome densities are correlated with gene expression variations during development. Mean-normalized ChIP-seq densities of 1-bp resolution (y-axis) were plotted within a 4-kb region flanking the TSS or the TTS (x-axis). Genes with the top 10% expression levels (dark gray plots), the lowest 10% expression levels (black plots), and the top 10% highest expression variance (light gray plots), across 63 developmental stages. Genes that were up-regulated in athd1-t1 leaves (blue plots) and flowers (pink plots).
Chromatin modifications in gene expression groups
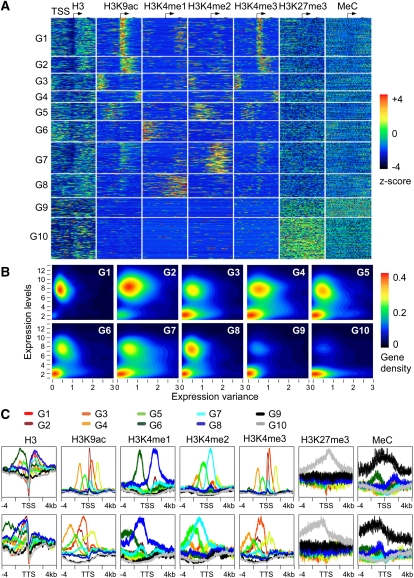
H3K27me3 and H3K4me3 are found in bivalent domains of the genes in mouse embryonic stem (ES) cells (Mikkelsen et al. 2007) and multipotent human hematopoietic stem cells (Cui et al. 2009). Moreover, a recent study suggests a role of chromatin modifications in regulating circadian-mediated gene expression networks in Arabidopsis hybrids and allopolyploids (Ni et al. 2009). These findings led us to test if the specific chromatin modifications or in combination play a part in gene expression changes in biological and cellular functions during plant development. We used the Partitioning Around Medoids (PAM) algorithm of Cluster Analysis to minimize the sum of dissimilarities within a group but maximize dissimilarities among groups (Kaufman and Rousseeuw 1990). The unsupervised PAM algorithm was used to analyze histone modification patterns around the TSS of the selected genes (∼18,000) in microarray data sets and cluster these genes into 10 groups (G1–G10) (Fig. 3; Supplemental Table 3). Each group was classified by a combination of distinctive histone modifications and their genomic locations. Consistent with the above data, H3 was void near the TSS of genes in most groups (Fig. 3A,C), due to binding of transcriptional machinery (Hassan et al. 2001). H3K9ac distributions in G1 to G5 genes peaked within a specific distance (∼500 bp) from the TSS, which corresponded to respective H3K4me3 peaks (Fig. 3A). Distinct H3K4me1 and H3K4me2 patterns were also found for genes in G6/G8 and G7, respectively (Fig. 3A,C). In contrast, H3K27me3 and DNA methylation patterns of G9 and G10 genes were scattered in broad regions with relatively low levels of H3K9ac and H3K4 methylation. These data suggest correlative and anti-correlative relationships among histone modifications with distinct genomic locations.
Figure 3.
Distinctive patterns of chromatin modifications around the TSS are associated with transcription levels and expression variation of genes in Arabidopsis. (A) Clustering of genes into 10 groups (G1 to G10) using the unsupervised PAM algorithm and chromatin modification patterns around TSS. Standardized mean densities of histone modifications flanking the TSS in each cluster group were plotted at 1-bp resolution. The occupancy density of each chromatin modification was standardized to yield a distribution with mean = 0 and variance = 1 using ChIP-seq (H3, H3K9ac, and H3K4me3) and ChIP-chip (H3K4me1, H3K4me2, H3K27me3, and MeC) data. All genes were aligned around the TSS and sorted by groups after clustering analysis based on their chromatin modification densities in a 100-bp window using PAM (k = 10). (B) The kernel density (gene density from 0 to 0.4) of the genes in each group (G1 to G10) was plotted using gene expression levels in leaves (y-axis) versus gene expression variance in 63 developmental stages (x-axis). (C) Gene expression variation was associated with chromatin modifications. Ten groups (G1 to G10) of genes with distinct chromatin modification patterns were plotted within a 4-kb region flanking the TSS or the TTS.
Different histone modifications confer distinct gene expression patterns. H3K9ac and H3K4me3 are related to temporal and spatial regulation of gene expression, depending on the location of these modifications relative to the TSS. For example, among the highly expressed genes (G1 and G2), the H3K9ac and H3K4me3 levels peaked immediately downstream from the TSS (Fig. 3A,B), suggesting that H3K9ac and H3K4me3 levels within the 500-bp region of TSS are associated with transcription activities. G1 genes with the highest levels of H3K9ac and H3K4me3 tended to have low expression variance and constantly high expression levels in various developmental stages (Fig. 3B). G2 genes had a wider distribution of H3K9ac and H3K4me3 at the 5′ end and showed an increased range of gene expression levels and variance, indicating differential expression in various developmental stages and in response to environmental changes. A wide range of H3K4me3 and H3K9ac distributions in developmentally regulated genes may suggest recruitment of histone acetyltransferases and methyltransferases through interaction among transcription factors, leading to developmental regulation of gene expression. Unlike G1 and G2 genes, G10 genes showed high levels of H3K27me3, and expression of these genes was low but highly variable, suggesting developmental repression, as observed in animal and human stem cells (Bernstein et al. 2006; Mikkelsen et al. 2007; Cui et al. 2009). In addition to the high levels of H3K27me3, G9 genes displayed high levels of DNA methylation, suggesting that DNA methylation is associated with constitutive repression and a low level of gene expression variation within this group (Fig. 3B).
Distinctive patterns of histone modifications with respect to gene functions
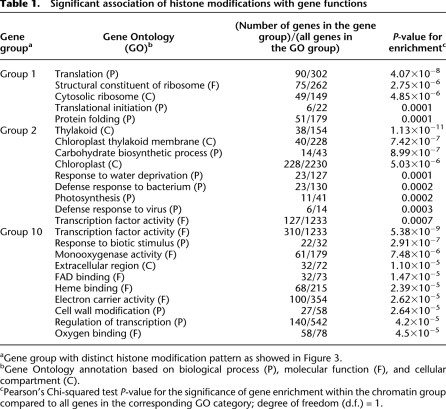
The distinctive landscape of chromatin modifications is associated with functional groups based on Gene Ontology (GO) classifications (Berardini et al. 2004). Several chromatin modifications are significantly enriched in specific GO classes (Table 1). G1 genes with high levels of H3K9ac and H3K4me3 and constitutive expression belong to the GO classes of translation initiation, translation, and ribosome constituents, which are abundantly expressed during growth and development (Supplemental Fig. S4). Translation initiation factor 3 H1 (TIF3H1) and eukaryotic initiation factor 2 (the EIF2) gamma subunits are components of translation initiation complexes and show the characteristic chromatin modification profile of G1 (Supplemental Fig. S4). G2 is dominated by genes involved in photosynthesis, chloroplast, and thylakoid membrane activities, which are most important biological processes specialized in leaves. These genes have a wide range of H3K9ac and H3K4me3 densities and are highly expressed in leaves, with a high expression variance. Examples include beta-amylase 3 (BAM3) and protochlorophyllide oxidoreductase B (PORB), which are involved in maltose biosynthesis and chlorophyll biosynthesis, respectively. G10 genes are associated with H3K27me3 and tissue-specific repression and are highly enriched in transcription factors and DNA binding proteins that are important to developmental regulation. For example, PLETHORA 1 (PLT1) and PLETHORA (PLT2) are essential transcription factors for the formation of stem cells, and their expression is co-repressed by TPL (TOPLESS) (Smith and Long 2010). PLT1 and PLT2 show high densities of scattered H3K27me3 across their genic regions with low levels of H3K4me3 and H3K9ac (Supplemental Fig. S5). H3K27me3 is associated with repression of PLT1 and PLT2 in leaves.
Table 1.
Significant association of histone modifications with gene functions
aGene group with distinct histone modification pattern as showed in Figure 3.
bGene Ontology annotation based on biological process (P), molecular function (F), and cellular compartment (C).
cPearson's Chi-squared test P-value for the significance of gene enrichment within the chromatin group compared to all genes in the corresponding GO category; degree of freedom (d.f.) = 1.
G3, G4, G5, G6, G7, G8, and G9 groups do not show a significant enrichment in specific GO classifications, probably because these groups contain the genes with both high and low abundance and moderate expression variances. The results support the notion that the distribution patterns of H3K9ac and H3K4me3 at the 5′ ends of genes are associated with gene expression for essential biological processes. Therefore, explicit arrangements of modified histones are coordinated with tissue-specific expression changes that are required for specialized functions or morphologies.
Association of histone deacetylase activity with gene expression variation
The association of H3K9ac and H3K4me3 with developmental gene expression led us to test if histone acetylation and deacetylation affect transcriptional regulation in response to developmental changes and environmental cues (Chen and Tian 2007). In A. thaliana, ATHD1 (or ATHDA19) encodes an important histone deacetylase that is expressed throughout development (Tian and Chen 2001). In ATHD1 T-DNA insertion mutant (athd1-t1), 2714 and 2010 genes were differentially expressed in leaves and flowers, respectively. The up-regulated genes in athd1-t1 display significantly high expression variance across developmental stages (Fig. 4A), suggesting a dynamic gene expression change during development (Wilcoxon rank-sum test, P < 10−8), and this change is significantly associated with the genes whose expression is altered in the histone deacetylase mutant in leaves (blue line) and flowers (pink line) (R = 0.22, P < 10−22) (Fig. 2). In addition, the differentially expressed genes in the athd1-t1 mutant were significantly correlated with the genes that have high levels of expression variation in response to various abiotic stresses in 63 microarray data sets (Fig. 4B; Ha et al. 2007) (R = 0.21, P < 10−22). Furthermore, the majority of genes that show onefold to twofold changes in expression had higher levels of H3K9ac than all other genes (Supplemental Fig. 8). The genes with a higher than twofold expression change in the ATHD1 mutant had lower levels of H3K9ac than all other genes. These data suggest that ATHD1 is associated with gene expression changes at specific developmental stages or in response to environmental stimuli, but other factors may affect gene expression variation during development.
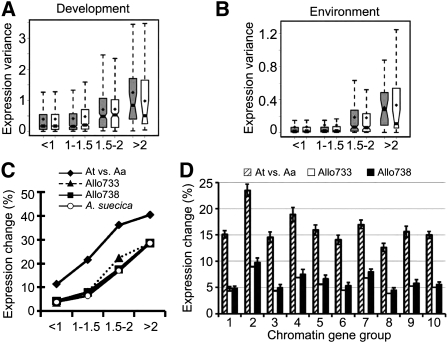
Figure 4.
Histone deacetylase affects gene expression variation within and between Arabidopsis species. (A) Genes whose expression was affected by ATHD1 in leaves and flowers showed a high level of expression diversity among development stages. Diamond points indicate mean values of the expression variance. Notches of the boxplots indicate 95% confidence intervals of median values of the expression variance among 63 developmental stages. Genes up-regulated in athd1-t1 leaves (gray boxplots) and flowers (open boxplots). Numbers under the x-axis represent the fold changes of gene expression in athd1-t1 compared to the wild type. (B) Genes highly affected by ATHD1 defect in leaves and flowers of A. thaliana show a high expression variance in response to various environmental changes. The expression variance of each gene (y-axis) was calculated from 63 combinations of environmental changes and time points after treatments. Genes up-regulated in athd1-t1 leaves (gray boxplots) and flowers (open boxplots) with a range of expression fold changes (x-axis). (C) Histone acetylation and deacetylation target genes tend to change expression levels between closely related species and in allotetraploids. The logistic regression analyses were conducted for gene expression changes between A. thaliana and Arabidopsis arenosa (diamond), between a resynthesized allotetraploid (Allo733) and mid-parent value (MPV) (triangle), between Allo738 and MPV (square), and between the natural allotetraploid Arabidopsis suecica and MPV (circle), relative to the expression fold changes (x-axis). (D) Boxed bars represent the proportion of genes differentially expressed between A. thaliana and A. arenosa (slashed), Allo733 and MPV (open), and Allo738 and MPV (filled). Standard deviations were calculated by 10,000 bootstrapping replicates.
Gene expression changes between A. thaliana and A. arenosa and in their allotetraploids are associated with genetic and epigenetic mechanisms (Chen 2007). The gene expression diversity between species and in allotetraploids was compared with the genes whose expression is affected in the athd1 mutant. The differentially expressed genes in athd1 were significantly correlated with the genes that displayed expression changes between species and in the allotetraploids (Fig. 4C). Genes up-regulated in athd1 are more differentially expressed between species and nonadditively expressed (from the mid-parent values) in the allotetraploids. Among them, 40% of the genes that displayed more than twofold up-regulation in athd1 were also differentially expressed between A. thaliana and A. arenosa. A simple logistic regression analysis reveals significant correlation between ATHD1-dependent gene expression changes and the gene expression changes between species (At4 vs. Aa, β1 = 0.2, P = 1.2 × 10−5) or in the allotetraploids (Allo733 vs. MPV, β1 = 0.3, P = 4.7 × 10−6; and Allo738 vs. MPV, β1 = 0.2, P = 6.6 × 10−5). The data suggest an association of ATHD1 with gene expression divergence between species and nonadditive gene expression in allopolyploids.
Histone modification patterns confer expression differentiation between species
Are chromatin modifications around the TSS responsible for expression divergence between A. thaliana and A. arenosa and in their allotetraploids? We found that distinct chromatin modification groups were associated with expression divergence between A. thaliana and A. arenosa and had the same trend as the expression variability observed at different developmental stages in A. thaliana (Fig. 4D). G1 genes had the low tendency of expression divergence between species and in allotetraploids. However, they were constitutively expressed at high levels across developmental stages. G2 genes had dense and broad distribution of H3K9ac and H3K4me3 at the 5′ end of the genes and also showed the highest tendency of expression divergence between the two species and in the allotetraploids, which is consistent with up-regulation of photosynthetic and starch metabolic activities and growth vigor in the allotetraploids and hybrids (Ni et al. 2009). The results suggest that the locations and overall levels of these histone modifications near the TSS are associated with gene expression variation within and between Arabidopsis species.
Discussion
In this study, we generated ChIP-seq data of H3, H3K4me3, and H3K9Ac in leaves of A. thaliana and compared the ChIP-seq data with ChIP-chip data of H3K4me1, H3K4me2, and H3K27me3 (Zhang et al. 2007, 2009; Oh et al. 2008) and DNA methylation (Zilberman et al. 2007). The data from other sources are derived from aerial parts and seedlings. We comprehensively analyzed these data from these slightly different tissues and technologies including ChIP-seq and ChIP-chip. In spite of the heterogeneity of data sources, the correlative and exclusive relationships among histone modifications and DNA methylations are highly significant and reproducible (Supplemental Figs. 6, 7). Using the data generated in leaves and aerial tissues would potentially increase the level of variation. If the data sets from the same tissue or in the same condition were used, the amount of variation across platforms and developmental stages would be reduced. It is expected that the trend of coordinated distributions among chromatin modifications in the same tissues would be even higher. This does not preclude that specific modifications of some (e.g., tissue-specific) genes may be different among different data sets. The increased level of variation may also affect weak correlative data such as bivalent domains (H3K4me3 and H3K27me3).
Comprehensive genome-wide analyses of gene expression and chromatin modification data suggest interlinks betweenH3K9ac, H3K4me3, and H3K27me3 modifications, on the one hand, and temporal and spatial gene expression variations in related Arabidopsis species and their hybrids, on the other. Significant correlations are observed between H3K4me3 and H3K9ac in highly expressed genes, and the location and distribution of H3K9ac and H3K4me3 affect transcriptional activity and expression variability in response to developmental and environmental changes. Chromatin regions with H3K4me3 are targeted for histone acetylation by histone acetyltransferases (Wang et al. 2009), and these modifications may reinforce the activity of constitutively expressed genes such as translation initiation and protein synthesis that are important to all stages of growth and development in an organism. Moreover, maintaining a dynamic regulation for a subset of tissue-specific or developmentally regulated genes through distribution of H3K9ac and H3K4me3 over coding sequences is essential for important physiological pathways such as photosynthesis in plants. Localizations of two repressive marks, H3K27me3 and DNA methylation, are exclusively in the genes with low transcript abundance. These findings imply coordinated regulation of histone modifications on gene expression, in addition to previous findings that DNA methylation and histone variant H2A.Z are mutually exclusive (Zilberman et al. 2008), and both histone variants H3.3 and H2A.Z occupy nucleosome free regions (Jin et al. 2009). As in animals, H3K27me3 in Arabidopsis is a repressive mark for developmentally regulated genes (Bernstein et al. 2006; Barski et al. 2007; Hammoud et al. 2009). The correlation between H3K27me3 and gene repression in A. thaliana was not obvious in a previous study (Zhang et al. 2007), probably because chromatin density plots and gene expression groups were not comparatively analyzed. Unlike in animal cells, H3K4me3 is not associated with H3K27me3 in A. thaliana (Supplemental Fig. S5), which may suggest different strategies between animal and plant cells with respect to cell differentiation and organ development. Plant cells are pluripotent and totipotent and contain essentially identical and cell-autonomous properties during cell differentiation and development. A poised state of H3K4me3 may not be required for the genes repressed by H3K27me3 prior to differentiation. Chromatin structure plays a role in pluripotent and totipotent plant stem cells (Verdeil et al. 2007). The interchangeability between totipotency and cell differentiation in plants remains an interesting subject. As the genes that carry broad distributions of H3K9ac and H3K4me3 at the coding region, the genes whose expression is affected by histone deacetylation mutation are associated with expression differentiation after species divergence and genome merger. This suggests that activities of histone-modifying enzymes exemplified by ATHD1 maintain many expression differences between closely related Arabidopsis species by regulating histone modifications. Also, nonadditive expression of orthologous genes in the new allotetraploid species is associated with active histone marks. It is likely that different chromatin modifications are associated with expression variation of orthologous genes between A. thaliana and A. arenosa, leading to changes in gene expression and differences in growth and development. These species-specific histone modifications are remodeled in the new allopolyploids species, resulting in nonadditive gene expression (Chen 2007).
The above conclusion drawn from genome-wide analysis of chromatin modifications and gene expression is supported by experimental data. As examples, early flowering is inhibited by a high level of FLC expression in A. thaliana (Michaels and Amasino 1999). FLC is expressed at higher levels in late flowering A. arenosa than in early flowering A. thaliana, and the expression levels are directly correlated with histone marks such as H3K9ac and H3K4me2 (Wang et al. 2006). In the allopolyploids, the A. thaliana FLC allele is highly expressed and associated with high levels of H3K9ac and H3K4me2, whereas its homologous A. arenosa FLC allele is repressed with relatively low levels of H3K9ac and H3K4me2. In another study, active but not repressive histone marks are associated with the altered expression levels of circadian clock genes, including CCA1, LHY, and TOC1, in Arabidopsis hybrids and allotetraploids (Ni et al. 2009). In addition, reducing DNA methylation in the met1-RNAi lines of the natural allotetraploid Arabidopsis suecica reactivates transposons but does not affect expression variation of protein-coding genes (Chen et al. 2008). These data suggest that histone acetylation and deacetylation coupled with histone methylation have an important role in gene expression variation between closely related species and in allopolyploids. A. thaliana HDT1 (a plant-specific histone deacetylase) is required for H3K9 deacetylation and subsequent H3K9 methylation. Mutation of HDT1 leads to reactivation of silenced rRNA genes that are subjected to nucleolar dominance (Lawrence et al. 2004). In yeasts, genes with high expression variability are also affected by histone acetyltransferase mutations (Steinfeld et al. 2007; Choi and Kim 2008). In human cells, both histone acetyltransferases and deacetylases are recruited to actively transcribed genes (Wang et al. 2009). Therefore, control of histone acetylation distribution by histone acetyltransferases and deacetylases is an important regulatory mechanism for gene regulation in most eukaryotes (Berger 2007; Li et al. 2007) and probably for expression of homoeologous genes in allopolyploids.
Methods
Plant materials
A. thaliana, A. arenosa, and allotetraploid plants were grown in vermiculite mixed with 30% soil in a growth chamber with growth conditions of 22°/18°C (day/night) and 16 h of illumination per day. Rosette leaves prior to bolting were collected at noon for the analysis of chromatin modifications in this study and for gene expression analysis in the previous work (Tian et al. 2005; Ha et al. 2009).
ChIP and template preparation for Illumina sequencing analysis
Chromatin immunoprecipitation (ChIP) was performed using mature leaves (∼3 g) of A. thaliana (Col) following the published protocols (Saleh et al. 2008; http://www.natureprotocols.com/2009/01/08/chromatin_immunoprecipitation_2.php). For immunoprecipitation, 2, 4, and 3.6 μg of antibodies against H3-core (Ab1791; Abcam), H3K4me3 (Ab8580), and H3K9ac (Ab10812), respectively, were added to a 600-μL solution containing diluted chromatin extracts and incubated overnight at 4°C with gentle rotation. Immunoprecipitated chromatin-DNA (IP-DNA) and 50 μL of input chromatin-DNA (without IP) were subjected to reverse cross-link and purified for sequencing libraries preparation. ChIP-seq libraries were constructed according to protocols described in the Illumina ChIP sequencing kit (Illumina Inc.). DNA fragments with a range of 250–300 bp were excised and amplified for subsequent cluster generation and massively parallel sequencing.
Sequence read mapping and identification of nucleosomes in A. thaliana genome
The sequence reads (40–60 bp) were aligned to the A. thaliana genome (version TAIR9), allowing no mismatch or gap. To exclude any ambiguous signal, sequence reads aligned perfectly to only one region of the genome were used for estimating densities of nucleosomes and histone modifications. We analyzed 7,441,484, 5,219,184, and 7,425,084 sequence reads from H3, H3K4me3, and H3K9ac ChIP-seq, respectively. Out of the total reads, 4,481,737, 1,260,434, and 3,047,219 reads were mapped to the A. thaliana genome. The density or frequency of sequencing reads covering the locus was normalized by mean coverage of total sequence reads in the whole genome. The peaks of read starting points at two opposing strands normally displayed average ∼150-bp intervals, equivalent to the DNA length in a nucleosome, which was used as the mean DNA fragment length. The nucleosome density of 1-bp resolution was estimated by the number of extended reads covering each nucleotide within an ∼150-bp region. The correlation coefficient of nucleosome densities in two DNA strands was used to test consistency. In three experiments, the correlation coefficients between H3, H3K4me3, and H3K9ac densities were >0.99, suggesting similar distributions of modified nucleosomes in both strands. Thus, nucleosome reads of both strands were summed up to generate nucleosome maps with 1-bp resolution. The number of reads covering each base was normalized by dividing the average read number per base for the whole genome. The ChIP-seq data were deposited in GEO (accession no. GSE22276).
Genome-wide chromatin modification data from ChIP-chip
The normalized ChIP-chip data of H3K4 mono-, di-, and tri-methylation were obtained from Oh et al. (2008) and Zhang et al. (2009). Using the log ratio of the average H3K4 methylation over H3, the individual single-channel results of H3K4 methylation and H3 (Oh et al. 2008) were linearly scaled to produce the same mean and the same variance. The TileMap Statistic ranging from 0 to 1 within 25-nt probes with a 35-nt interval was used to normalize H3K4 ChIP-chip data (Oh et al. 2008; Zhang et al. 2009). The data of H3K27me3 methylation and DNA methylation were obtained from ChIP-chip (Zhang et al. 2006; Zilberman et al. 2007) and ChIP-seq (Cokus et al. 2008; Lister et al. 2008) assays, respectively. The ChIP-seq results of H3 and H3K4me3 reported in this study were validated and comparatively analyzed using the ChIP-chip data sets.
Histone modification densities within and around genes
The annotation (version TAIR9, June 2009) of genes was downloaded from The Arabidopsis Information Resource (TAIR; ftp://ftp.arabidopsis.org/home/tair/Genes/TAIR9_genome_release/). To estimate position-specific mean densities of histone modifications, we calculated mean values of standardized sequence read frequencies in each base or a 100-bp window around transcription start sites.
Clustering analysis of chromatin modifications around genes
Histone modification densities around the transcription start sites were generated using a 100-bp window in the matrix form. For each chromatin modification, the intensities were standardized into a Z-score so that the total mean is 0 and the standard deviation is 1 across all windows. The correlation coefficient of 1 was chosen as the dissimilarity matrix for PAM clustering with 10,000 iterations and k = 10 (Kaufman and Rousseeuw 1990). In PAM, once the dissimilarity matrix among genes is calculated, a representative set of medoids was determined, and the data were aggregated around the medoids.
Gene expression changes in A. thaliana histone deacetylase (ATHD1) mutants
The raw microarray data of gene expression changes were obtained from leaves and flowers of A. thaliana ecotype Ws (ATHD1/ATHD1, +/+) versus athd1-t1/athd1-t1 (−/−) plants, respectively (Tian et al. 2005). Two large dye-swap experiments were performed using four biological replications and eight slide hybridizations. ANOVA was used for replicated microarray data. To test the gene expression difference between WT and athd1/athd1, t-tests were used for individual genes (per-gene variance). The type I error rate of multiple testing was controlled to be below 0.05 using the false discovery rate (FDR) approach described by Benjamini and Hochberg (1995). The microarray data were deposited in GEO (accession no. GSE22276).
Estimation of gene expression variance
The Affymetrix ATH1 expression array data were obtained from the AtGenExpress expression atlas at TAIR (http://www.arabidopsis.org/info/expression/ATGenExpress.jsp). We compiled microarray data sets from 63 different developmental stages and 63 environmental conditions (Supplemental Table 2). The relative gene expression values at each condition were estimated using the GC-RMA method (Wu and Irizarry 2004; Wu et al. 2004), which considers GC content and position-specific base effects from the probe sequences. The gcrma package implemented in R was used. The normalized expression levels were used for calculating expression variance (Ha et al. 2007). The expression variance of individual genes in developmental stages and environmental responses was estimated separately, using expression data obtained from 63 different developmental stages or tissue-types and 63 sets of treatment and time-course combinations under abiotic stress. If the variance is 0, the gene is expressed at a constant level in various conditions. If the variance is large, the expression levels of the genes are highly variable among comparisons.
The microarray data of gene expression change between progenitors and in the allopolyploids were obtained from four sets of comparisons between (1) A. thaliana and A. arenosa (GSME341923), (2) Allo733 and mid-parent value (MPV, an artificial mixture of RNA from two parents; GSME342087), (3) Allo738 and MPV (13) (GSME342029), and (4) A. suecica and MPV (GSME334287) (Ha et al. 2009).
Cluster analysis of functional enrichment in gene groups
The Gene Ontology for A. thaliana was adopted from The Arabidopsis Information Resource (TAIR) (ftp://ftp.arabidopsis.org/home/tair/Ontologies/Gene_Ontology/) released in July 2009.
Acknowledgments
We thank colleagues in the Chen lab for valuable suggestions to improve the manuscript. This work is supported by grants from the National Institutes of Health (GM067015 to Z.J.C. and GM30998 to W.-H.L.) and from NSC (NSC 99-2321-B-001-041-MY2, Taiwan) to W.-H.L.
Authors' contributions: M.H. designed and initiated the research. D.W.N. generated ChIP-seq libraries and analyzed experimental data. M.H. performed computational analysis. M.H. and Z.J.C. analyzed the data and wrote the draft manuscript. M.H., D.W.N., W.-H.L., and Z.J.C. revised the manuscript.
Footnotes
[Supplemental material is available for this article. The ChIP-seq data and microarray data have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE22276.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.116467.110.
References
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y 1995. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57: 289–300 [Google Scholar]
- Berardini TZ, Mundodi S, Reiser L, Huala E, Garcia-Hernandez M, Zhang P, Mueller LA, Yoon J, Doyle A, Lander G, et al. 2004. Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol 135: 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL 2007. The complex language of chromatin regulation during transcription. Nature 447: 407–412 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Boeger H, Griesenbeck J, Strattan JS, Kornberg RD 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol Cell 11: 1587–1598 [DOI] [PubMed] [Google Scholar]
- Chen ZJ 2007. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol 58: 377–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Tian L 2007. Roles of dynamic and reversible histone acetylation in plant development and polyploidy. Biochim Biophys Acta 1769: 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Ha M, Lackey E, Wang J, Chen ZJ 2008. RNAi of met1 reduces DNA methylation and induces genome-specific changes in gene expression and centromeric small RNA accumulation in Arabidopsis allopolyploids. Genetics 178: 1845–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JK, Kim YJ 2008. Epigenetic regulation and the variability of gene expression. Nat Genet 40: 141–147 [DOI] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE 2008. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452: 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui K, Zang C, Roh T-Y, Schones DE, Childs RW, Peng W, Zhao K 2009. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell 4: 80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Li WH, Chen ZJ 2007. External factors accelerate expression divergence between duplicate genes. Trends Genet 23: 162–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Kim ED, Chen ZJ 2009. Duplicate genes increase expression diversity in closely related species and allopolyploids. Proc Natl Acad Sci 106: 2295–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR 2009. Distinctive chromatin in human sperm packages genes for embryo development. Nature 460: 473–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan AH, Neely KE, Workman JL 2001. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell 104: 817–827 [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD 2001. Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G 2009. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet 41: 941–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman L, Rousseeuw PJ 1990. Finding groups in data: An introduction to cluster analysis. Wiley, New York [Google Scholar]
- Lawrence RJ, Earley K, Pontes O, Silva M, Chen ZJ, Neves N, Viegas W, Pikaard CS 2004. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol Cell 13: 599–609 [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL 2007. The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR 2008. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DG, Grimes DE, Baetz K, Howe L 2006. Methylation of histone H3 mediates the association of the NuA3 histone acetyltransferase with chromatin. Mol Cell Biol 26: 3018–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM 1999. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Kim ED, Ha M, Lackey E, Liu J, Zhang Y, Sun Q, Chen ZJ 2009. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457: 327–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Park S, van Nocker S 2008. Genic and global functions for Paf1C in chromatin modification and gene expression in Arabidopsis. PLoS Genet 4: e1000077 doi: 10.1371/journal.pgen.1000077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke H, Horz W 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol Cell 11: 1599–1607 [DOI] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Avramova Z 2008. An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Smith ZR, Long JA 2010. Control of Arabidopsis apical–basal embryo polarity by antagonistic transcription factors. Nature 464: 423–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinfeld I, Shamir R, Kupiec M 2007. A genome-wide analysis in Saccharomyces cerevisiae demonstrates the influence of chromatin modifiers on transcription. Nat Genet 39: 303–309 [DOI] [PubMed] [Google Scholar]
- Tamaru H, Selker EU 2001. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414: 277–283 [DOI] [PubMed] [Google Scholar]
- Tian L, Chen ZJ 2001. Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc Natl Acad Sci 98: 200–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Fong MP, Wang JJ, Wei NE, Jiang H, Doerge RW, Chen ZJ 2005. Reversible histone acetylation and deacetylation mediate genome-wide, promoter-dependent and locus-specific changes in gene expression during plant development. Genetics 169: 337–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdeil JL, Alemanno L, Niemenak N, Tranbarger TJ 2007. Pluripotent versus totipotent plant stem cells: dependence versus autonomy? Trends Plant Sci 12: 245–252 [DOI] [PubMed] [Google Scholar]
- Wang J, Tian L, Lee HS, Chen ZJ 2006. Nonadditive regulation of FRI and FLC loci mediates flowering-time variation in Arabidopsis allopolyploids. Genetics 173: 965–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K 2009. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138: 1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA 2004. Preprocessing of oligonucleotide array data. Nat Biotechnol 22: 656–658 [DOI] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F 2004. A model-based background adjustment for oligonucleotide expression assays. J Am Stat Assoc 99: 909–917 [Google Scholar]
- Zhang X 2008. The epigenetic landscape of plants. Science 320: 489–492 [DOI] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al. 2006. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126: 1189–1201 [DOI] [PubMed] [Google Scholar]
- Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, Jacobsen SE 2007. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol 5: e129 doi: 10.1371/journal.pbio.0050129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Bernatavichute YV, Cokus S, Pellegrini M, Jacobsen SE 2009. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol 10: R62 doi: 10.1186/gb-2009-10-6-r62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wang X, He K, Charron J-B, Elling A, Deng X 2010. Genome-wide profiling of histone H3 lysine 9 acetylation and dimethylation in Arabidopsis reveals correlation between multiple histone marks and gene expression. Plant Mol Biol 72: 585–595 [DOI] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S 2007. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet 39: 61–69 [DOI] [PubMed] [Google Scholar]
- Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S 2008. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 456: 125–129 [DOI] [PMC free article] [PubMed] [Google Scholar]