Abstract
We present whole genome profiling (WGP), a novel next-generation sequencing-based physical mapping technology for construction of bacterial artificial chromosome (BAC) contigs of complex genomes, using Arabidopsis thaliana as an example. WGP leverages short read sequences derived from restriction fragments of two-dimensionally pooled BAC clones to generate sequence tags. These sequence tags are assigned to individual BAC clones, followed by assembly of BAC contigs based on shared regions containing identical sequence tags. Following in silico analysis of WGP sequence tags and simulation of a map of Arabidopsis chromosome 4 and maize, a WGP map of Arabidopsis thaliana ecotype Columbia was constructed de novo using a six-genome equivalent BAC library. Validation of the WGP map using the Columbia reference sequence confirmed that 350 BAC contigs (98%) were assembled correctly, spanning 97% of the 102-Mb calculated genome coverage. We demonstrate that WGP maps can also be generated for more complex plant genomes and will serve as excellent scaffolds to anchor genetic linkage maps and integrate whole genome sequence data.
Physical clone maps are indispensable tools that form the intermediate layer between local (gene) sequences, genetic maps, and whole genome sequences. Physical maps are widely used for a range of purposes including positional (map-based) cloning (Bakker et al. 2003), anchoring chromosomes using fluorescence in situ hybridization (FISH) (Islam-Faridi et al. 2002), repeat classification (Cardle et al. 2000), draft genome sequence assembly (Sasaki et al. 2005), local marker development (van der Vossen et al. 2000), and analysis of structural variation in the genome (Kidd et al. 2008). Despite advances in next-generation sequencing (NGS) technologies which have accelerated (re)sequencing complete genomes (Hillier et al. 2008; Wheeler et al. 2008), the need for high-quality physical maps remains (Lewin et al. 2009). For example, de novo sequencing and assembly of complex genomes containing large regions of repeated sequences will not be easily addressed by NGS alone and require additional procedures to provide anchor points to link sequence contigs and bridge large repeat regions. An efficient way to provide these anchor points is by construction of a whole genome physical map from bacterial artificial chromosome (BAC) clones (Shizuya et al. 1992; Rounsley et al. 2009), in combination with BAC-end sequencing (Nelson and Soderlund 2009). BAC insert clones are relatively easy to generate and store and have proven to be effective for genome-wide physical map construction (Gregory et al. 1997; Marra et al. 1997; Klein et al. 2000; Wu et al. 2004). Hence, BAC-based physical maps combined with BAC-end sequencing have formed the basis of several whole genome sequencing projects (Sasaki et al. 2005; Wei et al. 2007).
In the current gold standard for physical mapping, SNaPshot (Luo et al. 2003) and alternative methods such as amplified fragment length polymorphism (AFLP) (Vos et al. 1995), BACs are characterized by means of DNA fingerprinting (Srinivasan et al. 2003; Borm 2008). The general principle behind these approaches is the characterization of individual BACs by means of specific tags, such as restriction fragments of specific lengths. These fragments are visualized by gel- or capillary-electrophoresis, and fingerprint patterns are scored based on the size of unique bands, with an assumption that bands of identical length represent identical fragments. To provide sufficient power for distinction and correct assembly, around 100 fragments are typically scored per BAC, and BACs originating from the same region of the genome will be linked into a contig based on shared fragments. One of the assumptions for contig building is that the majority of these fragments are uniquely identifiable, i.e., that they represent a single location in the genome. However, in practice, this is not always the case because the assessment of fragment length is known to suffer from both scoring inaccuracy as well as occasional comigration of nonidentical or duplicated fragments (Koopman and Gort 2004). The latter may lead to false linkage between BACs. For example, in the maize high information content fingerprinting (HICF) map (Nelson et al. 2005), the average number of erroneously shared bands was reported to be 10.8 from an average of 98 bands per clone, i.e., a random overlap of 11% of bands (Nelson and Soderlund 2009). This observation underscores the need to apply stringent assembly criteria to prevent formation of contigs comprising noncontiguous BAC clones when using DNA fingerprint data for physical map building. As an alternative, optical mapping (Schwartz et al. 1993) has been described for constructing ordered restriction maps and has been used in sequencing projects of several whole genomes (e.g., Church et al. 2009; Zhou et al. 2009). In contrast to SNaPshot, it preserves the order of the restriction fragments in a given DNA fragment. However both methods share the inaccuracy of restriction fragment (length) calling and are not sequence-based. Other known methods for BAC clone mapping which are sequence-based, such as short tagged pooled genomic indexing using Sanger reads (ST-PGI [Milosavljevic et al. 2005]) and end-sequence profiling using Sanger BAC end sequencing (ESP [Volik et al. 2003]), are specifically designed for comparative genomics and require reference genome sequence data and cannot be used for de novo physical mapping of BAC clones. Finally, the Clone-Array Pooled Shotgun Sequencing (CAPPS) (Cai et al. 2001) method provides a large-scale genome sequencing strategy based on BAC pools requiring only minimal computational power for genome assembly, but does not provide genome-wide physical mapping of BAC clones.
AFLP is a robust complexity-reduction technology which uses restriction enzyme digestion and consecutive adapter-ligated restriction fragment amplification, without the need for prior sequence information. These features are shared with the restriction-site associated DNA (RAD) technique which has been described as an integrated platform for SNP discovery and genotyping (Baird et al. 2008). Sample preparation for short read NGS platforms such as the Illumina Genome Analyzer (GA) uses similar steps. Expanding on AFLP, we envisaged the use of NGS to uniquely characterize restriction enzyme fragments not by their size but by their unique sequence content. Hence, we developed WGP as a high resolution sequence-based physical mapping technology based on short read NGS with sequence tags placed along the entire BAC clone. WGP incorporates the use of sample identification tags (also known as barcodes) and a two-dimensional (2D) BAC clone pooling strategy to make optimal use of sequencing capacity and reduce costs for BAC DNA preparation. WGP tags uniquely characterize individual BACs. Shared WGP tags are consecutively used for BAC contiging (Fig. 1; Supplemental Fig. S1).
Figure 1.
Overview of whole genome profiling technology. (A) BAC library: BAC clones are available in a 384-wells plate format. (B) BAC pooling and DNA extraction: DNA is extracted after pooling each row (24 BACs) and each column (16 BACs). (C) Template preparation and sequencing: Pooled BAC DNA is digested (EcoRI/MseI) and amplified after barcoded adapters are ligated for pool identification after sequencing on the Illumina GA platform. (D) Deconvolution: Sequence tags (30–50 per BAC) are assigned to individual BACs based on presence in one row and one column pool. (E) Contiging: Overlapping (sets of) sequence tags from individual BAC clones generate contigs. Together, these contigs represent a sequence-based physical map of the genome.
To provide proof of concept of the WGP technology, a wet-lab experiment was conducted involving genome-wide assembly of an Arabidopsis thaliana ecotype Columbia BAC library. The WGP results were validated using the publicly available reference genome sequence of this ecotype. Subsequent application of WGP in other plant genomes and a simulation study performed in maize demonstrate the scalability of WGP to complex plant genomes.
Results
In silico analysis
To investigate the feasibility of the WGP approach and the requirements for the WGP tags, the complete Arabidopsis thaliana Columbia sequence was EcoRI/MseI-digested in silico. This produced 69,275 fragments representing 64,059 unique sequences flanking the EcoRI sites (Table 1). The length of the sequence tag influenced its uniqueness and showed a saturation point around 20 nt, where over 87% of all restriction fragments were represented by a single unique tag (Supplemental Fig. S2). Such tags can be easily generated with short read NGS platforms such as Illumina's Genome Analyzer (GA) or Life Technologies' SOLiD.
Table 1.
Overview of the number of sequence tags derived from the Arabidopsis WGP experiment and the in silico simulation analysis
a6 nt EcoRI + 20 nt fragment sequence.
In silico WGP simulation analysis on Arabidopsis chromosome 4 and the complete genome of maize
Two WGP simulations—one on Arabidopsis chromosome 4 and another on the entire genome of maize—were carried out to substantiate the hypothesis that short sequence tags adjacent to restriction enzyme sites can be used for unique identification of BAC clones and subsequent BAC-based whole genome physical mapping. Both analyses demonstrated that pooling of BACs in a 2D format allowed satisfactory deconvolution of tags to individual BACs and the ability to use such information to generate high quality physical maps ranging from relatively simple single chromosomes up to complex large genomes such as maize. For details, see supplemental information.
WGP tag generation and deconvolution
An existing six-genome equivalent library of Arabidopsis thaliana ecotype Columbia was subjected to WGP. The BAC library of 6144 clones was divided over sixteen 384-well plates which were each pooled by row (24 clones) and column (16 clones) to result in 40 pools per plate. DNA isolated from these pools was digested with EcoRI and MseI, barcoded adapters were ligated, and restriction fragments were amplified. Amplified fragments were subsequently sequenced from the EcoRI restriction site end using the Illumina GA. In total this yielded 30.3 million passed filter reads with 31-nt read length (NCBI Sequence Read Archive: SRA026464). Of these reads, 93% (28.2 M) contained a valid (100% matching) sample identification tag and EcoRI restriction site sequence. In Table 2 the data are specified for each of the eight lanes of the experiment, which demonstrate the high consistency of the data quality and percentages of reads that could be used for assignment to individual BACs.
Table 2.
Number of reads, deconvolutable reads, tags, and BACs for the eight Illumina lanes
Next, the reads were analyzed with custom-made Perl scripts. Using these scripts, the sample (pool) identification tag and the restriction site sequence were identified and removed after binning in the appropriate pool. Clustering on 100% identity was done to obtain unique WGP tags. Only those WGP tags which were present in a single row and a single column pool per plate were used for further analysis, since they could be unequivocally assigned to a single BAC. In total, this encompassed 12.1 M reads corresponding to 43% of the sequence read data (% deconvolution; Table 2). With these reads 4599 BACs were identified having at least one WGP tag. An average of 40 WGP tags was identified per BAC, each WGP tag being sequenced on average 66 times (Table 2; Fig. 2).
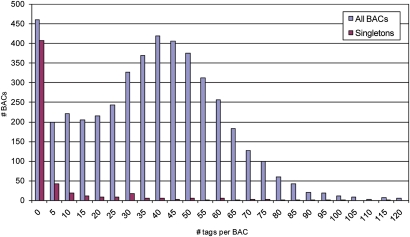
Figure 2.
Distribution of the number of tags per BAC, specified for all 4599 deconvoluted BACs and for the subset of 551 BACs that were not assembled in contigs (singleton BACs).
Experimental data confirms the uniqueness of 20-nt tag length
To substantiate the in silico analysis of tag length required for uniqueness, an analysis of required read length was performed on the data derived from the Illumina GA reads. Reads were trimmed to lengths ranging from 11 to 26 nt. The number of resulting deconvolutable tags showed a saturation point around 20 nt, similar to the in silico data (Supplemental Fig. S2). This suggests that generating sequence tag reads longer than 20 nt will not improve the WGP map and confirms the practical use of short read lengths as provided by the Illumina GA sequencer for WGP in Arabidopsis thaliana.
Verification of WGP tag quality
The total of 183,366 tags that were assigned to single BACs represented 65,734 different sequences (Tables 1 and 2). These sequences were compared to the Arabidopsis genome sequence and only tags with sequences that were 100% identical were retained for further comparison, yielding 61,638 hits in the genome from 56,513 different tags (82%; Table 1). The majority of these tags—54,271—showed a single hit to the genome (96%; Table 1). The remaining 2242 tags matched two or more genomic regions, likely indicating genuine low-copy repeats in the genome that were nevertheless deconvoluted. The average distance between the uniquely mapped tags equaled 2100 bp. As each EcoRI site can theoretically generate a forward and a reverse WGP tag, this is an accurate reflection of the ∼4.5-kb average distance between the EcoRI sites in the genome, taking into account that not all possible tags are indeed recovered. The in silico EcoRI/MseI digest of the Columbia genome that was generated for validation of the experimental data yielded 69,275 fragments. When trimmed to 26 nt to match the experimental WGP analysis, these fragments represented 60,676 different tags, 57,992 (96%) of which represented a unique position in the genome. Eighty-six percent (51,935 tags) of the theoretically possible 60,676 unique in silico tags were also retrieved in the wet-lab WGP experiment (Table 1).
Contig building using FPC and map quality assessment
A cut-off value of 1.0 × 10−6 and a consecutive DQ step was used in the FingerPrinted Contig (FPC) software (Soderlund et al. 1997) to generate the WGP map, as it produced a high level of BACs in contigs with a small number of questionable clones that may cause false overlaps. This resulted in 273 contigs representing 4048 BACs and 61,514 WGP tags. The distribution of contig sizes and number of BACs is shown in Supplemental Fig. S3. These contigs covered ∼101 Mb (78%) of the genome, using a 2100-bp average distance between WGP tags as a basis for the coverage calculation. Next to the contigs, 551 singleton BACs remained, including the majority of the clones containing less than five tags (Fig. 2). At the FPC settings used in this experiment, the minimum number of WGP tags on a contiged BAC was two, even though only a limited number of such BACs were incorporated.
To validate correctness of the BAC contigs that together form the whole genome physical map, the 65,734 WGP tag sequences were mapped to the Arabidopsis genome sequence using a perfect string match algorithm. Ninety-one percent of the 270 contigs containing two or more unique WGP tag hits mapped to contiguous regions of the genome and covered 82 Mb, representing 81% of the contig coverage (Table 3). However, 23 contigs mapped to two distinct regions and one contig mapped to three regions. These 24 contigs (9%) comprised 19% of the contig coverage. To address this issue, a generic identification and purging tool was applied to remove 230 potentially problematic BACs from the data set, including 177 BACs that contained more than 80 WGP tags. With this filtered BAC data set, another FPC run was performed using the same settings. This resulted in 3813 BACs assembled into 362 contigs and 556 singleton BACs (Table 3). The 362 contigs represented a calculated genome coverage of 102.4 Mb, which, in spite of the removal of 230 BACs, is nearly the same as the unfiltered data set (100.8 Mb), albeit split over more contigs. Of these 362 contigs, 357 contained two or more WGP tags with unique hits to the genome, of which 350 (98%) mapped to contiguous genome regions and covered 99 Mb (97%). The remaining seven contigs (3%) mapped to multiple regions and comprised 3% contig coverage (Table 3). Figure 3 shows the alignment of a representative contig mapping to chromosome 3.
Table 3.
FPC results for contig building of WGP Arabidopsis for the initial physical BAC map (all BACs) and after filtering problematic BACs (filtered BACs). Indicated is whether contigs map to a single region or have multiple hits
Parameter settings: tolerance = 0; cut-off = 1.0 × 10−6; DQ step 1
Figure 3.
Overview (A) and zoomed-in detail (B) of a typical BAC contig located on Arabidopsis chromosome 3. The sequence of the WGP tags, the chromosome number, and the base pair position in the chromosome of the first base of the WGP tag are shown. In each color-coded BAC, the presence of a WGP tag is indicated by “x.” Gaps in BACs represent missing tags due to insufficient deconvoluted reads.
Scalability of WGP to larger and more complex genomes
To further substantiate the robustness of the WGP method, four additional plant genomes were subjected to WGP, comprised of melon (Cucumis melo, 450 Mb genome size), tomato (Solanum lycopersicum, 950 Mb), the allotetraploid rape seed (Brassica napus, 1200 Mb), and lettuce (Lactuca sativa, 2600 Mb). Data presented in Supplemental Table S1 demonstrate that, as in Arabidopsis, these genomes were amenable to WGP and thus generated high- quality, high-coverage whole genome physical maps.
Discussion
In this manuscript, whole genome profiling is demonstrated as a novel approach to construct sequence-based genome-wide physical maps of complex genomes. Restriction fragment-based sequence data from pooled BAC clones were generated using the Illumina GA NGS platform and resulted in the assignment of an average of 40 unique WGP tags per BAC clone. Using these tags, a whole genome physical map of Arabidopsis thaliana was created using a 6× BAC library. Validation of the WGP map using the Columbia reference genome sequence confirmed that 350 BAC contigs (98%) were assembled correctly, which comprised 97% of 102 Mb calculated genome coverage, equaling ∼80% of the genome of Arabidopsis.
A first version of the Arabidopsis WGP map was produced using stringent cut-off settings in the FPC software. As the actual position of the WGP tags in the released genome sequence of Arabidopsis could easily be established, the quality of the contigs produced by WGP was investigated in detail. This demonstrated that tags belonging to the vast majority of the contigs originated from a single region in the genome (see e.g., Fig. 3). However, tags present in 24 of the 273 contigs mapped to more than one region in the genome. Careful inspection of these contigs showed that, in all these cases, single BACs were responsible for the merger of two distinct genomic regions. The Arabidopsis BAC library that was used in this study contained either a chimeric BAC or multiple BACs per well in less than 0.6% of the plate wells. However, due to the integrating power of the FPC assembly, this had a profound effect on the WGP map, as nearly 9% of the contigs obtained from the first round of FPC using unfiltered BACs was not contiguous and represented 19% of the contig coverage. In order to minimize chimeric contig formation, a generic method was developed that does not rely on a reference genome sequence to recognize and eliminate problematic BACs. The selection of these BACs was based on two observations: first, that many of the disturbing BACs produced a significantly higher number of tags compared to the genome fraction they contained, suggesting the presence of two separate genome regions; and second, chimeric contigs showed a typical tag distribution, which allowed pinpointing of potentially chimeric contigs, identification of BACs causing the chimerism, and a purge of these BACS from the data set.
As a result, the final physical map based on the filtered BAC data set retained its coverage but was split into more contigs. Seven contigs, containing 130 BACs, were still identified as split over two genome regions. However, this covered no more than 3% of the genome, a marked improvement over the 19% coverage of chimeric contigs prior to filtering (Table 3). It should be noted that the genome coverage of this experiment (6×) is relatively low and that contig building with FPC of deeper coverage BAC libraries under high stringency settings will prevent formation of chimeric contigs more easily, particularly in combination with the purge tool.
A number of general features of the WGP technology are worth addressing in more detail, such as the influence of sequence read length, pooling schemes, restriction enzyme choice, and genome size on map resolution and cost effectiveness.
In silico data, confirmed by experimental data, demonstrated that a tag length of 20 nt was sufficient to define more than 90% of the unique sequence tags in the Arabidopsis genome. This is in agreement with results from previous studies on the effect of read length on determining unique positions in the genome (Whiteford et al. 2005; Chaisson et al. 2009). A 20-nt tag length, even when adding 6 nt for a pool identifier, is well within the sequence length output of contemporary high-throughput sequencing machines. Additional analyses on other (partially) sequenced genomes and our simulation of the maize genome demonstrated that, even for significantly larger genomes, tag lengths between 26 and 31 nt are sufficient for WGP.
The 2D pooling strategy that was chosen to process the limited number of 6144 Arabidopsis BACs was a trade-off between costs of sample preparation and sequencing using the Illumina GA and genome size. Since sample preparation of single BACs was deemed too costly, it was decided to pool BACs prior to DNA isolation and then apply the restriction, ligation, and fragment amplification to pooled DNAs in combination with deconvolution of sequencing results. The backbone of the contiging strategy is the occurrence of unique WGP tags in two or more BACs that share the same portion of the genome. If, however, these two BACs or fragments thereof appear in the same pooling set (“deconvolution space”), the WGP tag will be lost in the deconvolution process, as it will show up in three or more BAC pools. The 6× BAC library we used was divided over sixteen 384-well plates. A pooling set size of a full plate (384 BACs with a 125 kb average insert size) thus covers ∼48 Mb (40%) of the Arabidopsis genome. The chance of finding multiple BACs that contain fragments from the same genomic region within a single pool is, therefore, relatively high. In general, the probability of encountering the same genomic region in a single pool will depend on the size of the genome related to the complexity of the pools. Thus, WGP analyses on small genomes require less complex pools, whereas large genomes allow larger numbers of BACs per pool. Furthermore, for each WGP tag on each BAC, a minimum redundancy is required to compensate for random sampling variation. This implies that a 2D pooling set-up requires twice the number of sequence reads per BAC compared to individual BAC clone sequencing (1D), whereas a 3D pooling scheme requires three times as many reads. Depending on relative costs of sequencing compared to the costs of sample preparation, an optimal pooling scheme can be designed. As the cost of sequencing per nucleotide is rapidly dropping in the NGS systems used to date, it is likely that higher-level pooling schemes will become cost effective in the near future.
A particular advantage of using BAC DNA pooling and the consecutive deconvolution procedure in WGP is the selection against tags derived from repetitive regions, which often hamper building accurate contigs in fingerprint-based physical mapping methods. As repeat tags are likely to occur in multiple BACs within a pooling set, the majority will not be assigned to BACs by deconvolution and excluded from the input file for contig building. As a result, only unique or low-copy regions of BACs are used for contig building, and this is beneficial for utility of the map for integrating sequence scaffolds or marker development. On the other hand, this also implies that BAC clones containing predominantly repeated sequences may end up with too few unique WGP tags for contiging and will not be represented in the WGP map. Indeed, it was our observation that singleton BACs were characterized by far fewer WGP tags per BAC (Fig. 2).
Earlier studies have reported on building BAC-based physical maps from pooled BAC libraries by using Sanger sequence tags called ST-PGI (Milosavljevic et al. 2005) or mapping Sanger end sequences of BAC clones called ESP (Volik et al. 2003). The main difference between these approaches is that ST-PGI and ESP require a sequenced reference genome for mapping and alignment of sequence reads. Without a reference genome template, ST-PGI cannot deconvolute the BAC clone addresses (plate and well position). However, WGP uses an AFLP-based reduced representation library, which enables the direct deconvolution of all BAC clone addresses into highly reproducible WGP tags and thus enables the generation of an ultra-high-density physical map on a de novo genome (no previous sequence information is needed with WGP).
EcoRI/MseI was used for the Arabidopsis WGP, but the use of amplified restriction fragments as a starting point in the WGP concept is flexible in the choice of restriction enzyme(s). Commonly used restriction enzymes such as EcoRI or HindIII typically generate between 30 and 50 fragments per BAC clone. Depending on the abundance of restriction sites in a given species, the use of alternative enzymes will allow additional fine tuning in the number or distribution of WGP tags per BAC. Depending on the complexity of a target genome, the use of specific restriction enzymes in combination with pooling complexity assures an optimal use of sequencing capacity to further improve cost effectiveness. In silico analysis of available sequence data can aid in providing the required information for novel WGP projects.
Additional results of WGP applied to melon, tomato, rape seed, and lettuce substantiate the robustness of this new technology (Supplemental Table S1). These data are complemented with the results from the in silico maize analysis (Supplemental data). Results show that, even for large and complex genomes that are known to contain a high proportion of repeated regions, WGP is efficient in tagging 70%–90% of the BAC clones and yields high coverage physical maps.
In conclusion, whole genome profiling was demonstrated to be an efficient novel NGS-based technique for whole genome physical map construction. In addition, WGP maps are an excellent scaffold to complement de novo whole genome sequencing efforts, as they consist of densely spaced unique sequence tags. To date, random shotgun whole genome sequencing (WGS) efforts in particular rely on high sequencing redundancies in combination with paired-end sequencing strategies to achieve sufficiently large sequence contigs and scaffolds. WGP maps, as they consist of sequence-tagged contiged BACs, provide multiple anchor points over distances that by far exceed those that can be bridged by current paired-end sequencing strategies. It can thus be expected that WGP will contribute to significantly improved genome sequence assembly metrics and cost reduction for WGS efforts, because very high sequencing redundancy levels are no longer needed to obtain large scaffolds. In addition, like fingerprint-based physical methods, WGP maps provide a “minimum tiling path” of BAC clones for targeted sequencing of genome regions of particular interest and direct access to individual BAC clones. These features of WGP and the widespread availability of BAC libraries underscore the power of WGP to advance genome analysis in a broad range of species.
Methods
In silico analysis
The complete Arabidopsis thaliana ecotype Columbia sequence (TAIR8; http://www.arabidopsis.org) was digested in silico with EcoRI/MseI to obtain all possible restriction fragments. An analysis was performed to obtain the number of unique WGP tags after trimming them to varying read lengths from 10 to 26 nt from the EcoRI site. Data were compared to the number of tags resulting from the in vitro WGP experiment.
BAC library and pooling strategy
A 6144 clone BAC library of the Arabidopsis thaliana Col-1 ecotype (available at the ABRC stock center, stock # CS6000) and two-dimensionally (2D) pooled BAC DNAs were made available for this project by Amplicon Express, Inc. (Pullman, WA, USA). The 2D pooling strategy was based on a simple 384-well plate by rows and by columns pooling strategy implemented with a liquid handling robot (Beckman Coulter, Biomek 2000, Brea, CA, USA). The robot pooled 75 μl of freshly grown bacterial culture in 2× YT media from all of the clones from each row (24 BACs) on each plate and 112 μl from all of the clones from each column (16 BACs). The set of 16-row pools and 24-column pools is termed a SuperPool (SP). The DNA tray format allows culture liquid to be collected from two individual 384-well library plates (two SPs) and have all 80 wells processed independently for high quality DNA extraction in a 96-well deep block format. The collected culture fluid is centrifuged to pellet the cells and discard the supernatant. The blocks of cell pellets are then frozen at -20°C until they are processed with a modified alkaline lysis DNA extraction protocol (Maniatis et al. 1982). The average yield of plasmid DNA is ∼200 ng per well of high-quality, Illumina sequencing-grade DNA.
Sequencing sample preparation
AFLP templates were prepared from the pooled BAC clone DNA as described by Vos and co-workers (Vos et al. 1995). Twenty ng of pooled BAC DNA was digested using 5 units EcoRI and 2 units MseI for at least 1 h at 37°C. Next, adapter-ligation using a universal P7 MseI adapter and a sample-specific tagged EcoRI P5 adapter was carried out for 3 h at 37°C. PCR was performed in 20 μl and contained 5 μl 10-fold diluted restriction ligation mixture, 30 ng Illumina P5 primer (5′-AATGATACGGCGACCACCG-3′), 30 ng Illumina P7 primer (5′-CAAGCAGAAGACGGCATACGA-3′), 0.2 mM dNTPs, 0.4 U AmpliTaq (Applied Biosystems) and 1× AmpliTaq buffer. PCR was performed with the following profile: 2 min at 72°C, followed by 22 cycles of 30 sec each at 94°C, 60 sec at 56°C, 60 sec at 72°C, and finally held at 4°C. Next, equal amounts of SP samples were purified using the QIAquick PCR Purification Kit (Qiagen). All 80 sample-specific EcoRI P5 adapters included a unique 5-nt sample identification tag adjacent to the EcoRI restriction site overhang for identification of individual BACs by deconvolution.
Sequencing
Two Illumina Genome Analyzer runs were performed, a titration run and a full scale run. Each run was done on a flow cell divided into eight lanes for physical separation of samples, such that the same set of sample tags was used for each lane. The first run contained only four SPs, with two sets of two SPs replicated on three lanes, each in a range of increasing concentration (1.5, 2.0, and 2.5 pM). The 2.5 pM lanes were selected as the optimal DNA concentration, and a second GA run was performed comprising six lanes with Arabidopsis WGP samples each covering two SPs represented in 80 row- or column pools. A total of 16 SPs were sequenced equaling 16 × 384 = 6144 BACs.
The Illumina pipeline software (GA pipeline, v0.3) was used to analyze images into sequence reads of 31-nt length. An additional quality filter was applied to select only those reads with all base calls being at least zero on the Illumina GA scale. All sequence data have been deposited in the NCBI Sequence Read Archive (SRA) SRA026464.
Deconvolution
Sequence reads were split into three parts to enable assignment of unique tags to pools and to allow for consecutive deconvolution into individual BACs: The first 5 nt represent the sample (i.e., BAC pool) identification tag or barcode; the next 6 nt match the EcoRI restriction site of the adapter, and the remaining 20 nt define the WGP tag of Arabidopsis genomic DNA sequence from BAC clone inserts. The assignment of unique WGP tags to individual BACs was based on the following criteria: (1) A specific WGP tag must occur in two pools to indicate its unique position on the plate; one column and one row pool with both being represented by at least two reads; and (2) if WGP tags were inadvertently observed in a third or fourth pool, the number of reads in these other pools must be less than a tenth of those in the smallest (correct) pool. All WGP tags not matching these criteria were discarded. Perl scripts were used to recognize and trim the sample identification tags and the restriction site part of the sequence reads and to perform the deconvolution. Unique WGP tags were defined by grouping them in 100% identical read sets. The output of this procedure consisted of a list of all WGP tags, the corresponding number of reads, and the identification number of the unique BAC to which they were assigned.
Contig building
Contiging was performed using the FingerPrinted Contig (FPC) program (v9.4; Soderlund et al. 1997; http://www.agcol.arizona.edu/software/fpc/). This software tool was originally developed for analyzing BAC fingerprint data—restriction fragments determined by their length. The WGP tags were adapted for use in FPC by converting each unique sequence tag into a number, yielding pseudo restriction fragment sizes for which the FPC software was originally designed. As the WGP tags are uniquely defined by their sequence composition, FPC could be used at the highest stringency setting of tolerance (value = 0). Normally this setting compensates for variation in band mobilities of restriction fragments, which is not an issue here. Different cut-off values were tested, specifying the threshold on the probability of BAC coincidence, i.e., the likelihood that different BACs share overlapping WGP tags, while not originating from the same genomic region. A DQ analysis was performed to further split problematic contigs with so-called Q-clones, i.e., clones which cause potential false overlap. The output of FPC consisted of a list of contigs and the corresponding order of BACs within each contig.
Map validation
WGP tag data, including the adjacent EcoRI restriction site, were mapped to the Arabidopsis genome sequence. All hits with a 100% identity and full-length alignment were recorded, and the corresponding WGP tags were subsequently characterized by their chromosome number and base pair position. Visual alignments were made from the sorted position of the WGP tags, based on their location in the genome compared to their orientation on the BAC contigs. To check the quality of the resulting FPC contigs, all tags were included which showed a single hit to the genome reference. For each contig with two or more of such unique tags, the tag positions were verified in the following way: The median genome position was calculated for the set of tags, and tags were classified as outliers if their position was more distant from the median than a given threshold (the number of unique tags in this contig multiplied by 5000 bp, approximately twice the mean distance between tags). Contigs with more than five outliers were identified as hybrid contigs, mapping to multiple regions on the genome and checked as such. All other contigs with two or more unique tags were defined as good-quality contigs.
Problematic BAC identification and purging
To overcome false (chimeric) contig formation caused by problematic BACs, e.g., BACs with chimeric inserts or two BACs which were accidentally deposited in the same well of a 384-well plate, a three-step procedure was developed. First, BACs containing more than twice the mean number of tags were excluded from the analysis, as they represented a large fraction of problematic clones relative to their total number. Second, FPC contig building was performed, and the resulting contigs were scored on two metric parameters which correlated with chimeric contig features. These metrics are the fraction of BAC pairs within a contig sharing at least one WGP tag (C1) and the average tag density in a contig as defined by the number of unique WGP tags in the contig divided by the number of BACs in the contig (C2). Empirically, the square of C1 divided by C2 provided a value that effectively discriminated chimeric contigs from contiguous contigs at a threshold of 0.003 and lower. The rationale for C1 was that chimeric contigs were expected to represent two proper contigs that were incorrectly linked by a single problematic BAC, causing the fraction of BAC pairs that share at least one WGP tag (C1) to drop. At the same time, the total number of WGP tags represented in the contig increases compared to the genome coverage it represents (C2). Third, to identify the problematic BACs within putatively chimeric contigs, an iterative approach of removing individual BACs from these contigs based on transitivity clustering was performed for all BACs. BACs were eliminated from the data set if their removal led to breaking up the contig. The filtered BAC data set was used for a final FPC analysis using the same stringency settings.
All scripts used to perform the WGP experiments as described are freely available after registering at http://www.keygene.com/research/WGPpublication.php.
Acknowledgments
This work was supported by Enza Zaden, Rijk Zwaan, Vilmorin & Cie, and Takii & Co. We are very grateful to Drs. William Nelson and Carol Soderlund of the University of Arizona for adapting the FPC program for WGP data processing. The AFLP and Whole Genome Profiling (WGP) technologies are covered by patents and patent applications owned by Keygene N.V. AFLP and KeyGene are registered trademarks of Keygene N.V.
Footnotes
[Supplemental material is available for this article. The sequence data from this study have been submitted to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession no. SRA026464.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.112094.110.
References
- Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One 3: e3376 doi: 10.1371/journal.pone.0003376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker E, Butterbach P, Rouppe van der Voort JN, van der Vossen EA, van Vliet J, Bakker J, Goverse A 2003. Genetic and physical mapping of homologues of the virus resistance gene Rx1 and the cyst nematode resistance gene Gpa2 in potato. Theor Appl Genet 106: 1524–1531 [DOI] [PubMed] [Google Scholar]
- Borm TJA 2008. “Construction and use of a physical map of potato.” PhD thesis, Wageningen University, The Netherlands [Google Scholar]
- Cai W-W, Chen R, Gibbs RA, Bradley A 2001. A Clone-Array Pooled Shotgun Strategy for sequencing large genomes. Genome Res 11: 1619–1623 [DOI] [PubMed] [Google Scholar]
- Cardle L, Ramsay L, Milbourne D, Macaulay M, Marshall D, Waugh R 2000. Computational and experimental characterization of physically clustered simple sequence repeats in plants. Genetics 156: 847–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson MJ, Brinza D, Pevzner PA 2009. De novo fragment assembly with short mate-paired reads: Does the read length matter? Genome Res 19: 336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church DM, Goodstadt L, Hillier LW, Zody MC, Goldstein S, She X, Bult CJ, Agarwala R, Cherry JL, DiCuccio M, et al. 2009. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol 7: e1000112 doi: 10.1371/journal.pbio.1000112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory SG, Howell GR, Bentley DR 1997. Genome mapping by fluorescent fingerprinting. Genome Res 7: 1162–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier LW, Marth GT, Quinlan AR, Dooling D, Fewell G, Barnett D, Fox P, Glasscock JI, Hickenbotham M, Huang W, et al. 2008. Whole-genome sequencing and variant discovery in C. elegans. Nat Methods 5: 183–188 [DOI] [PubMed] [Google Scholar]
- Islam-Faridi MN, Childs KL, Klein PE, Hodnetta G, Menz MA, Klein RR, Rooney WL, Mullet JW, Stelly DM, Price HJ 2002. A molecular cytogenetics map of sorghum chromosome 1: FISH analysis with mapped BACs. Genetics 161: 345–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, et al. 2008. Mapping and sequencing of structural variation from eight human genomes. Nature 453: 56–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein PE, Klein RR, Cartinhour SW, Ulanch PE, Dong J, Obert JA, Morishige DT, Schlueter SD, Childs KL, Ale M, et al. 2000. A high-throughput AFLP-based method for constructing integrated genetic and physical maps: Progress towards a sorghum genome map. Genome Res 10: 789–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman WJM, Gort G 2004. Significance tests and weighted values for AFLP similarities, based on Arabidopsis in silico AFLP fragment-length distributions. Genetics 167: 1915–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin HA, Larkin DM, Pontius J, O'Brien SJ 2009. Every genome sequence needs a good map. Genome Res 19: 1925–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo MC, Thomas C, You FM, Hsiao J, Ouyang S, Buell CR, Malandro M, McGuire PE, Anderson OD, Dvorak J 2003. High-throughput fingerprinting of bacterial artificial chromosomes using the snapshot labeling kit and sizing of restriction fragments by capillary electrophoresis. Genomics 82: 378–389 [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J 1982. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- Marra MA, Kucaba TA, Dietrich NL, Green ED, Brownstein B, Wilson RK, McDonald KM, Hillier LW, McPherson JD, Waterston RH 1997. High throughput fingerprint analysis of large-insert clones. Genome Res 7: 1072–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milosavljevic A, Harris RA, Sodergren EJ, Jackson AR, Kalafus KJ, Hodgson A, Cree A, Dai W, Csuros M, Zhu B, et al. 2005. Pooled genomic indexing of rhesus macaque. Genome Res 15: 292–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson W, Soderlund C 2009. Integrating sequence with FPC fingerprint maps. Nucleic Acids Res 37: e36 doi: 10.1093/nar/gkp034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson WM, Bharti AK, Butler E, Wei F, Fuks G, Kim H, Wing RA, Messing J, Soderlund C 2005. Whole-genome validation of high-information-content fingerprinting. Plant Physiol 139: 27–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounsley S, Marri PR, Yu Y, He R, Sisneros N, Goicoechea JL, Lee SJ, Angelova A, Kudrna D, Luo M, et al. 2009. De novo next generation sequencing of plant genomes. Rice 2: 35–43 [Google Scholar]
- Sasaki T, Matsumoto T, Yamamoto K, Sakata K, Baba T, Katayose Y, Wu J, Niimura Y, Cheng Z, Nagamura Y, et al. 2005. The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Schwartz DC, Li X, Hernandez L, Ramnarain SP, Huff EJ, Wang YK 1993. Ordered restriction maps of Saccharomyces cerevisiae chromosomes constructed by optical mapping. Science 262: 110–114 [DOI] [PubMed] [Google Scholar]
- Shizuya H, Birren B, Kim UJ, Mancino V, Slepak T, Tachiiri Y, Simon M 1992. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci 89: 8794–8797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderlund C, Longden I, Mott R 1997. FPC: A system for building contigs from restriction fingerprinted clones. Comput Appl Biosci 13: 523–535 [DOI] [PubMed] [Google Scholar]
- Srinivasan J, Sinz W, Jesse T, Wiggers-Perebolte L, Jansen K, Buntjer J, van der Meulen M, Sommer RJ 2003. An integrated physical and genetic map of the nematode Pristionchus pacificus. Mol Gen Genet 269: 715–722 [DOI] [PubMed] [Google Scholar]
- van der Vossen EA, van der Voort JN, Kanyuka K, Bendahmane A, Sandbrink H, Baulcombe DC, Bakker J, Stiekema WJ, Klein-Lankhorst RM 2000. Homologues of a single resistance-gene cluster in potato confer resistance to distinct pathogens: A virus and a nematode. Plant J 23: 567–576 [DOI] [PubMed] [Google Scholar]
- Volik S, Zhao S, Chin K, Brebner JH, Herndon DR, Tao Q, Kowbel D, Huang G, Lapuk A, Kuo WL, et al. 2003. End-sequence profiling: Sequence-based analysis of aberrant genomes. Proc Natl Acad Sci USA 100: 8794–8797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, et al. 1995. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res 23: 4407–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F, Coe E, Nelson W, Bharti AK, Engler F, Butler E, Kim H, Goicoechea JL, Chen M, Lee S, et al. 2007. Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet 3: e123 doi: 10.1371/journal.pgen.0030123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, et al. 2008. The complete genome of an individual by massively parallel DNA sequencing. Nature 452: 872–876 [DOI] [PubMed] [Google Scholar]
- Whiteford N, Haslam N, Weber G, Prügel-Bennett A, Essex JW, Roach PL, Mark Bradley M, Cameron Neylon C 2005. An analysis of the feasibility of short read sequencing. Nucleic Acids Res 33: e171 doi: 10.1092/nar/gni171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CC, Nimmakayala P, Santos FA, Springman R, Scheuring C, Meksem K, Lightfoot DA, Zhang HB 2004. Construction and characterization of a soybean bacterial artificial chromosome library and use of multiple complementary libraries for genome physical mapping. Theor Appl Genet 109: 1041–1050 [DOI] [PubMed] [Google Scholar]
- Zhou S, Wei F, Nguyen J, Bechner M, Potamousis K, Goldstein S, Pape L, Mehan MR, Churas C, Pasternak S, et al. 2009. A single molecule scaffold for the maize genome. PLoS Genet 5: e1000711 doi: 10.1371/journal.pgen.1000711 [DOI] [PMC free article] [PubMed] [Google Scholar]