Abstract
Small proteins (10–200 amino acids [aa] in length) encoded by short open reading frames (sORF) play important regulatory roles in various biological processes, including tumor progression, stress response, flowering, and hormone signaling. However, ab initio discovery of small proteins has been relatively overlooked. Recent advances in deep transcriptome sequencing make it possible to efficiently identify sORFs at the genome level. In this study, we obtained ∼2.6 million expressed sequence tag (EST) reads from Populus deltoides leaf transcriptome and reconstructed full-length transcripts from the EST sequences. We identified an initial set of 12,852 sORFs encoding proteins of 10–200 aa in length. Three computational approaches were then used to enrich for bona fide protein-coding sORFs from the initial sORF set: (1) coding-potential prediction, (2) evolutionary conservation between P. deltoides and other plant species, and (3) gene family clustering within P. deltoides. As a result, a high-confidence sORF candidate set containing 1469 genes was obtained. Analysis of the protein domains, non-protein-coding RNA motifs, sequence length distribution, and protein mass spectrometry data supported this high-confidence sORF set. In the high-confidence sORF candidate set, known protein domains were identified in 1282 genes (higher-confidence sORF candidate set), out of which 611 genes, designated as highest-confidence candidate sORF set, were supported by proteomics data. Of the 611 highest-confidence candidate sORF genes, 56 were new to the current Populus genome annotation. This study not only demonstrates that there are potential sORF candidates to be annotated in sequenced genomes, but also presents an efficient strategy for discovery of sORFs in species with no genome annotation yet available.
In recent years, individual experiments have demonstrated that small proteins (<200 amino acids [aa] in length), encoded by short open reading frames (sORF), play a major role in plant and animal development, e.g., the TAL protein (11 aa) influencing fruit fly development (Galindo et al. 2007), the Cg-1 protein (<33 aa) controlling the tomato–nematode interaction (Gleason et al. 2008), the CLE family proteins (75–140 aa) (Fletcher et al. 1999; Trotochaud et al. 2000; Muller et al. 2008; Oelkers et al. 2008) involved in Arabidopsis meristem development, the galectin-1 protein (∼130 aa) associated with the malignant human tumor progression (Camby et al. 2006), the lipid-binding protein AZI1 (161 aa) involved in priming plant defenses (Jung et al. 2009), and the FLOWERING LOCUS T (FT) protein (175 aa) acting as a long-range signal regulating flowering (Notaguchi et al. 2008).
Although small proteins play important roles in regulation of biological processes, genome-wide identification and characterization of sORFs has been limited. Typically, an arbitrary minimum open reading frame (ORF) cutoff (e.g., 100 aa) is applied in gene annotation algorithms to reduce the likelihood of falsely categorizing non-protein-coding RNAs (ncRNAs) as mRNAs (Dinger et al. 2008). As a result, sORF genes are under-represented in many current genome annotations. By searching for ORFs, Lease and Walker (2006) identified 33,809 unannotated Arabidopsis ORFs encoding small proteins between 25 and 250 aa in length, out of which 10,247 (30%) had expression evidence from genome-wide tiling hybridization experiments. Hanada et al. (2007) performed a large-scale search for sORFs encoding proteins of 30–100 aa in the intergenic regions of the Arabidopsis genome using a simple gene-finding method. They identified 7159 sORF candidates, of which 3241 had either transcriptional evidence or indication of purifying selection. Based on this research, Hanada et al. (2010) developed a program package, sORF Finder, for identifying sORFs according to the nucleotide composition bias among coding sequences and the potential functional constraint at the amino acid level through evaluation of synonymous and nonsynonymous substitution rates. However, only 2% of unannotated sORFs predicted by Hanada et al. (2007) were confirmed by the Arabidopsis proteomic data (Castellana et al. 2008). The sORF-finding approaches used by Lease and Walker (2006) and Hanada et al. (2007) are solely in silico gene predictions based on genomic DNA sequence, with the assumption that small proteins are encoded by intronless (i.e., single-exon) genes. In silico prediction of full-length transcripts based on genomic sequences is challenging and has low accuracy (sensitivity ranging from 41% to 68% and specificity from 20% to 53%) (http://augustus.gobics.de/accuracy). Thus, an alternative strategy is needed for identifying protein-coding sORFs.
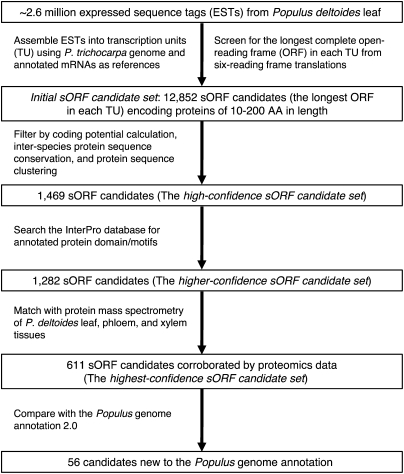
Here we report the outcome of an integrative procedure based on transcriptomics, proteomics, and computational biology for the discovery of sORFs that encode small proteins <200 aa in length in Populus deltoides. Our strategy for large-scale discovery of small proteins is outlined in Figure 1. Briefly, a three-step approach was used to reconstruct transcription units (TU) using expressed sequence tags (EST) obtained from deep sequencing of the P. deltoides leaf transcriptome. Since a true protein-coding transcript is more likely to have a long and high-quality ORF compared with a non-coding transcript (Kong et al. 2007), we established an initial sORF candidate set by selecting the longest ORF-encoding protein sequence of <200 aa in length for each TU. Then we applied three computational approaches sequentially to enrich for protein-coding sORFs: (1) coding-potential prediction based on known protein sequences, (2) evolutionary conservation between P. deltoides and other plant species, and (3) protein sequence clustering within P. deltoides.
Figure 1.
The strategy for large-scale discovery of small proteins in Populus deltoides.
The efficiency of our sORF discovery strategy was validated by both bioinformatics (e.g., protein domain-scanning) and experimental approaches (e.g., protein mass spectrometry). This study not only demonstrates that there are many potential sORF candidates yet to be annotated in sequenced genomes, but also presents an efficient strategy for sORF discovery in species with as yet unannotated genomes.
Results
Establishment of an initial sORF candidate set
We sequenced the transcriptome of six Populus deltoides leaf samples and generated ∼2.6 million ESTs with a median length of ∼240 nucleotides. We comparatively examined the representation of 100 annotated P. trichocarpa gene models encoding proteins <200 aa in length (http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html) using the ∼634,000 ESTs from one of the six leaf samples. Twenty-five of these selected gene models were found in the EST data, with 80% (= 20/25) of them having full-length or alternative splicing coverage (Supplemental Fig. 1; Supplemental Table 1), indicating that our EST data provided an appreciable full-length coverage of transcripts encoding proteins <200 aa in length. To minimize the number of truncated TU reconstructed from all ESTs, a three-step approach was utilized to create full-length TUs: (1) high-stringency de novo assembly, followed by (2) genome-location-based assembly, and then (3) medium-stringency assembly. Since there was only ∼1% divergence between P. trichocarpa and P. deltoides at the genomic sequence level (data not shown), the genomic resources (genome sequence and annotated mRNA sequences) in P. trichocarpa were used as references for P. deltoides EST assembly. As such, the P. deltoides ESTs, pooled with the annotated P. trichocarpa mRNA sequences (Tuskan et al. 2006), were assembled into TUs that each contained at least three ESTs. From these TUs, an initial sORF candidate set (Fig. 1) encoding 12,852 proteins of 10–200 aa in length was created by including the longest possible complete ORF that contained start and stop codons in six-frame translations from each TU.
Enrichment for protein-coding sORFs from the initial sORF candidate set
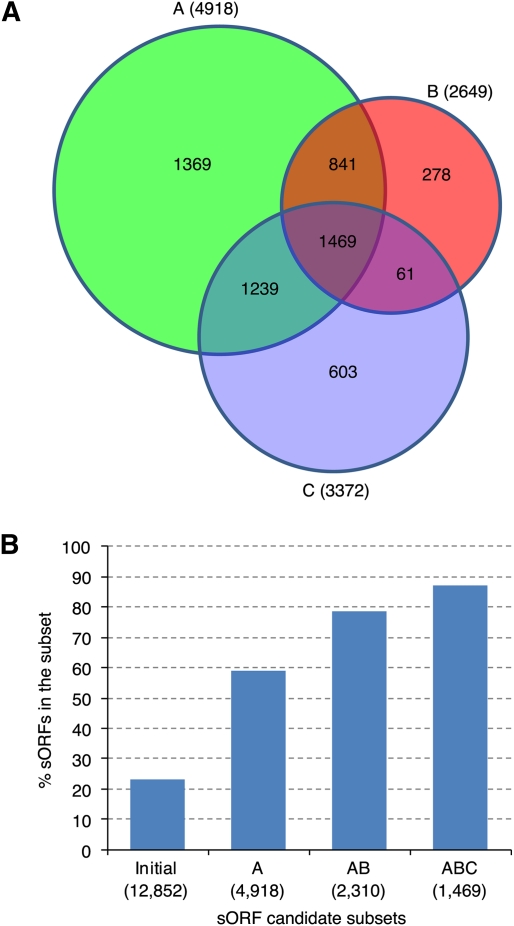
Three computational approaches were used to enrich for protein-coding sORFs to address the challenge of identifying protein-coding genes from a large number of short TUs assembled directly from ESTs. First, we interrogated the initial sORF candidate set using the Coding Potential Calculator (Kong et al. 2007) trained with protein sequences obtained from the UniProt database (The UniProt Consortium 2009). This approach identified 4918 sORF candidates with high protein-coding potential, designated as Subset A (Fig. 2A). Second, we compared the initial sORF candidate set derived from P. deltoides with 14 additional plant genome sequences ranging from algae to angiosperm species (Supplemental Fig. 2) and identified 2649 sORFs that are conserved between P. deltoides and at least one other plant species, designated as Subset B (Fig. 2A). The number of conserved sORFs between P. deltoides and the 14 tested species ranged between 300 and 2076 and as expected, the number of conserved sequences was inversely proportional to evolutionary distance (Supplemental Fig. 2). Finally, we performed a clustering analysis of the initial sORF candidate set and detected 3372 sORFs that clustered into families with 3–51 members, designated as Subset C (Fig. 2A). The 1469 sORF candidates shared by Subsets A, B, and C were designated as the high-confidence sORF candidate set (Figs. 1, 2A).
Figure 2.
P. deltoides small protein-coding candidate genes enriched from transcription units. (A) Number of genes in different sORF candidate subsets. (B) Proportion of the sORF subsets having known protein domains detected by InterProScan. Subset A contains the sORF candidates with high protein-coding potential predicted using known proteins as training sequences. Subset B contains sORF candidates conserved between P. deltoides and at least one other plant species. Subset C contains sORF candidates clustered into families. (Initial) The initial sORF candidate set (Fig. 1). (AB) The intersection of Subsets A and B. (ABC) (i.e., the high-confidence sORF candidate set) The intersection of Subsets A, B, and C. The value in parentheses represents the number of sORFs in each individual subset.
Length distribution of protein sequences in the high-confidence sORF set
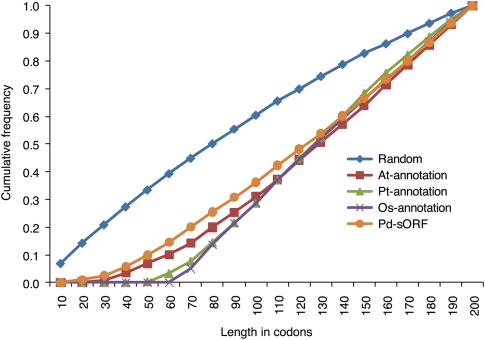
We examined the protein length distribution to assess whether the putative small protein sequences occurred more often than expected by chance alone. The frequency of protein sequences <100 aa in length in the high-confidence sORF candidate set was, as expected, lower than that in the random sequence set (Fig. 3), suggesting that sORFs in the high-confidence sORF candidate set are likely not randomly generated as a result of assembly errors. Moreover, the length distribution of the high-confidence sORF candidate set was similar to that of the small protein set (<200 aa) in the current Arabidopsis genome annotation (v9) (Fig. 3).
Figure 3.
Length distribution of predicted protein sequences. (Random) The random sORFs; (At-annotation) the small proteins in Arabidopsis genome annotation (v9); (Pt-annotation) the small proteins in Populus genome annotation (v2.0); (Os-annotation) the small proteins in rice genome annotation (v6.1); (Pd-sORF) the high-confidence sORF candidate set (Fig. 1) shared by Subsets A, B, and C in Figure 2.
The most recent annotations of the Arabidopsis genome (v8–9) include more small proteins relative to the earlier versions (v5–7) (Supplemental Fig. 3A). Out of the 1694 new protein sequences added to Arabidopsis genome annotation v8, 1079 (64%) gene models encode proteins of <200 aa in length (Supplemental Fig. 3B), indicating that incorporation of a large number of new small protein sequences is a key feature of the improved annotation. Similarly, the frequency of sequences <120 aa in length in the high-confidence sORF candidate set was greater than that found in the current annotation of Populus (Fig. 3), suggesting that sORFs are under-represented in the current annotation data sets in Populus, particularly for those proteins between 30 and 120 aa in length. Similar results were found within the rice genome (Fig. 3).
To evaluate the possibility that the sORF sequences were not full-length (i.e., truncated), we cloned full-length TUs for 15 sORFs (Supplemental Table 2) in the high-confidence sORF candidate set using the Rapid Amplification of cDNA Ends (RACE) technology. All of the 15 tested sORFs were confirmed as full-length sequences. Thus, it is likely that the sORFs in the high-confidence sORF candidate set are predominantly full-length transcript sequences.
Validation of the high-confidence sORF candidate set by analysis of protein domain and mass spectrometry data
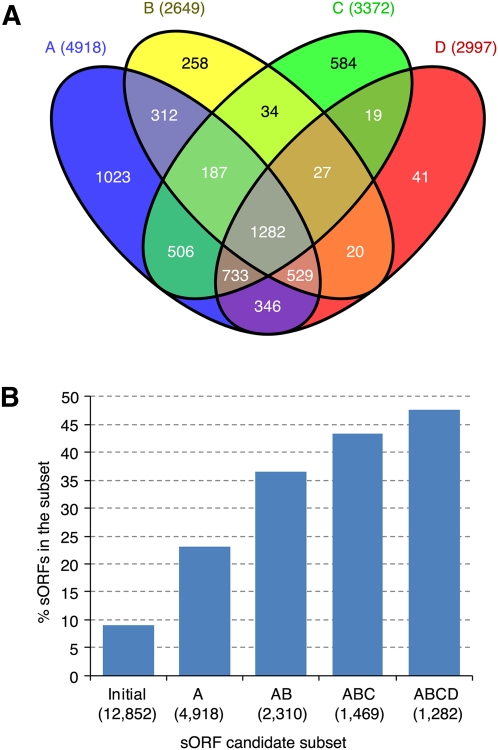
We subsequently surveyed the high-confidence sORF candidate set for known protein domain(s) using InterProScan, which integrates results obtained from searching 14 databases (Mulder and Apweiler 2007). Approximately 23% of the initial sORF candidate set had known protein domains. The protein domain discovery rate was increased by sequential filtering using coding potential prediction, interspecies conservation, and protein sequence clustering (Fig. 2B). We detected known protein domains in 1282 (87%) sORFs in the high-confidence sORF candidate set (Fig. 4A). These 1282 sORF candidates were designated as a higher-confidence sORF candidate set (Fig. 1).
Figure 4.
Protein domain annotation of sORF candidates. (A) Venn diagram showing the number of sORF candidates in four different subsets and their intersections. (B) Proportion of the sORF subsets having protein mass spectrometry data support. The “Initial” set, Subsets A, B, C, AB, and ABC are as described in Figure 2. Subset D contains sORF candidates with known protein domains detected by InterProScan. (ABCD) (i.e., the higher-confidence sORF candidate set) The intersection of Subsets A, B, C, and D. The value in parentheses represents the number of sORFs in each individual subset.
To further evaluate the validity of the sORF candidates, we surveyed existing P. deltoides xylem (Kalluri et al. 2009) as well as new xylem, phloem, and leaf protein mass spectrometry (MS) data (Supplemental Tables 3, 4). The MS analysis led to identification of 4943 different tryptic peptides, which were assembled into 1158 sORF-encoded proteins (Supplemental Table 3). Unique peptides were detected in one or more experiments for 307 sORF-encoded proteins (see “distinct” [DS] or “differentiable” [DF] sets in Supplemental Table 3). Only 9% (= 1158/12,852) of the initial sORF candidate set had proteomics matches, with the length of matched proteins ranging from 20 to 200 aa (Supplemental Fig. 4). The size of the sORF subset containing known protein domains was three times that of the sORF subset with proteomics support (Subset D vs. Subset P in Supplemental Fig. 5), indicating the possibility that deeper proteome coverage would provide additional evidence supporting more sORFs.
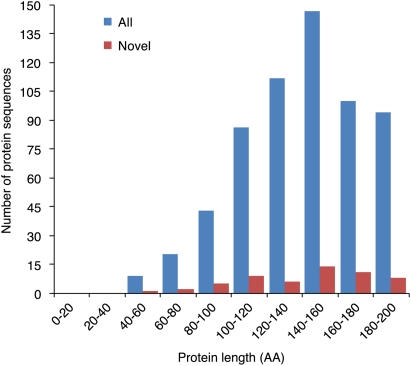
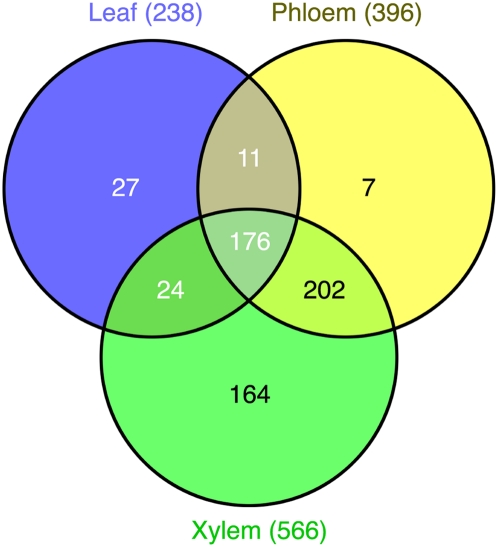
The proteomics-matching rate was increased by sequential filtering using coding potential prediction followed by analysis of interspecies conservation and protein sequence clustering (Fig. 4B). Our proteomics analyses revealed that ∼43% of the high-confidence sORF candidate set matched the xylem, phloem, or leaf protein MS data (Fig. 4B). Filtering of the high-confidence sORF candidate set by InterProScan search for known protein domains increased the proteomics-matching rate to 48%, with 611 sORFs in the higher-confidence sORF candidate set having proteomics support (Fig. 4B). These 611 proteomics-supported ORF candidates were designated as the highest-confidence sORF candidate set (Fig. 1), with protein length ranging from 40 to 200 aa (Fig. 5). Furthermore, 373 small proteins encoded by sORFs in the highest-confidence sORF candidate set were detected in proteomics measurements of conductive tissues (i.e., in phloem or xylem), but not in leaf (Fig. 6). Approximately 9% (56 protein sequences) of the highest-confidence sORF candidate set were misannotated in or missing from the P. trichocarpa genome annotation (v2.0; http://www.phytozome.net/) (Supplemental Table 5).
Figure 5.
Size distribution of the 611 sORF-encoded proteins in the highest confidence set. (All) All of the 611 proteins. (Novel) The 56 sORF-encoded small proteins new to the Populus genome annotation (v2.0; http://www.phytozome.net/).
Figure 6.
Venn diagram showing the number of sORFs from the 611-sORF set with the highest confidence that were detected in P. deltoides leaf, phloem, and xylem tissue based on analysis of the trypsin-digested whole proteome using two-dimensional HPLC interfaced with tandem mass spectrometry.
Possibility of sORF candidates as non-protein-coding RNA
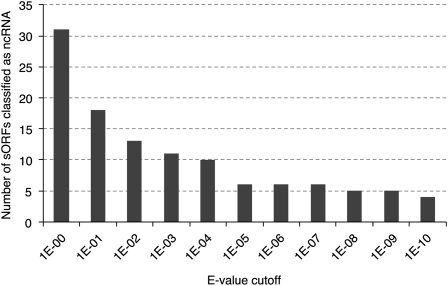
Many short RNA sequences have been identified as ncRNAs, as documented in the Rfam database (Gardner et al. 2009). To determine whether the high-confidence sORF candidate set contains potential ncRNAs, we conducted an Rfam-based search with all of its 1469 TU sequences using the Infernal program (Nawrocki et al. 2009) and found that only 0.3%–2.1% (4–31 TUs) of the high-confidence sORF candidate set were potential ncRNAs (Fig. 7), suggesting a low probability of ncRNAs (Rfam database) in high-confidence sORF candidate set.
Figure 7.
Number of sORFs in the high-confidence sORF candidate set classified as potential ncRNAs by an Rfam database search. The e-value cutoff was used in the Rfam search.
Discussion
Although small proteins have been shown to play important roles in various biological processes, they have largely escaped detection because it is difficult to predict sORFs (Kastenmayer et al. 2006; Dinger et al. 2008). Previous large-scale ab initio discovery of sORFs (Lease and Walker 2006; Hanada et al. 2007, 2010) identified thousands of single-exon genes directly from genomic sequences. Interestingly, nearly 50% of the annotated Arabidopsis genes encoding small proteins <100 aa in length contain introns (unpublished observation). For example, the Arabidopsis RCI2A gene encoding a small protein of 54 aa in length contains two introns (http://www.arabidopsis.org). The major limitation in these previous in silico sORF prediction efforts is that their sORF predictions were not designed to detect multiple-exon sORF genes. As a result, only 155 (∼2%) of 7442 sORFs predicted by Hanada et al. (2007) were verified by proteomics data (Castellana et al. 2008). Our approach integrates experimental data (transcriptome), coding potential prediction, evolutionary conservation, and gene family clustering. We reconstructed full-length transcription units (i.e., mRNAs) directly from the large volume of EST sequences obtained from deep sequencing of the transcriptome. In other words, experimental evidence provided the initial candidate set for our predictions. The sORF candidates predicted in this study had a relatively high rate of proteomics support. In our high-confidence sORF candidate set, ∼43% have protein MS data support. This rate is similar to the Arabidopsis whole-genome annotation, in which 40% of the gene models have protein MS support (Castellana et al. 2008).
In this study, we reconstructed TU directly from EST sequences, avoiding the uncertainty caused by ab initio prediction from genomic sequences. The EST sequences obtained from deep transcriptome sequencing provided numerous full-length transcripts (Supplemental Fig. 1; Supplemental Table 1) and all 15 RACE-tested TUs from the high-confidence sORF candidate set were shown to be full-length messages (Supplemental Table 2). This high-quality reconstruction of full-length TUs suggests that the majority of the predicted sORF-encoded proteins are not false positive predictions of truncated portions of long protein sequences.
Our first computational filter, prediction of protein-coding potential of transcript sequences based on known protein sequences, markedly increased the proteomics-matching rate for the sORFs. Still, the remaining two filtering approaches, based on interspecific conservation and protein family clustering, respectively, identified additional sORF candidates with protein support. These data suggest that small protein sequences are under-represented in the current protein databases. Thus, we anticipate that coding potential prediction can be improved as additional information is deposited in public protein databases.
It is well known that many genes are conserved among species across different evolutionary distances (Kriventseva et al. 2008; Ostlund et al. 2010). We identified sORFs that encode protein sequences conserved between P. deltoides and 14 other plant species (Supplemental Fig. 2), suggesting that interspecific conservation is a valid approach to enrich for protein-coding genes. As additional plant genome sequences become available, the interspecific conservation approach should become more useful in small protein discovery.
Matching sORF candidate sequences with proteomics data can provide direct evidence for small protein discovery. In this study, we demonstrated that ∼43% of the high-confidence sORF candidate set had supportive MS data. However, the number of sORFs having experimental proteomics support was lower than the number of sORFs with predicted protein domains. sORFs with protein MS data support are limited by protein sampling depth. Our analysis showed that the vast majority of protein MS data were represented by protein domain data (Supplemental Fig. 5), suggesting that computational validation of sORFs using a protein domain could be complementary to the more expensive experimental validation approach based on protein MS analysis.
By using EST data obtained from deep transcriptome sequencing, this study revealed more sORFs than predicted in the current Populus annotation (Fig. 3). One possible reason for this bias against small proteins in the current annotation of Populus is that the EST sequences were obtained by traditional Sanger sequencing of cloned cDNA libraries, in which cDNAs smaller than 400–500 bp were typically eliminated by size selection (Lease and Walker 2006). A key feature of recent improvements in the Arabidopsis genome annotation is the incorporation of a large number of short protein sequences (Supplemental Fig. 3). Small proteins are proportionately under-represented in the current Populus genome annotation compared with the most recent Arabidopsis annotation (Fig. 3), which reflects the more mature nature of the Arabidopsis annotation (v9.0) relative to Populus (v2.0). The length distribution of our predicted sORFs is similar to that of Arabidopsis (Fig. 3), indicating that our prediction offers a potential improvement in small protein annotation in Populus.
Some small proteins have been reported to be involved in cell-to-cell communications in plants. For example, a small protein of 94 aa called CAPRICE (CPC) is a transcription factor involved in intercellular signal transduction associated with root hair development in Arabidopsis (Kurata et al. 2005). It was also recently demonstrated that a membrane-associated thioredoxin (140 aa) moves from cell to cell, suggestive of a role in intercellular communication (Meng et al. 2010). Our proteomics measurements identified hundreds of sORF-encoded proteins in P. deltoides in phloem or xylem, but not in leaf. These sORFs represent a candidate pool of putative proteins that may be mobile molecules mediating intercellular signal transduction.
We have been able to demonstrate that deep RNA sequencing can be used in combination with computational approaches to predict high-likelihood protein-encoding sORFs that have not typically been annotated in most plant genomes. These results, supported by protein domain and proteomics evidence, suggest that the integrative approach used in this study to create the high-confidence sORF candidate set is effective in identifying protein-coding sORFs.
Methods
Plant material and RNA extraction and sequencing
Total RNA was isolated from leaf tissue of 6-mo-old Populus deltoides plants grown under normal and drought conditions using a Sigma Spectrum Plant Total RNA kit. RNA was extracted from 10 biological replicates. Equal amounts of total RNA from each of the biological replicates were pooled. The samples were run on an Experion (BioRad) to verify RNA quality and a Nanodrop (Thermo Fisher Scientific) to determine sample concentration. A total of 500 μg of total RNA from each sample was sent to the Joint Genome Institute (JGI), where transcriptome sequencing was performed using the Roche 454 Genome Sequencer FLX System (GS FLX). The raw expressed sequence tag (EST) sequences generated by transcriptome sequencing were trimmed for vector, adaptor/linker, poly(A) or T tails. The trimmed ESTs were edited for length and ESTs <100 nt were removed. ESTs with low complexity sequence greater than the threshold (default = 50%) were also removed. The EST sequences were then blasted against the GenBank nucleotide database in order to identify and eliminate contaminants. ESTs found to match nontarget sequences (e.g., non-nuclear) were removed.
Transcription unit assembly
Transcription units (TU) were created through three rounds of assembly using the P. trichocarpa genome and annotated mRNAs as references. Whole-genome resequencing of P. deltoides revealed that there was only ∼1% divergence between P. trichocarpa and P. deltoides at the genomic sequence level (data not shown). Thus, the reference P. trichocarpa transcript sequences (GeneCatalog_frozen20080522; ftp://ftp.jgi-psf.org/pub/JGI_data/Populus_trichocarpa/v1.1/) were pooled with the filtered P. deltoides EST sequences obtained from the 454 sequencing data and clustered using sclust implemented within the tgicl software (Pertea et al. 2003) using 97% identity and 80% sequence coverage criteria. Sequence clusters were then assembled using the CAP3 software (Huang and Madan 1999) to form consensus sequences with an overlap length cutoff of 40 and an overlap identity of 97%. The second-round assembly was alignment based. The consensus sequences were aligned onto the P. trichocarpa genome sequence version 1.1 (http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html) using BLAT (Kent 2002) with a minimum coverage (i.e., minimum fraction of query that must be aligned) of 80% and a minimum identity of 92%. Only the “best match” position was selected as the genomic location for each query consensus sequence. The genomic locations (i.e., GFF) of the annotated gene models were obtained from http://genome.jgi-psf.org. The consensus sequences and/or the JGI gene models that have overlapping genomic locations were reassembled using the CAP3 software with an overlap length cutoff of 30 and an overlap identity of 75%. In the final round of assembly the contigs obtained from the second-round assembly were mixed with the P. trichocarpa genome annotation v2.0 (http://www.phytozome.net/) mRNA sequences and assembled using the CAP3 software with an overlap length cutoff of 30 and an overlap identity of 92%. We empirically examined the influence of overlap identity (75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, and 98%) on CAP3 assembly and found that with 92% or 93% identity we were able to distinguish gene duplications while being capable of tolerating sequencing error or single-nucleotide polymorphisms. Similarly, Masoudi-Nejad et al. (2007) found that 92% was an appropriate overlap identity for CAP3 assembly of plant EST sequences.
sORF/small protein analysis
The TUs assembled from EST and JGI gene models were translated in 6-frame using the Emboss package (Rice et al. 2000). An initial sORF candidate set encoding proteins of 10–200 aa in length was created by including the longest possible complete ORF that contained start and stop codons in six-frame translations from each TU.
Clustering of protein sequences
All-vs-all BLASTp (Altschul et al. 1997) of the protein sequences were performed with an e-value cutoff of 10. The BLASTp data was then used to cluster the protein sequences into groups using the FORCE program (Wittkop et al. 2007) with a cutoff of −3.4.
Assessment of protein coding or non-coding potential
The assessment of coding potential of the sORF candidate set was conducted using the Coding Potential Calculator (Kong et al. 2007) based on a training set of proteins obtained from the UniProt database (The UniProt Consortium 2009). To determine whether the sORFs correspond to the non-coding RNAs, the ORF sequences were used as queries to search against the Rfam database (Griffiths-Jones et al. 2005; Gardner et al. 2009) using Infernal (Nawrocki et al. 2009).
Protein motif analysis
Protein sequences were scanned for domains using blastprodom, coils, gene3d, hmmpanther, hmmpir, hmmpfam, hmmsmart, hmmtigr, fprintscan, patternscan, profilescan, superfamily, seg, signalp, and tmhmm implemented in InterPro (Zdobnov and Apweiler 2001; Mulder and Apweiler 2007).
Conservation analysis of protein sequences between species
The putative P. deltoides small protein sequences were used as queries to search against the genome sequences of Arabidopsis lyrata (http://www.phytozome.net/), A. thaliana (http://www.arabidopsis.org/), Brachypodium distachyon (http://www.brachypodium.org/), Carica papaya (http://asgpb.mhpcc.hawaii.edu/papaya/), Chlamydomonas reinhardtii (http://www.phytozome.net/), Cucumis sativis (http://www.phytozome.net/), Glycine max (http://www.phytozome.net/), Medicago truncatula (http://www.medicago.org/), Oryza sativa (http://rice.plantbiology.msu.edu/), Physcomitrella patens (http://www.phytozome.net/), Selaginella moellendorffii (http://www.phytozome.net/), Sorghum bicolor (http://genome.jgi-psf.org/Sorbi1), Vitis vinifera (http://www.genoscope.cns.fr), and Zea mays (http://maizesequence.org/) using BLAT (Kent 2002) with a minimum coverage (i.e., minimum fraction of query that must be aligned) of 80% and a minimum identity of 60%.
Generation of random sORF sequences
Random coding sequences were generated using GenRGenS (Ponty et al. 2006). Specifically, a Markov model was first constructed based on the P. trichocarpa coding sequences (GeneCatalog_frozen20080522; ftp://ftp.jgi-psf.org/pub/JGI_data/Populus_trichocarpa/v1.1/) with an order of 2 and a phase of 3. Then, 20,000 random sequences of 600 bp starting with ATG (the start codon in protein-coding sequences) were generated. Finally, the complete coding sequences containing the first start codon (ATG) and a stop codon (TAA, TAG, or TGA) were selected as the random sORFs.
Mass spectrometry analysis of proteins
Protein was extracted from fully expanded P. deltoides leaves following the method of Lee et al. (2009). Tissues from each plant were ground separately under liquid N2 and stored at −80°C. Approximately 600 mg of leaf powder from each plant was suspended in 2.5 mL of lysis buffer (100 mM Tris HCl at pH 8.5; 5 mM DTT; 1 mM EDTA; 1 mM PMSF; 0.1 μg/mL leupeptin) and homogenized using a glass dounce tube. Each homogenate was centrifuged at 1000g for 10 min; each supernatant was further centrifuged at 30,000g for 60 min. Protein concentration in each final supernatant was measured by the Lowry method (Lowry et al. 1951) and equal protein amounts from the separate supernatants were combined to yield three pooled extracts, each containing a total of 3 mg of protein. Proteins were precipitated using 25% trichloroacetic acid; the resulting pellets were washed with acetone and resolubilized in 6 M guanidine/100 mM Tris HCl (pH 8.5) with sonication. Aliquots corresponding to ∼1 mg of protein were reduced by incubation with 10 mM DTT for 20 min and carboxyamidomethylated by incubation with 100 mM iodoacetamide for 15 min in the dark, both at ambient temperature. Samples were diluted to decrease guanidine concentration to 1 M with 50 mM Tris HCl/10mM CaCl2. Proteins were digested by incubating overnight at 37°C with trypsin (10 μg/mg protein; Promega sequencing grade), followed by the addition of a second identical amount of trypsin and an additional 4-h incubation. Digests were desalted (SepPak Lite C18, Waters) and analyzed in triplicate using two-dimensional HPLC interfaced with tandem mass spectrometry as described previously (Kalluri et al. 2009).
Protein extraction and quantification from xylem and phloem tissue was performed using a method essentially identical to that recently applied for proteomic analysis of xylem tissue (Kalluri et al. 2009). An additional centrifugation step (3000g for 10 min) was performed following trypsin digestion to remove cellular debris from solution. Digests (100-μg aliquots, based on protein measurement) were analyzed using two-dimensional HPLC interfaced with tandem mass spectrometry.
Tryptic peptide identifications were extracted from the tandem mass spectra from leaf, xylem, and phloem tissues, as well as from previously published data on P. deltoides xylem proteins (Kalluri et al. 2009) using Sequest. Peptide identifications were filtered and compiled using DTASelect to provide protein identifications; a protein required evidence from two or more tryptic peptides per protein, or identification of a single tryptic peptide in two or more charge states. The protein database for the Sequest searches contained protein sequences in P. trichocarpa annotation v2.0 (www.phytozome.net), the initial sORF candidate set (12,852 sequences) (Fig. 1), a sequence-reversed analog of each protein for estimation of false discovery rates, and commonly observed contaminant proteins. Sequest was executed with no enzyme specificity, and non-tryptic peptide identifications were subsequently removed from the data set using DTASelect. False discovery rates among the remaining tryptic peptides were typically 1% or less.
Parsimony analysis was performed to identify tryptic peptides shared among several proteins (Yang et al. 2004). Further details are provided in the caption for Supplemental Table 4.
The proteomics data have been deposited in Proteome Commons Tranche repository, https://proteomecommons.org/tranche/ (hashes Fv9zgC97mv0bld5KMOM7ww9mP24qhchGLvS7Cx4ddYCJgm28KiUsD5xp0UQCglivuBSz59Tdes8+auDPWYDOleix/vUAAAAAAAAMhA== and /WZkinVg1kkkYxvkqAQKAXSW5ujyhPCjp9WFBdzXoLAh6qnH50N+Tl1sekqV9XVWLCssVOGS63e9OyhkAvCLG49wAeQAAAAAAAAOlA==).
Full-length cDNA cloning using Rapid Amplification of cDNA Ends
Full-length cDNA was synthesized from total RNA using a GeneRacer Kit (Invitrogen). The resulting cDNA template was used in PCR reactions to amplify both the 5′ and 3′ end of each gene of interest. 5′ ends were amplified using a GeneRacer 5′ primer and a reverse gene-specific primer (GSP). 3′ ends were amplified using a 3′ GeneRacer primer and a forward GSP. GSPs (Supplemental Table 2) were designed according to specifications provided in the GeneRacer protocol. After PCR amplification the 5′ and 3′ fragments were run on an agarose gel, excised out, purified using a Minelute Gel Purification kit (Qiagen), and sequenced. Sequences were aligned using Sequencher version 4.5.
Comparison of sORF-encoded proteins with the Populus genome annotation
The sORF coding sequences were mapped to the P. trichocarpa genome v2.0 (http://www.phytozome.net/) by BLAT (Kent 2002) with a minimum coverage (i.e., minimum fraction of query that must be aligned) of 80% and a minimum identity of 92%. Only the “best match” position was selected as the genomic location for each query sequence. The genomic locations (i.e., GFF) of the gene models in annotation v2.0 were obtained from http://www.phytozome.net/. In cases where there were overlapping genomic locations between sORF CDS and the annotated Populus gene models, the sORF CDS sequences were compared with annotated CDS using the MAFFT alignment program (Katoh et al. 2002, 2005).
Acknowledgments
We thank S.D. Wullschleger and D.J. Weston for thoughtful and insightful comments on the manuscript. Transcriptome sequencing was supported by the U.S. Department of Energy Joint Genome Institute Laboratory Science Program project with X.Y. and T.J.T. The work conducted by the U.S. Department of Energy Joint Genome Institute is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. Proteomics and bioinformatics analysis was supported by the U.S. DOE Office of Biological and Environmental Research, Genomic Science Program and the U.S. DOE BioEnergy Science Center. The BioEnergy Science Center is a U.S. Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science. Oak Ridge National Laboratory is managed by UT-Battelle, LLC for the U.S. Department of Energy under Contract Number DE–AC05–00OR22725.
Footnotes
[Supplemental material is available for this article. The sequence data from this study have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/Genbank/) under accession nos. HP451655–HP451687, HP451690–HP451709, and HP451711–HP451725. Mass spectrometry data have been uploaded to the Proteome Commons Tranche repository (https://proteomecommons.org/tranche/).]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.109280.110.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camby I, Le Mercier M, Lefranc F, Kiss R 2006. Galectin-1: A small protein with major functions. Glycobiology 16: 137R–157R [DOI] [PubMed] [Google Scholar]
- Castellana NE, Payne SH, Shen Z, Stanke M, Bafna V, Briggs SP 2008. Discovery and revision of Arabidopsis genes by proteogenomics. Proc Natl Acad Sci 105: 21034–21038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Pang KC, Mercer TR, Mattick JS 2008. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput Biol 4: e1000176 doi: 10.1371/journal.pcbi.1000176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM 1999. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914 [DOI] [PubMed] [Google Scholar]
- Galindo MI, Pueyo JI, Fouix S, Bishop SA, Couso JP 2007. Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol 5: e106 doi: 10.1371/journal.pbio.0050106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, et al. 2009. Rfam: Updates to the RNA families database. Nucleic Acids Res 37: D136–D140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason CA, Liu QL, Williamson VM 2008. Silencing a candidate nematode effector gene corresponding to the tomato resistance gene Mi-1 leads to acquisition of virulence. Mol Plant Microbe Interact 21: 576–585 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A 2005. Rfam: Annotating non-coding RNAs in complete genomes. Nucleic Acids Res 33: D121–D124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Zhang X, Borevitz JO, Li WH, Shiu SH 2007. A large number of novel coding small open reading frames in the intergenic regions of the Arabidopsis thaliana genome are transcribed and/or under purifying selection. Genome Res 17: 632–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Akiyama K, Sakurai T, Toyoda T, Shinozaki K, Shiu SH 2010. sORF finder: A program package to identify small open reading frames with high coding potential. Bioinformatics 26: 399–400 [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A 1999. CAP3: A DNA sequence assembly program. Genome Res 9: 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HW, Tschaplinski TJ, Wang L, Glazebrook J, Greenberg JT 2009. Priming in systemic plant immunity. Science 324: 89–91 [DOI] [PubMed] [Google Scholar]
- Kalluri UC, Hurst GB, Lankford PK, Ranjan P, Pelletier DA 2009. Shotgun proteome profile of Populus developing xylem. Proteomics 9: 4871–4880 [DOI] [PubMed] [Google Scholar]
- Kastenmayer JP, Ni L, Chu A, Kitchen LE, Au WC, Yang H, Carter CD, Wheeler D, Davis RW, Boeke JD, et al. 2006. Functional genomics of genes with small open reading frames (sORFs) in S. cerevisiae. Genome Res 16: 365–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Miyata T, Toh H 2005. Improvement in the accuracy of multiple sequence alignment program MAFFT. Genome Inform 16: 22–33 [PubMed] [Google Scholar]
- Kent WJ 2002. BLAT–the BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G 2007. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 35(Web Server issue): W345–W349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriventseva EV, Rahman N, Espinosa O, Zdobnov EM 2008. OrthoDB: The hierarchical catalog of eukaryotic orthologs. Nucleic Acids Res 36: D271–D275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata T, Ishida T, Kawabata-Awai C, Noguchi M, Hattori S, Sano R, Nagasaka R, Tominaga R, Koshino-Kimura Y, Kato T, et al. 2005. Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development 132: 5387–5398 [DOI] [PubMed] [Google Scholar]
- Lease KA, Walker JC 2006. The Arabidopsis unannotated secreted peptide database, a resource for plant peptidomics. Plant Physiol 142: 831–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Feng J, Campbell KB, Scheffler BE, Garrett WM, Thibivilliers S, Stacey G, Naiman DQ, Tucker ML, Pastor-Corrales MA, et al. 2009. Quantitative proteomic analysis of bean plants infected by a virulent and avirulent obligate rust fungus. Mol Cell Proteomics 8: 19–31 [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ 1951. Protein measurement with the folin phenol reagent. J Biol Chem 193: 265–275 [PubMed] [Google Scholar]
- Masoudi-Nejad A, Goto S, Jauregui R, Ito M, Kawashima S, Moriya Y, Endo TR, Kanehisa M 2007. EGENES: Transcriptome-based plant database of genes with metabolic pathway information and expressed sequence tag indices in KEGG. Plant Physiol 144: 857–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Wong JH, Feldman LJ, Lemaux PG, Buchanan BB 2010. A membrane-associated thioredoxin required for plant growth moves from cell to cell, suggestive of a role in intercellular communication. Proc Natl Acad Sci 107: 3900–3905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder N, Apweiler R 2007. InterPro and InterProScan: Tools for protein sequence classification and comparison. Methods Mol Biol 396: 59–70 [DOI] [PubMed] [Google Scholar]
- Muller R, Bleckmann A, Simon R 2008. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 20: 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, Kolbe DL, Eddy SR 2009. Infernal 1.0: Inference of RNA alignments. Bioinformatics 25: 1335–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notaguchi M, Abe M, Kimura T, Daimon Y, Kobayashi T, Yamaguchi A, Tomita Y, Dohi K, Mori M, Araki T 2008. Long-distance, graft-transmissible action of Arabidopsis FLOWERING LOCUS T protein to promote flowering. Plant Cell Physiol 49: 1645–1658 [DOI] [PubMed] [Google Scholar]
- Oelkers K, Goffard N, Weiller GF, Gresshoff PM, Mathesius U, Frickey T 2008. Bioinformatic analysis of the CLE signaling peptide family. BMC Plant Biol 8: 1 doi: 10.1186/1471-2229-8-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostlund G, Schmitt T, Forslund K, Kostler T, Messina DN, Roopra S, Frings O, Sonnhammer EL 2010. InParanoid 7: New algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res 38: D196–D203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea G, Huang X, Liang F, Antonescu V, Sultana R, Karamycheva S, Lee Y, White J, Cheung F, Parvizi B, et al. 2003. TIGR Gene Indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 19: 651–652 [DOI] [PubMed] [Google Scholar]
- Ponty Y, Termier M, Denise A 2006. GenRGenS: Software for generating random genomic sequences and structures. Bioinformatics 22: 1534–1535 [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A 2000. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet 16: 276–277 [DOI] [PubMed] [Google Scholar]
- Trotochaud AE, Jeong S, Clark SE 2000. CLAVATA3, a multimeric ligand for the CLAVATA1 receptor-kinase. Science 289: 613–617 [DOI] [PubMed] [Google Scholar]
- Tuskan GA, DiFazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. 2006. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- The UniProt Consortium 2009. The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res 37: D169–D174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkop T, Baumbach J, Lobo FP, Rahmann S 2007. Large-scale clustering of protein sequences with FORCE - A layout based heuristic for weighted cluster editing. BMC Bioinformatics 8: 396 doi: 10.1186/1471-2105-8-396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Dondeti V, Dezube R, Maynard DM, Geer LY, Epstein J, Chen X, Markey SP, Kowalak JA 2004. DBParser: Web-based software for shotgun proteomic data analyses. J Proteome Res 3: 1002–1008 [DOI] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R 2001. InterProScan – An integration platform for the signature-recognition methods in InterPro. Bioinformatics 17: 847–848 [DOI] [PubMed] [Google Scholar]