Summary
Tumor cells exhibit aberrant metabolism characterized by high glycolysis even in the presence of oxygen. This metabolic reprogramming, known as the Warburg effect, provides tumor cells with the substrates required for biomass generation. Here, we show that the mitochondrial NAD-dependent deacetylase SIRT3 is a crucial regulator of the Warburg effect. Mechanistically, SIRT3 mediates metabolic reprogramming by destabilizing hypoxia-inducible factor-1α (HIF1α), a transcription factor that controls glycolytic gene expression. SIRT3 loss increases reactive oxygen species production, leading to HIF1α stabilization. SIRT3 expression is reduced in human breast cancers, and its loss correlates with the upregulation of HIF1α target genes. Finally, we find that SIRT3 overexpression represses glycolysis and proliferation in breast cancer cells, providing a metabolic mechanism for tumor suppression.
Introduction
Otto Warburg first noted in the 1920s that cancer cells undergo glycolysis even in the presence of ample oxygen (Warburg, 1956). This preferential use of aerobic glycolysis, termed the Warburg effect, has emerged as a metabolic hallmark of many cancers. As a result, there is much interest in understanding the pathways that regulate the potential survival and proliferative advantages conferred by increased glucose uptake and catabolism, and this topic has been extensively reviewed (Tennant et al., 2010; Vander Heiden et al., 2009). Accruing evidence suggests that aerobic glycolysis is used to support the rapid proliferation of tumor cells. Enhanced catabolism of glucose contributes to the raw materials needed to synthesize the nucleotides, amino acids and lipids necessary for cellular proliferation and can provide a distinct growth advantage for cells with elevated aerobic glycolysis (Tong et al., 2009). Constitutive upregulation of aerobic glycolysis can also provide a survival advantage for tumor cells because limitations in tumor vascularization result in periods of intermittent hypoxia that require a cell to rely on glycolysis (Gatenby and Gillies, 2004). In support of this notion, the transition from premalignant lesions to invasive cancer is often accompanied by increased tumor glucose uptake (Gatenby and Gillies, 2004).
Metabolic reprogramming in cancer cells is regulated by several oncogenic cues, including the PI3K/Akt, Myc or hypoxia-inducible transcription factor (HIF) pathways that serve to increase glucose uptake, glycolysis, angiogenesis and stress resistance (Kaelin and Ratcliffe, 2008; Semenza, 2010; Tennant et al., 2010; Tong et al., 2009). Recently, mutations in mitochondrial enzymes have emerged as important drivers of altered tumor cell metabolism. For example, gain- or loss-of-function mutations in tricarboxylic acid (TCA) cycle enzymes regulate HIF1 activity and promote carcinogenesis (Gottlieb and Tomlinson, 2005; Zhao et al., 2009). In turn, identification of new mitochondrial regulators of global cellular reprogramming could provide important insight into the contribution of altered metabolism to tumorigenesis.
Sirtuins are a conserved family of NAD-dependent ADP-ribosyltransferases and/or protein deaceytlases involved in metabolism, stress response and longevity (Finkel et al., 2009). Mammals express seven sirtuins (SIRT1-7), three of which (SIRT3-5) are localized to the mitochondrion. SIRT3 is a major mitochondrial deacetylase that targets many enzymes involved in central metabolism, resulting in the activation of many oxidative pathways (Verdin et al., 2010). For example, SIRT3 deacetylates complex I and complex II to activate electron transport (Ahn et al., 2008; Cimen et al., 2010). SIRT3 induces fatty acid oxidation during fasting in hepatocytes via deacetylation of LCAD (Hirschey et al., 2010). SIRT3 also targets the mitochondrial enzymes IDH2 and MnSOD (Qiu et al., 2010; Schlicker et al., 2008; Someya et al., 2010; Tao et al., 2010), which function in part to maintain reactive oxygen species (ROS) homeostasis. SIRT3 loss increases ROS levels and contributes to numerous age-related pathologies, including hearing loss and tumorigenesis (Kim et al., 2010; Someya et al., 2010). Previously, it was shown that SIRT3 functions as a tumor suppressor by decreasing ROS and maintaining genomic stability (Kim et al., 2010). In this study it was also demonstrated that SIRT3 null cells display increased glucose uptake and altered mitochondrial oxidation, but the direct contribution of SIRT3-mediated metabolic regulation on tumor growth remains a central question. Due to the pivotal role of SIRT3 as a regulator of ROS and multiple mitochondrial pathways, we sought to specifically probe the role of SIRT3 in regulating tumor cell metabolism and growth.
Results
SIRT3 promotes cellular metabolic reprogramming
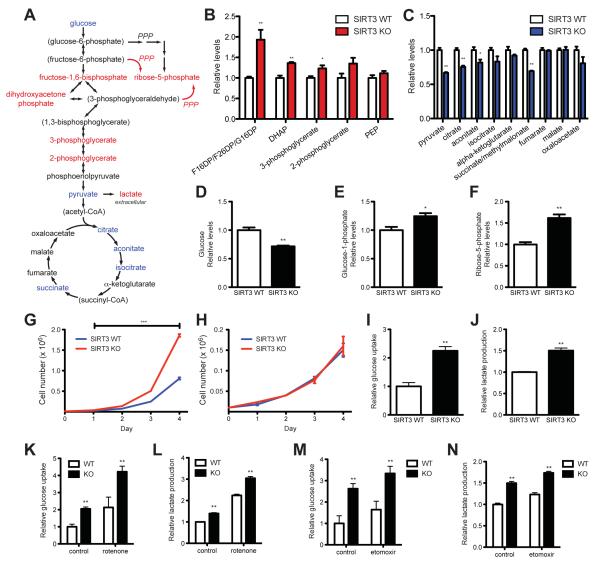
Since SIRT3 activates enzymes involved in mitochondrial fuel catabolism (Verdin et al., 2010), and SIRT3 loss increases glucose uptake (Kim et al., 2010), we hypothesized that SIRT3 could serve as an important regulator of the balance between glycolytic and anabolic pathways and mitochondrial oxidative metabolism to regulate tumor cell growth. To test this idea, we first examined the influence of SIRT3 loss on metabolites from mouse embryonic fibroblasts (MEFs) using liquid chromatography-mass spectrometry (LC-MS). The metabolic profile of SIRT3 null (KO) MEFs demonstrated a clear shift towards glycolytic metabolism when compared with SIRT3 wild-type (WT) counterparts (Figure 1A), similar to the metabolic shift reported for transformed cells in culture (Lu et al., 2010) and for cancer cells in vivo (Denkert et al., 2008; Hirayama et al., 2009). In SIRT3 KO cells, intermediates of glycolysis were elevated, while TCA cycle metabolites were reduced (Figure 1B and 1C). Consistent with a pattern of increased glucose usage, SIRT3 KO cells had lower levels of intracellular glucose (Figure 1D), while levels of glucose-1-phosphate, a product of glycogenolysis, were increased (Figure 1E). Glucose-1-phosphate can be converted by phosphoglucomutase into glucose-6-phosphate (G6P) to provide substrates for glycolysis or the oxidative arm of the pentose phosphate pathway (PPP), generating NADPH and ribose. The nonoxidative arm of the PPP forms ribose-5-phosphate from fructose-6-phosphate or glyceraldehyde-3-phosphate (Figure 1A). Importantly, G6P, glycolytic intermediates and ribose-5-phosphate were all increased in SIRT3 KO cells (Figure 1A and 1F), suggesting that glucose metabolites were diverted into the PPP in order to provide the ribose necessary for nucleic acid synthesis. Notably, the pattern and the magnitude of metabolomic changes caused by SIRT3 loss were similar to those observed comparing tumors to nearby normal tissue (Hirayama et al., 2009).
Figure 1.
Metabolic profiles of SIRT3 KO MEFs reflect an increase in glycolytic pathways and a decrease in mitochondrial oxidative metabolism. (A) Schematic illustrating the metabolites that are increased (red) or decreased (blue) in SIRT3 KO MEFs compared to SIRT3 WT MEFs (n = 4), p < 0.1. Metabolites in parentheses were not measured. PPP, pentose phosphate pathway. The nonoxidative (red) and oxidative (black) arms of the PPP are shown. Levels of glycolytic intermediates (B), TCA cycle intermediates (C), glucose (D), glucose-1-phosphate (E) and ribose-5-phosphate (F). Growth curves of SIRT3 WT and KO MEFs (n = 3) cultured in media containing glucose (G) or galactose (H). Error bars, ±SD. (I-N) Glucose uptake and lactate production in SIRT3 WT and KO MEFs (n = 6). (I) Relative glucose uptake and (J) lactate production in SIRT3 WT and KO MEFs. (K) Relative glucose uptake and (L) relative lactate production in SIRT3 WT and KO MEFs incubated with or without 100 nM rotenone. (M) Glucose uptake and (N) lactate production in SIRT3 WT and KO MEFs cultured in the presence or absence of 50 μg/ml etomoxir. Cells were treated with drugs for 24 hours before measuring glucose uptake and lactate. All error bars (except growth curves), ±SEM. (*) p < 0.05; (**) p < 0.01, (***) p < 0.001. See also Figure S1.
Increased metabolites involved in glycolysis and the PPP suggested that, like many cancer cells, SIRT3 KO MEFs might be using glucose to support increased proliferation by directing glucose away from the TCA cycle towards biosynthetic processes. Indeed, SIRT3 KO cells grew significantly faster than WT cells (Figure 1G). To test whether this increased growth rate required aerobic metabolism of glucose, we grew cells in media containing galactose instead of glucose, thereby reducing glycolytic flux and forcing the cell to rely on mitochondrial oxidative phosphorylation (Marroquin et al., 2007). Under these conditions, WT and KO cells grew at the same rate, demonstrating that the increased proliferation of KO cells required enhanced glucose catabolism (Figure 1H).
To confirm that the metabolite patterns reflected an increase in glycolysis, we measured glucose uptake and lactate production. As expected, SIRT3 KO MEFs consumed more glucose and extruded more lactate into the media than did WT cells (Figure 1I and 1J). This effect was not specific to MEFs; HEK293T cells in which SIRT3 expression was stably reduced by lentiviral expression of shRNA against SIRT3 also showed an increase in glucose uptake and lactate production (Figure S1A and S1B). Interestingly, the effect of SIRT3 loss on glucose uptake and lactate production was similar to the effect of pyruvate kinase M2 isoform overexpression or mTOR activation (Christofk et al., 2008; Duvel et al., 2010). These data suggest that loss of SIRT3 redirects cellular metabolism in favor of glycolysis, and as a result, cells with low levels of SIRT3 display features similar to the Warburg effect apparent in many cancer cells.
Previous studies have found that SIRT3 loss increases glucose uptake (Kim et al., 2010), yet the specific mechanism involved has not been elucidated. To test whether SIRT3 upregulates glycolysis as part of a compensatory response due to diminished oxidative capacity, we examined glucose uptake and lactic acid secretion in the presence of a mitochondrial respiratory inhibitor, rotenone, or an inhibitor of mitochondrial fatty acid oxidation, etomoxir. In WT cells, glycolysis is increased in the presence of both rotenone (Figure 1K and 1L) and etomoxir (Figure 1M and 1N). Strikingly, glucose uptake and lactate production remain elevated in the SIRT3 KO cells even in the presence of oxidation inhibitors (Figure 1K-1N). These data demonstrate that upregulated glycolysis in SIRT3 null cells does not result solely from nonspecific compensation for decreased mitochondrial oxidative functions. Instead, these data indicate that, surprisingly, SIRT3 may regulate glycolysis via activation of a specific signaling pathway.
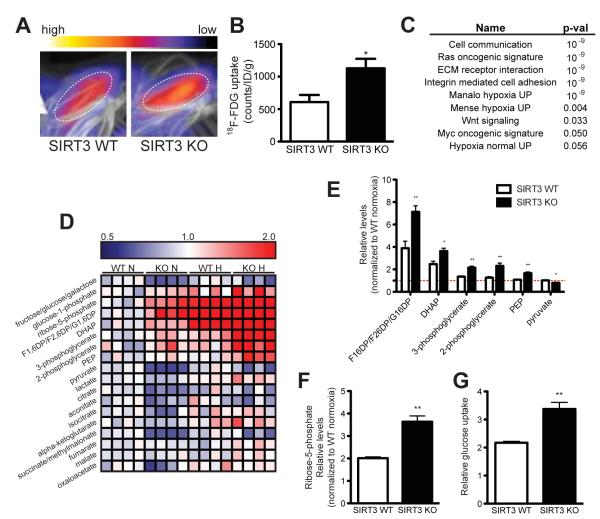
We next asked whether SIRT3 KO mice exhibited signs of increased glucose usage. We injected mice with 18F-fluordeoxyglucose (18F-FDG) and scanned animals using positron emission tomography-computed tomography (PET/CT) in order to monitor glucose uptake. We looked specifically in brown adipose tissue (BAT), which exhibits high glucose uptake (Cannon and Nedergaard, 2004). In line with our cellular studies, we found that SIRT3 KO mice had a increased in 18F-FDG uptake in brown adipose tissue (BAT) compared with WT mice (Figure 2A and 2B), even though the mass of BAT in SIRT3 KO mice was not larger than in WT mice (Figure S2A). Glucose uptake in BAT is regulated by the β-adrenergic pathway and is thus dramatically increased by cold exposure (Shimizu et al., 1991). We measured 18F-FDG uptake in BAT of SIRT3 WT and KO after a 6 hour cold challenge and found that SIRT3 KO mice have higher 18F-FDG uptake both at room temperature and at 4°C (Figure 2B and S2B), illustrating that SIRT3 WT and KO mice have a similar increase in BAT glucose uptake in response to β-adrenergic signaling. These differences in BAT glucose uptake occur independently of obvious changes in whole-body glucose homeostasis: we did not detect changes in blood glucose levels (Figure S2C) as reported previously (Lombard et al., 2007).
Figure 2.
SIRT3 KO mice have elevated glucose uptake and hypoxic signatures in vivo. 18F-fluorodeoxyglucose (18F-FDG) uptake in the brown adipose tissue (BAT) of SIRT3 WT and KO mice was measured using positron emission tomography-computed tomography (PET/CT). (A) Representative scans with color scale bar indicating relative levels of uptake from low (black) to high (white). (B) Quantification of BAT 18F-FDG uptake normalized to body weight (n = 6). (C) Gene set enrichment analysis of canonical pathways with the ranked genes list from most up- to most down-regulated in SIRT3 KO BAT. (D) Heat map comparing metabolite patterns of SIRT3 deletion and hypoxia. Red and blue indicate up- or down-regulation, respectively. SIRT3 WT and KO MEFs (n = 4) were cultured in 21% O2 (normoxia, N) or 1% O2 for 12 hours (hypoxia, H) and metabolites were analyzed by LC-MS. Relative levels of glycolytic intermediates (E) and ribose-5-phosphate (F). (G) Glucose uptake of MEFs cultured under hypoxia for 6 hours. Error bars, ±SEM. (*) p < 0.05; (**) p < 0.01. See also Figure S2.
To examine mechanisms underlying increased glucose uptake in SIRT3 KO BAT, we performed genome-wide expression profiling on RNA isolated from BAT and performed gene set enrichment analysis (GSEA) using the ranked gene list from most up- to most down-regulated in SIRT3 KO mice in order to identify the biological pathways most significantly altered by SIRT3 loss. As SIRT3 is a mitochondrial deacetylase, we expected to see compensatory up-regulation of pathways involving mitochondrial function or energy production. To our surprise, SIRT3 loss up-regulated pathways important in tumorigenesis. Strikingly, of the nine gene sets most significantly overrepresented in SIRT3 KO BAT, three were independently defined as gene sets induced by exposure to hypoxia (Figure 2C and S2B). Hypoxia itself increases 18F-FDG uptake (Clavo et al., 1995) and is associated with many transcriptional changes that result in increased glucose uptake and utilization (Brahimi-Horn et al., 2007). Thus, we hypothesized that the increase in glucose uptake in SIRT3 KO BAT could be explained by upregulation of the hypoxia response.
The similarity between gene signatures of SIRT3 KO mice and hypoxic cells was particularly notable because hypoxia induces a metabolic shift similar to that caused by loss of SIRT3, including a decrease in mitochondrial substrate oxidation and an increase in glycolysis (Semenza, 2010). To test the role of SIRT3 in hypoxia-induced metabolic reprogramming, we analyzed metabolites isolated from MEFs cultured at 21% O2 (normoxia) or 1% O2 (hypoxia) for 12 hours. Strikingly, we observed that the increase in glycolytic intermediates caused by hypoxia was similar to the effects of SIRT3 deletion (Figure 2D). Furthermore, hypoxia and SIRT3 loss had additive effects: while intermediates of glycolysis, glycogenolysis and the PPP were elevated by hypoxia, levels of these metabolites were dramatically higher in SIRT3 KO MEFs under these conditions (Figure 2E, 2F and S2E). Consistent with the metabolite profiles, hypoxia increased glucose uptake in both cell lines, and SIRT3 KO or knock-down cells consumed even more glucose than control cells (Figure 2G and S2F). Taken together, these data illustrate that SIRT3 loss and hypoxia result in similar metabolic shifts and implicate dysregulated activation of the hypoxia pathway as a cause of the metabolic reprogramming of SIRT3 null cells.
SIRT3 opposes the Warburg effect by destabilizing HIF1α
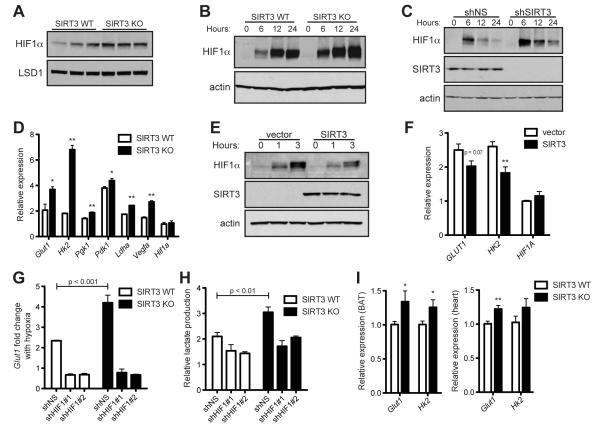
HIF1, comprised of the heterodimer HIF1α and HIF1β, is the primary driver of increased glycolysis and lactate production during hypoxia (Gordan and Simon, 2007; Hu et al., 2003; Seagroves et al., 2001). Under conditions of low oxygen, HIF1α is stabilized and promotes transcription of many genes crucial for the cellular response to hypoxia (Kaelin and Ratcliffe, 2008). Consequently, cells lacking HIF1α fail to upregulate glycolytic enzymes and lactic acid production in response to hypoxia (Seagroves et al., 2001). Given the in vivo hypoxic gene signature of SIRT3-null BAT, in addition to the striking similarity between the mitochondrial-independent glycolytic profiles of SIRT3 KO MEFs and hypoxic cells, we reasoned that the mechanism by which SIRT3 regulates glycolysis involves HIF1α. To test this idea, we first investigated whether SIRT3 directly modulates HIF1α stability under normoxic conditions. In the presence of high oxygen, HIF1α is rapidly degraded and difficult to measure from cell lysates, but HIF1α is detectable from isolated nuclei. Indeed, nuclei isolated from SIRT3 deficient cells during normoxia demonstrated elevated levels of HIF1α relative to WT cells (Figure 3A). Likewise, when MEFs were cultured under 1% O2, HIF1α was stabilized earlier and to a higher degree in SIRT3 KO cells compared to WT cells in whole cell lysates (Figure 3B). We obtained comparable results in HEK293T cells in which SIRT3 expression was stably reduced by lentiviral expression of shRNA against SIRT3 (Figure 3C). Importantly, SIRT3 also regulates expression of HIF1α target genes. Both the glucose transporter Glut1 and hexokinase II (Hk2)—HIF1α target genes that are critical for increased glucose uptake and catabolism via aerobic glycolysis or the PPP and are strongly implicated in tumorigenesis (O’Donnell et al., 2006; Tennant et al., 2010)—were elevated during hypoxia in SIRT3 KO MEFs and SIRT3 knock-down cells relative to control cells (Figure 3D and S3A). Furthermore, the HIF1α targets pyruvate dehydrogenase kinase 1 (Pdk1), lactate dehydrogenase A (Ldha), phosphoglycerate kinase (Pgk1) and vascular endothelial growth factor A (Vegfa) were significantly elevated in SIRT3 KO cells compared to WT cells during hypoxia (Figure 3D). Similar to the pattern we saw with metabolic intermediates of glycolysis, many of these genes were moderately elevated by SIRT3 loss under basal conditions (Figure S3B), and SIRT3 deletion and hypoxia had additive effects on expression of HIF1α target genes (Figure 3D).
Figure 3.
SIRT3 regulates HIF1α stability. (A) Immunoblots of nuclear extracts from SIRT3 WT and KO MEFs cultured at 21% O2. Immunoblots of MEFs (B) or HEK293T cells expressing control shRNA (shNS) or shRNA targeted against SIRT3 (C) cultured at 1% O2 for the indicated times. (D) HIF1α target genes in SIRT3 WT and KO MEFs after 6 hours of hypoxia were measured by qRT-PCR and shown as a ratio of SIRT3 WT normoxia levels. (E) Immunoblots of HEK293T cells stably overexpressing empty vector or SIRT3 cultured at 1% O2 for the indicated times. (F) Expression of HIF1α target genes in HEK293T control and SIRT3-overexpressing cells after 6 hours of hypoxia. (G) SIRT3 WT and KO MEFs expressing shNS or shRNA against HIF1α (shHIF1#1,2) were cultured in normoxia or hypoxia (6 hours) and the fold change in Glut1 levels was measured by qRT-PCR. (H) Lactate produced by SIRT3 WT and KO MEFs expressing shNS or shHIF1α after 6 hours of hypoxia expressed as a ratio of SIRT3 WT shNS normoxic controls. Significance was assessed by two-way ANOVA. (I) Expression of Glut1 and Hk2 in the brown adipose tissue (left) and heart (right) of SIRT3 WT and KO mice (n =5-7) was measured by qRT-PCR. β-2-microglobulin or Rps16 were used as endogenous controls for qRT-PCR. Error bars, ±SEM (n = 4-6). (*) p <0.05; (**) p <0.01. See also Figure S3.
To test whether SIRT3 directly represses HIF1α, we examined the levels of HIF1α and its target genes in cells overexpressing SIRT3. SIRT3 overexpression clearly and reproducibly reduced the extent of HIF1α stabilization in hypoxic cells (Figure 3E). Importantly, the induction of GLUT1 and HK2 during hypoxia was blunted by SIRT3 overexpression, demonstrating that SIRT3 directly inhibits HIF1α function (Figure 3F). SIRT3 catalytic activity was required for the full repression of HIF1α target genes: expression of a SIRT3 catalytic mutant did not significantly reduce hypoxic GLUT1 expression (Figure S3C). Furthermore, using primary MEFs, we found that two SIRT3 KO lines exhibited increased Glut1 expression relative to two WT lines, suggesting that SIRT3 can regulate HIF1α activity in primary cell lines (Figure S3D). Taken together, the data show that SIRT3 controls the stabilization of HIF1α and the induction of crucial HIF1α target genes that coordinate aerobic glucose consumption.
Next, to examine the requirement for HIF1α in the glycolytic shift observed in SIRT3 null cells, we used two separate shRNA constructs against HIF1α to generate SIRT3 WT and KO MEFs with HIF1α levels stably reduced (Figure S3E). We measured normoxic and hypoxic Glut1 expression in these cell lines and found, as predicted, that control (shNS) SIRT3 KO MEFs demonstrated an exaggerated response to hypoxia, measured as the fold change in Glut1 expression, compared to control WT MEFs (Figure 3G). In contrast, WT and SIRT3 KO MEFs expressing either shRNA against HIF1α had comparable responses to hypoxia (Figure 3G and S3F). Importantly, the increase in lactate production caused by SIRT3 deletion required HIF1α both in normoxia and hypoxia (Figure 3H and S3G). Together, these data demonstrate that SIRT3 regulates aerobic glycolysis through HIF1α.
To probe for evidence of increased HIF1α activation in vivo, we measured levels of HIF1α and HIF1α target genes from tissues of SIRT3 WT and KO mice. Levels of HIF1α protein and many HIF1α target genes involved in glycolysis were significantly elevated in the BAT of SIRT3 KO mice (Figures 3I and S3H-J), consistent with our studies demonstrating increased glucose uptake in SIRT3 KO BAT. Similarly, several HIF1α target genes showed a trend of increased expression in SIRT3 KO heart (Figure 3I and S3K).
The regulation of HIF1α is complex and not completely understood (Kaelin and Ratcliffe, 2008). During normoxia, HIF1α is hydroxlylated at two proline residues by a family of oxygen-dependent prolyl hydroxylases (PHD1-3), enabling the tumor suppressor von Hippel-Lindau (VHL) to bind and target HIF1α for ubiquitination and proteasomal degradation (Kaelin and Ratcliffe, 2008). As we did not detect changes in HIF1α mRNA levels (Figures 3D and 3F), we tested whether SIRT3 exerted a post-translational effect on HIF1α stability. SIRT1 binds HIF1α and regulates its activity through direct deacetylation (Lim et al., 2010). To test whether SIRT3 might act through a similar mechanism, we immunoprecipitated SIRT1 or SIRT3 and probed for interactions with HIF1α. SIRT1, but not SIRT3, pulled down HIF1α (Figure S4A), suggesting that SIRT3 does not interact with HIF1α directly.
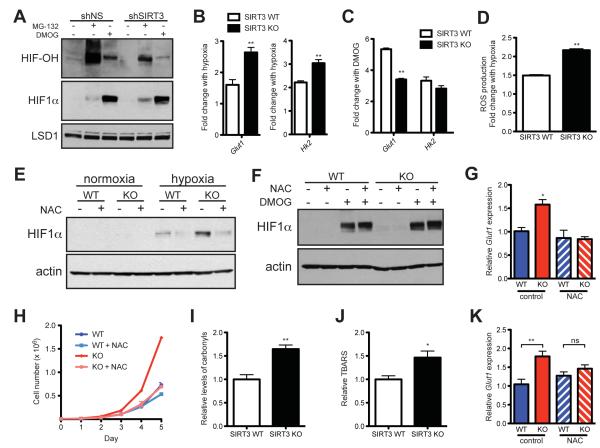
We next hypothesized that SIRT3 regulates HIF1α stability by modulating PHD activity by measuring the extent of HIF1α hydroxylation. We assessed PHD activity in control and SIRT3 knock-down HEK293T cells by treating cells with the proteasomal inhibitor MG-132 (to prevent hydroxylated HIF1α from being degraded) or with DMOG (dimethyloxaloylglycine, to inhibit PHDs). Although SIRT3 knock-down cells accumulated more HIF1α during MG-132 treatment, they had significantly less hydroxylated HIF1α, indicating that PHD activity is lower in SIRT3 knock-down cells (Figure 4A). Similarly, SIRT3 WT MEFs demonstrated higher levels of HIF1α hydroxylation than KO MEFs (Figure S4B). As a potent PHD inhibitor, we predicted that DMOG would overcome any differences between HIF1α levels SIRT3 WT and KO MEFs if SIRT3 acts at the level of the PHDs. Indeed, we observed that at every time point examined, SIRT3 WT and KO MEFs have equal levels of HIF1α stabilized in response to DMOG treatment (Figure S4C).
Figure 4.
SIRT3 regulates HIF1α stability through ROS. (A) Nuclear extracts from shNS and shSIRT3 HEK293T cells treated with or without 10 μM MG-132 for 1 hour or 1 mM DMOG for 4 hours as indicated were immunoblotted with antibodies specific to hydroxylated HIF1α (HIF-OH) or total HIF1α. (B) Fold induction of HIF1α target genes in response to hypoxia (n = 6) measured by qRT-PCR. The ratio of hypoxic to normoxic gene expression is shown. (C) Fold induction of Glut1 and Hk2 in response to DMOG treatment was measured by qRT-PCR and the ratio of untreated to DMOG-treated gene expression is shown (n = 6). (D) The increase in ROS production with hypoxia was calculated as the fold change in ROS in hypoxic cells relative to normoxic controls. (E) Immunoblots of SIRT3 WT and KO MEFs incubated with 10 mM N-acetylcysteine (NAC) and cultured under hypoxia. (F) Immunoblots of SIRT3 WT and KO MEFs cultured at 21% O2 with 10 mM NAC or 1 mM DMOG as indicated. (G) Glut1 expression was measured by qRT-PCR in SIRT3 WT and KO MEFs (n = 5) that were incubated with 10 mM NAC and cultured under hypoxia. Significance was assessed by one-way ANOVA. (G) Growth curves of SIRT3 WT and KO MEFs (n = 3) cultured in standard media or media supplemented with 10 mM NAC. Error bars, ±SD. Protein carbonyls (I) and lipid peroxidation (J) were measured in brown adipose tissue (BAT) of SIRT3 WT and KO mice (n = 7). (K) qRT-PCR analysis of Glut1 expression in BAT of SIRT3 WT and KO mice treated with 40 mM NAC. β-2-microglobulin or Rps16 were used as endogenous controls. Error bars (except for growth curves), ±SEM. (*) p < 0.05; (**) p < 0.01. See also Figure S4.
To confirm that SIRT3 influences HIF1α through the PHDs, we performed a series of experiments comparing the effects of hypoxia and DMOG treatment on SIRT3 WT and KO MEFs. We observed that both hypoxia and DMOG stabilize HIF1α and induce expression of HIF1α target genes (Figures 4B and 4C). The relative responses of SIRT3 WT and KO MEFs to hypoxia and DMOG underscore the PHDs as the point of regulation by SIRT3. During hypoxia, HIF1α target genes are induced more strongly in SIRT3 KO cells, illustrating the physiological importance of SIRT3 in regulating the metabolic response to hypoxia (Figure 4B). In contrast, SIRT3 deletion represses the induction of HIF1α target genes in response to DMOG (Figure 4C). These data support a model whereby PHD activity is already reduced in SIRT3 KO cells. Consequently, when PHD activity is potently blocked by DMOG, SIRT3 KO cells have a smaller change in PHD activity and thus a smaller induction of HIF1α target genes. Together, these results point to reduced PHD activity as the mechanism of increased HIF1α expression in SIRT3 deficient cells.
Several intracellular signals, in addition to changes in oxygen concentration, are known to regulate PHD activity. Notably, reactive oxygen species (ROS) have been shown to inhibit the PHDs and stabilize HIF1α (Gerald et al., 2004; Kaelin and Ratcliffe, 2008). Moreover, hypoxia triggers an increase in ROS production that is required for the hypoxic activation of HIF1α (Chandel et al., 1998; Hamanaka and Chandel, 2009). Because SIRT3 is a well-known inhibitor of ROS (Kawamura et al., 2010; Kim et al., 2010; Kong et al., 2010; Sundaresan et al., 2009), we hypothesized that increased ROS in SIRT3-deficient cells would contribute to the inhibition of the PHDs. Thus, we tested whether SIRT3 loss would magnify the increase in ROS associated with hypoxia. We found that the hypoxia-triggered increase in ROS was significantly higher in SIRT3 KO MEFs (Figure 4D), providing a mechanistic explanation for why SIRT3 null cells have an exaggerated response to hypoxia.
Next, we treated cells with the anti-oxidant N-acetylcysteine (NAC) in order to probe the model that suppressing ROS could block the effects of SIRT3 deletion. Indeed, we observed that while SIRT3 KO MEFs had higher levels of HIF1α during hypoxia, NAC treatment reduced HIF1α to comparable levels in SIRT3 WT and KO MEFs (Figure 4E). In contrast, SIRT3 WT and KO MEFs have comparable levels of HIF1α induced by DMOG (Figure 4F), and NAC could no longer destabilize HIF1α in the presence of DMOG (Figure 4F and S4D). As predicted by the decrease in HIF1α observed in NAC-treated KO MEFs, NAC treatment restored Glut1 expression in KO MEFs to WT levels (Figure 4G). Finally, to test whether increased ROS could underlie the proliferative phenotype of SIRT3 KO MEFs, we cultured cells with NAC and measured growth rates. Strikingly, we found that NAC rescued the increased proliferation of SIRT3 KO MEFs, restoring their growth to WT levels (Figure 4H). Thus, regulation of ROS by SIRT3 plays an important role in stabilization of HIF1α and activation of glycolytic metabolism in SIRT3 null cells.
To examine the contribution of increased ROS to altered BAT metabolism in vivo, we first looked for evidence of increased ROS in SIRT3 KO tissues. We found that two measures of oxidative damage, protein carbonyls and lipid peroxidation, were significantly elevated in SIRT3 KO BAT (Figures 4I and 4J). Because antioxidant treatment rescued the HIF1α-driven gene expression in cultured cells, we hypothesized that NAC treatment would reverse the glycolytic signature in SIRT3 KO tissues. To test this idea, we treated mice with NAC for one month and measured expression of HIF1α target genes in BAT. Strikingly, we found that NAC repressed expression of HIF1α target genes in SIRT3 KO mice, but not in SIRT3 WT mice (Figures 4K, S4E,F). These data demonstrate that increased ROS production in vivo contribute to enhanced glycolytic gene expression in SIRT3 deficient mice.
SIRT3 loss increases glycolytic signatures in tumors
HIF1α activity and aerobic glycolysis are strongly implicated in the Warburg effect (Semenza, 2010), and so we reasoned that SIRT3 may exert its tumor suppressive activity by opposing the HIF1α-mediated activation of the Warburg effect. Previously, SIRT3 deletion was shown to increase colony formation in a soft agar colony growth assay (Kim et al., 2010). To investigate the contribution of HIF1α to this tumorigenic phenotype, we transformed primary MEFs by expressing the Ras and E1a oncogenes and then stably knocked down HIF1α. As previously shown (Kim et al., 2010), we found that SIRT3 loss increased colony formation (Figure 5A). Importantly, knock down of HIF1α rescued the increased colony formation of SIRT3 KO cells (Figure 5A). Furthermore, SIRT3 WT and KO MEFs formed colonies at equivalent rates when cultured in media containing galactose instead of glucose (Figure S5A), suggesting that colony formation required glucose metabolism. Taken together, these data suggest that the metabolic reprogramming mediated by SIRT3 via HIF1α could be an important contributor of the tumor-suppressive role of SIRT3.
Figure 5.
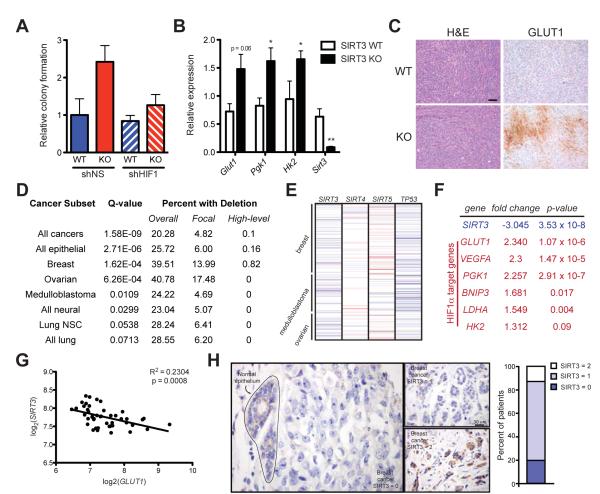
SIRT3 is significantly deleted in human breast cancer. (A) Soft agar assays using transformed SIRT3 WT and KO MEFs expressing shNS or shRNA against HIF1α (shHIF1) (n =4). (B) Quantitative RT-PCR on RNA isolated from xenograft tumors and normalized to expression of 36B4. (C) Hematoxylin and eosin (H&E) staining (left) and immunohistochemial analysis of GLUT1 expression (right) in xenograft tumors. One representative pair of contralateral tumors is shown. Scale bar, 50 μm. (D) Table summarizing SIRT3 deletion frequency across a panel of human tumors. (E) Schematic of copy number changes at the SIRT3-5 and TP53 loci. Amplifications are shown in red; deletions are shown in blue. (F) Expression levels of SIRT3 and several HIF1α target genes were determined using the Oncomine cancer microarray database (http://www.oncomine.org) in normal versus breast cancers. (G) Linear regression of SIRT3 and GLUT1 across the panel of normal and breast cancer samples described in (F). (H) Representative image of SIRT3 expression in normal breast epithelium and in breast tumor cells as assessed by immunohistochemistry. SIRT3 levels were classified as absent (0), weak scattered (1) or positive as strong (2) compared to normal epithelium and the percentage of patients classified in each category is depicted in histogram at right. Error bars, ±SEM (n = 4-6). (*) p <0.05; (**) p <0.01. See also Figure S5.
Next, we performed xenograft assays with the transformed MEFs in order to probe the metabolic status of SIRT3 null tumors. As has previously been shown (Kim et al., 2010), we found that tumors lacking SIRT3 had a growth advantage: tumors formed from 64% of KO injections but only 27% of WT injections and tumors lacking SIRT3 grew faster and were bigger than WT tumors (Figures S5B-F). As tumors are subject to intermittent hypoxia (Gatenby and Gillies, 2004), we examined expression of rate-limiting glycolytic genes in the xenograft tumors. Strikingly, HIF1α target genes were elevated in SIRT3 KO tumors (Figure 5B); SIRT3 KO tumors also showed higher levels of GLUT1 protein (Figure 5C). Taken together, these data suggest that increased levels of glycolytic enzymes, perhaps as part of a heightened response to hypoxia, provides a growth advantage for tumor cells lacking SIRT3 in vivo.
SIRT3 is deleted in many human cancers
Our data indicate that SIRT3 may regulate tumor cell metabolism and anabolic growth pathways. In order to determine the relevance of SIRT3 in human cancers, we first examined the copy-number variations of SIRT3 that are associated with the progression of multiple types of human cancer (Beroukhim et al., 2010). Strikingly, at least one copy of the SIRT3 gene is deleted in 20% of all human cancers and 40% of breast and ovarian cancers present in the dataset (Figure 5D). SIRT3 is significantly focally deleted (deletions of less than a chromosome arm) across all cancers, and focal deletions of SIRT3 were especially frequent in breast and ovarian tumors (Figure 5D). In contrast, SIRT4 and SIRT5 were not significantly focally deleted in any of the 14 subtypes analyzed (Figure 5E and data not shown). TP53, a tumor suppressor known to be frequently deleted in many human cancers, is included as a control (Fisher, 2001) (Figure 5E and S4G,H). Our analysis of copy-number changes at the SIRT3 locus revealed no evidence of focal amplifications across 14 types of cancer. Most of the genomic SIRT3 deletions are heterozygous, and SIRT3 deletion frequencies are similar to the well-known breast cancer tumor suppressors, BRCA1 and BRCA2, which are heterozygously deleted in 43% and 40% of human breast cancers, respectively (data not shown). Intriguingly, the peak region of deletion that includes SIRT3 (11p15.5) does not contain any known tumor suppressor (Beroukhim et al., 2010).
Because breast cancers exhibited exceptionally high frequency of SIRT3 deletions compared to other tumor types (Figure 5D) (Kim et al., 2010), we further examined SIRT3 in human breast cancers. Elevated HIF1α expression in breast carcinomas is associated with tumor aggressiveness and poor prognosis (Chaudary and Hill, 2006). Many breast cancer cells exhibit increased glycolysis, and expression of GLUT1 is a characteristic feature of may breast cancer biopsies (Rivenzon-Segal et al., 2003). In xenograft models, SIRT3 loss increases expression of HIF1α target genes and results in strong GLUT1 expression (Figure 5B and 5C). Thus, we looked for a relationship between SIRT3 loss and HIF1α targets in human breast cancer. Gene expression profiling of 7 normal breast samples and 40 ductal breast carcinomas revealed that SIRT3 expression is significantly reduced (p = 3.53e−8) in breast carcinomas (Richardson et al., 2006) (Figure 5F). Moreover, several HIF1α target genes—most notably GLUT1—were significantly increased in the same dataset (Figure 5F). We further analyzed the correlation between SIRT3 and GLUT1 expression in individual samples from this data set and found that SIRT3 is significantly inversely correlated with GLUT1 (p = 0.0008) (Figure 5G). Our results demonstrate that SIRT3 loss is associated with increased expression of HIF1α target genes in vivo and in human breast cancer and provide a metabolic link between SIRT3 deletion and breast cancer tumorigenesis.
To confirm that SIRT3 expression is reduced in human breast cancers, we analyzed SIRT3 protein levels by immunohistochemistry in normal breast epithelium in addition to a large panel of human breast cancer tissue. Out of 46 patient samples, only 6 demonstrated SIRT3 staining that was positive or as strong as SIRT3 staining in normal epithelium (Figure 5H). Strikingly, 87% of patients showed decreased or undetectable SIRT3 staining in adjacent cancer tissue and 20% of patients showed no detectable SIRT3 (Figure 5H). Similarly, gene expression profiling of an independent set of human breast cancer samples (Richardson et al., 2006) revealed that 25% of breast cancers exhibited at least a six-fold reduction in the mRNA of SIRT3 compared to normal breast epithelium (Figure S5I). This independent dataset provides additional validation for the observation that SIRT3 is deleted in human tumors (Figure 5D) (Beroukhim et al., 2010). Furthermore, an earlier high-resolution analysis of copy number variation in 171 human breast tumors similarly found significant reduction in SIRT3 copy number (Chin et al., 2007). These findings also support those of Kim et al. who first reported that SIRT3 KO mice develop mammary tumors and that SIRT3 levels were decreased in human breast cancer (Kim et al., 2010).
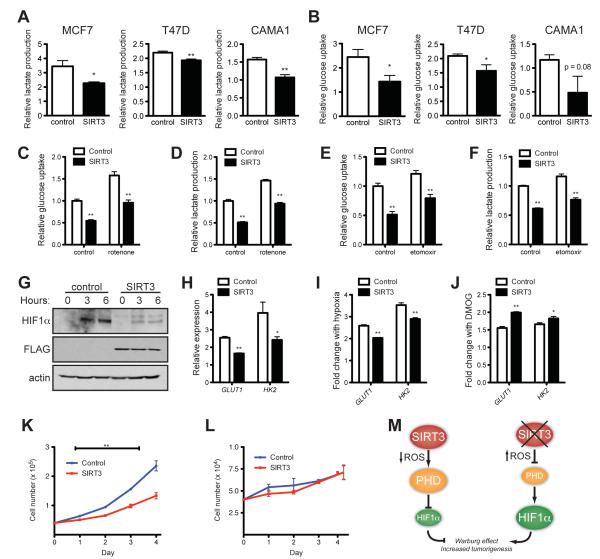
The studies of SIRT3 expression in human cancers suggest that SIRT3 may function as a tumor suppressor in part by preventing the metabolic shift that facilitates tumor growth. In order to examine whether SIRT3 can actively repress the Warburg effect in tumor cells, we stably overexpressed SIRT3 in three independent breast cancer cell lines: MCF7, T47D and CAMA1 (Figure S6A). We analyzed the glucose uptake and lactate secretion in cells during hypoxia in order to simulate the tumor microenvironment. We found that SIRT3 repressed both lactate production and glucose uptake in every cell line tested (Figures 6A and 6B). These data clearly demonstrate that overexpression of SIRT3 in tumor cells is sufficient to reverse the metabolic shift associated with the Warburg effect.
Figure 6.
SIRT3 suppresses the Warburg effect in human breast cancer cells. (A) Lactate production and (B) glucose consumption of MCF7, T47D and CAMA1 cells stably expressing empty vector or SIRT3 and cultured under hypoxia expressed as a ratio of empty-vector normoxic controls. (C) Relative glucose uptake and (D) relative lactate production in CAMA1 control or SIRT3 overexpressing cells incubated with or without 100 nM rotenone. (E) Glucose uptake and (F) lactate production in CAMA1 cell lines cultured in the presence or absence of 50 μg/ml etomoxir. (G) Immunoblots of CAMA1 cells stably expressing control vector or SIRT3-FLAG cultured at 1% oxygen for the indicated. (H) qRT-PCR of HIF1α target genes in CAMA1 cells cultured at 1% oxygen. (I) Induction of HIF1α target genes in response to hypoxia measured by qRT-PCR in CAMA1 cells. The ratio of normoxic to hypoxic gene expression in each cell line is shown. (J) Induction of HIF1α target genes in response to 1 mM DMOG treatment measured by qRT-PCR in CAMA1 cells. The ratio of untreated to DMOG-treated gene expression in each cell line is shown. Growth curves of CAMA1 cells (n =3) cultured in glucose (K) or galactose (L). Error bars, ±SD. (M) Schematic of the regulation of HIF1α and the Warburg effect by SIRT3. β-2-microglobulin was used as an endogenous control for qRT-PCR. Error bars (except for growth curves), ±SEM. (*) p <0.05; (**) p <0.01. See also Figure S6.
Because SIRT3 robustly suppressed glucose uptake and lactate production in the CAMA1 cells, we chose to further analyze these cell lines. To examine the contribution of complex I activity or fatty acid oxidation on these phenotypes, we measured glucose uptake and lactate production in the presence of rotenone and etomoxir. Both rotenone and etomoxir increased glucose uptake and lactate production to a similar degree in both control and SIRT3 overexpressing cell lines, indicating that the repression of glycolysis by SIRT3 is independent of the influence of SIRT3 on fatty acid oxidation or complex I activity (Figures 6C-F).
We next examined whether SIRT3 repressed HIF1α in CAMA1 cells. SIRT3 overexpression strongly reduced HIF1α protein levels and expression of HIF1α target genes in hypoxic cells (Figures 6G and 6H). Moreover, when we examined the fold change of HIF1α targets in response to hypoxia or DMOG treatment, we found the inverse of the results using SIRT3 KO MEFs. SIRT3 overexpression blunted the response to hypoxia (Figure 6I) while increasing the response to DMOG (Figure 6J). This is consistent with a model of elevated PHD activity in SIRT3 overexpressing cells and illustrates the importance of SIRT3 in regulating the physiological response to hypoxia at the level of the PHDs.
Next, we tested the hypothesis that SIRT3-mediated control of glucose metabolism could influence cancer cell proliferation. SIRT3 overexpression significantly repressed proliferation of CAMA1 cells cultured in high glucose (Figure 6K). Remarkably, control and SIRT3-expressing cells proliferated at similar rates when cultured in media containing galactose instead of glucose (Figure 6L). These data illustrate that SIRT3 regulates cancer cell growth by influencing the use of glucose for anabolic processes.
Discussion
In this study, we demonstrate that SIRT3 regulates cellular metabolism through HIF1α with important implications for tumor cell growth. Previously, it has been shown that SIRT3 is a mitochondrial deacetylase that activates multiple metabolic enzymes and promotes mitochondrial substrate oxidation and ATP production (Finkel et al., 2009; Verdin et al., 2010). Our study shows that SIRT3 additionally controls glycolytic metabolism (Figures 1 and 2) by regulating the stability and activity of HIF1α (Figure 3). We find that elevated ROS in SIRT3 null cells contributes to increased HIF1α stabilization and activity (Figure 4). Significantly, loss of SIRT3 in human tumor samples correlates with glycolytic gene expression, highlighting the potential importance of SIRT3-mediated metabolic reprogramming in human cancers (Figure 5). This idea is further validated by the finding that SIRT3 represses the Warburg effect in human breast cancer cell lines (Figure 6). Taken together, these data provide a mechanism whereby SIRT3 functions as a tumor suppressor by regulating glycolytic and anabolic metabolism (Figure 6M).
Our findings are consistent with previous work showing that SIRT3 functions as a tumor suppressor through regulation of ROS (Kim et al., 2010). Kim et al. found that elevated ROS in the absence of SIRT3 increased genomic instability, promoting a tumor-permissive environment. We propose that SIRT3 loss and increased ROS also promote tumorigenesis by altering global cellular metabolism. In this study, we demonstrate that elevated ROS stabilizes HIF1α, increasing glucose uptake and catabolism and thus providing the metabolic precursors necessary to fuel a high rate of proliferation. Importantly, we demonstrate that increased glycolysis is not simply compensation for reduced mitochondrial oxidative capacity. Rather, SIRT3 actively regulates cellular glucose metabolism by activating a specific signaling node (Figures 1K-N and 6C-F). Thus, taken together, our study and the Kim and al. study show that SIRT3 loss results in a double-edged sword for tumor cells – creating an environment of increased genome instability as well as HIF1α activation, enabling increased glycolysis and cellular growth.
Recent studies have shed light on the mechanism through which SIRT3 regulates cellular ROS. Several groups have provided evidence that SIRT3 can influence transcription of antioxidant genes through activation of FoxO3a (Kim et al., 2010; Sundaresan et al., 2009), although we did not find a difference in Sod2 expression under our culture conditions (Figure S6B). Additionally, SIRT3 can directly target IDH2, influencing cellular redox status, and SOD2, activating mitochondrial ROS scavenging (Qiu et al., 2010; Someya et al., 2010; Tao et al., 2010). As a result, SIRT3 can directly influence mitochondrial metabolism and ROS generation through deacetylation of multiple substrates. At the same time, the ROS byproduct of reduced SIRT3 activity acts as a retrograde signal to reprogram cellular metabolism.
Our studies reveal the profound impact of SIRT3 function on glycolysis and tumor cell metabolism. SIRT3 appears to be decreased in human breast cancers (Figure 5) (Beroukhim et al., 2010; Chin et al., 2007; Kim et al., 2010; Richardson et al., 2006). It will be important for future work to examine the impact of SIRT3 on each stage of tumorigenesis in mechanistic detail. Given the function of SIRT3 in breast cancer (Kim et al., 2010), in addition to our findings that SIRT3 can reprogram cellular metabolism (Figures 1 and 2) we propose that the glycolytic switch evident in cells lacking SIRT3 will contribute to tumorigenesis, particularly in breast cancers. In support of this idea, we show that SIRT3 can directly repress the Warburg effect in three independent breast cancer cell lines (Figure 6). In sum, our studies illustrate that SIRT3 functions as a tumor suppressor, in part by regulating cellular metabolism through HIF1α. These findings suggest that the regulation of tumor cell metabolism by SIRT3 could provide an important area for cancer diagnosis or therapeutic intervention.
Experimental Procedures
Metabolite profiling
Metabolites were extracted in ice-cold methanol and endogenous metabolite profiles were obtained using two liquid chromatography-tandem mass spectrometry (LC-MS) methods as described (Luo et al., 2007). Data were acquired using a 4000 QTRAP mass spectrometer (Applied Biosystems/Sciex). MultiQuant software (Applied Biosystems/Sciex) was used for analysis. Metabolite levels were normalized to protein content, which was determined by performing a Bradford assay (Bio-Rad) on a duplicate set of cells treated identically to the experimental cells.
Lactate and glucose measurements
Glucose and lactate levels in culture media were measured using the BioProfile FLEX analyzer (NOVA biomedical) and normalized to cell number or using the Lactate Reagent Kit (Trinity Biosciences). Fresh media was added to a 6-well plate of subconfluent cells and lactate concentration in the media was measured 30-60 minutes (Lactate Reagent Kit) or 6-24 hours (BioProfile analyzer) later and normalized to the number of cells in each well.
Reactive oxygen species measurement
Cellular ROS was measured according to published protocols (Eruslanov and Kusmartsev, 2010). Briefly, cells were washed with PBS and incubated with 5 μM CM-H2DCFDA (Invitrogen) for 30 minutes. Cells were trypsinized and mean FL1 fluorescence was measured by flow cytometry.
Oxidative damage
Protein carbonyl content was determined as previously described (Levine et al., 1994). Levels of lipid peroxidation were assayed using the thiobarbituric acid reactive substances (TBARS) were determined according to a modification of a published procedure (Ernster et al., 1968).
Animal studies
Animal studies were performed according to protocols approved by the Institutional Animal Care and Use Committee, the Standing Committee on Animals at Harvard. Male 129Sv SIRT3 WT and KO (Lombard et al., 2007) littermates (a generous gift from Dr. Fred Alt) fed a normal chow diet (PicoLab diet 5053) were used for all studies. PET/CT studies were performed at the Longwood SAIF (Boston, MA). 8-10 month old male mice were injected with 300 μCi 18F-FDG and imaged one hour later on PET/CT. For cold challenge, mice were injected after 6 hours at 4°C. Results were analyzed using InVivoScope software. For N-acetylcysteine studies (NAC), drinking water was supplemented with 40 mM NAC for four weeks prior to sacrifice. For xenograft studies, 5 × 106 or 7.5 × 106 SIRT3 WT or KO MEFs transformed by expression of the Ras and E1a oncogenes were mixed with matrigel (BD Biosciences) and injected subcutaneously into nude mice (6-9 week old males, Taconic Farms). Each mouse was injected with WT cells on one flank and KO cells on the other flank. Tumor size was measured every two days, and tumors were dissected and weighed after 4 weeks.
Immunohistochemistry
Tumors were fixed in 4% paraformaldehyde and embedded in paraffin. Sections were stained with hematoxylin and eosin (H&E) in accordance with standard procedures. Immunohistochemistry was performed using antibodies against GLUT1 (Alpha Diagnostic) according to manufacturer instructions. A tissue microarray (TMA) was constructed as previously published using a fully automated Beecher Instrument, ATA-27. The study cohort comprised of breast carcinoma, consecutively ascertained at the Memorial Sloan-Kettering Cancer Center (MSKCC) between 1993 and 2005. Use of tissue samples were approved with an Institutional Review Board Waiver and samples were de-identified prior to analysis. All breast cancer biopsies were evaluated at MSKCC, and the histological diagnosis was based on H&E staining. One to several cores contained normal breast duct epithelium. The TMA was stained with an antibody against SIRT3 (Cell Signaling) with pretreatment conditions including citrate buffer and microwave at 1:100 dilution. Cores were scored by a pathologist (JTF) and tumor staining intensity compared to normal breast duct epithelium as: 0=tumor showing no staining; 1=tumor weaker than normal epithelium; 2=tumor of equal or stronger intensity compared to normal ductal epithelium (Figure S5J). Histologic immunohistochemical images for Figure 4E were obtained with the Olympus AH2 microscope Camera from Center Valley, Pennsylvania. Image acquisition and processing software was performed using Adobe Photoshop 7.0 and Olympus DP12 software. Magnification: x400 (scale bar ~30 um).
Statistics
Unpaired two-tailed student’s t-tests were performed unless otherwise noted. All experiments were performed at least two to three times.
Significance.
The dysregulation of cell metabolism is a unique and defining feature of many tumor cells. Thus, identifying novel factors that regulate cancer cell metabolism will enhance our understanding of how tumors hijack metabolism for selective advantages and provide novel approaches for cancer therapy. We find that loss of SIRT3, a sirtuin with NAD-dependent deacetylase activity, stabilizes HIF1α and shifts cellular metabolism towards increased glycolysis, a common metabolic switch in cancer cells. Importantly, we also show that SIRT3 overexpression represses the Warburg effect. Our studies identify a high rate of SIRT3 deletion in human cancers and suggest that SIRT3 may be an important therapeutic target.
Supplementary Material
Acknowledgements
We thank Fred Alt for the SIRT3 KO mice. We thank Elaine Lunsford and the Longwood SAIF for the PET/CT imaging and analysis, Kristin Waraska and the Harvard Biopolymers Facility for the running the Taqman assays, Kelly Dakin and Bruce Yankner for use of their hypoxic incubator, Natalie German and Maren Shapiro for technical assistance, Ditte Lee for help with mouse work and Maria Jiao in the Laboratory of Comparative Pathology, Sloan Kettering Institute for immunohistochemical technical assistance. We thank Kevin Haigis, Carla Kim and Karen Cichowski for critical reading of the manuscript, and David Gius, Sandra Ryeom, Gaelle Laurent, Lenny Guarente and Eric Bell for helpful discussion. L.F. is supported by a National Science Foundation graduate research fellowship. M.H. is supported in part by NIH grant AG032375, Alexander and Margaret Stewart Trust, and funding from the Muscular Dystrophy Association.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession number. The SIRT3 BAT microarray has been deposited in a GEO database with accession number GSE27309.
References
- Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, Finkel T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci U S A. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahimi-Horn MC, Chiche J, Pouyssegur J. Hypoxia signalling controls metabolic demand. Curr Opin Cell Biol. 2007;19:223–229. doi: 10.1016/j.ceb.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- Chandel NS, Maltepe E, Goldwasser E, Mathieu CE, Simon MC, Schumacker PT. Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc Natl Acad Sci U S A. 1998;95:11715–11720. doi: 10.1073/pnas.95.20.11715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudary N, Hill RP. Hypoxia and metastasis in breast cancer. Breast Dis. 2006;26:55–64. doi: 10.3233/bd-2007-26105. [DOI] [PubMed] [Google Scholar]
- Chin SF, Teschendorff AE, Marioni JC, Wang Y, Barbosa-Morais NL, Thorne NP, Costa JL, Pinder SE, van de Wiel MA, Green AR, et al. High-resolution aCGH and expression profiling identifies a novel genomic subtype of ER negative breast cancer. Genome Biol. 2007;8:R215. doi: 10.1186/gb-2007-8-10-r215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Cimen H, Han MJ, Yang Y, Tong Q, Koc H, Koc EC. Regulation of succinate dehydrogenase activity by SIRT3 in mammalian mitochondria. Biochemistry. 2010;49:304–311. doi: 10.1021/bi901627u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavo AC, Brown RS, Wahl RL. Fluorodeoxyglucose uptake in human cancer cell lines is increased by hypoxia. J Nucl Med. 1995;36:1625–1632. [PubMed] [Google Scholar]
- Denkert C, Budczies J, Weichert W, Wohlgemuth G, Scholz M, Kind T, Niesporek S, Noske A, Buckendahl A, Dietel M, Fiehn O. Metabolite profiling of human colon carcinoma--deregulation of TCA cycle and amino acid turnover. Mol Cancer. 2008;7:72. doi: 10.1186/1476-4598-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernster L, Nordenbrand K, Orrenius S, Das ML. Microsomal lipid peroxidation. Hoppe Seylers Z Physiol Chem. 1968;349:1604–1605. [PubMed] [Google Scholar]
- Eruslanov E, Kusmartsev S. Identification of ROS using oxidized DCFDA and flow-cytometry. Methods Mol Biol. 2010;594:57–72. doi: 10.1007/978-1-60761-411-1_4. [DOI] [PubMed] [Google Scholar]
- Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher DE. The p53 tumor suppressor: critical regulator of life & death in cancer. Apoptosis. 2001;6:7–15. doi: 10.1023/a:1009659708549. [DOI] [PubMed] [Google Scholar]
- Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- Gerald D, Berra E, Frapart YM, Chan DA, Giaccia AJ, Mansuy D, Pouyssegur J, Yaniv M, Mechta-Grigoriou F. JunD reduces tumor angiogenesis by protecting cells from oxidative stress. Cell. 2004;118:781–794. doi: 10.1016/j.cell.2004.08.025. [DOI] [PubMed] [Google Scholar]
- Gordan JD, Simon MC. Hypoxia-inducible factors: central regulators of the tumor phenotype. Curr Opin Genet Dev. 2007;17:71–77. doi: 10.1016/j.gde.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb E, Tomlinson IP. Mitochondrial tumour suppressors: a genetic and biochemical update. Nat Rev Cancer. 2005;5:857–866. doi: 10.1038/nrc1737. [DOI] [PubMed] [Google Scholar]
- Hamanaka RB, Chandel NS. Mitochondrial reactive oxygen species regulate hypoxic signaling. Curr Opin Cell Biol. 2009;21:894–899. doi: 10.1016/j.ceb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama A, Kami K, Sugimoto M, Sugawara M, Toki N, Onozuka H, Kinoshita T, Saito N, Ochiai A, Tomita M, et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 2009;69:4918–4925. doi: 10.1158/0008-5472.CAN-08-4806. [DOI] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CJ, Wang LY, Chodosh LA, Keith B, Simon MC. Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol Cell Biol. 2003;23:9361–9374. doi: 10.1128/MCB.23.24.9361-9374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG, Jr., Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Kawamura Y, Uchijima Y, Horike N, Tonami K, Nishiyama K, Amano T, Asano T, Kurihara Y, Kurihara H. Sirt3 protects in vitro-fertilized mouse preimplantation embryos against oxidative stress-induced p53-mediated developmental arrest. J Clin Invest. 2010;120:2817–2828. doi: 10.1172/JCI42020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17:41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Wang R, Xue Y, Liu X, Zhang H, Chen Y, Fang F, Chang Y. Sirtuin 3, a new target of PGC-1alpha, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS One. 2010;5:e11707. doi: 10.1371/journal.pone.0011707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RL, Williams JA, Stadtman ER, Shacter E. Carbonyl assays for determination of oxidatively modified proteins. Methods Enzymol. 1994;233:346–357. doi: 10.1016/s0076-6879(94)33040-9. [DOI] [PubMed] [Google Scholar]
- Lim JH, Lee YM, Chun YS, Chen J, Kim JE, Park JW. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1alpha. Mol Cell. 2010;38:864–878. doi: 10.1016/j.molcel.2010.05.023. [DOI] [PubMed] [Google Scholar]
- Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Bennet B, Mu E, Rabinowitz J, Kang Y. Metabolomic changes accompanying transformation and acquisition of metastatic potential in a syngeneic mouse mammary tumor model. J Biol Chem. 2010 doi: 10.1074/jbc.C110.104448. doi: 10.1074/jbc.C1110.104448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B, Groenke K, Takors R, Wandrey C, Oldiges M. Simultaneous determination of multiple intracellular metabolites in glycolysis, pentose phosphate pathway and tricarboxylic acid cycle by liquid chromatography-mass spectrometry. J Chromatogr A. 2007;1147:153–164. doi: 10.1016/j.chroma.2007.02.034. [DOI] [PubMed] [Google Scholar]
- Marroquin LD, Hynes J, Dykens JA, Jamieson JD, Will Y. Circumventing the Crabtree effect: replacing media glucose with galactose increases susceptibility of HepG2 cells to mitochondrial toxicants. Toxicol Sci. 2007;97:539–547. doi: 10.1093/toxsci/kfm052. [DOI] [PubMed] [Google Scholar]
- O’Donnell JL, Joyce MR, Shannon AM, Harmey J, Geraghty J, Bouchier-Hayes D. Oncological implications of hypoxia inducible factor-1alpha (HIF-1alpha) expression. Cancer Treat Rev. 2006;32:407–416. doi: 10.1016/j.ctrv.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010;12:662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, Liao X, Iglehart JD, Livingston DM, Ganesan S. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Rivenzon-Segal D, Boldin-Adamsky S, Seger D, Seger R, Degani H. Glycolysis and glucose transporter 1 as markers of response to hormonal therapy in breast cancer. Int J Cancer. 2003;107:177–182. doi: 10.1002/ijc.11387. [DOI] [PubMed] [Google Scholar]
- Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol. 2008;382:790–801. doi: 10.1016/j.jmb.2008.07.048. [DOI] [PubMed] [Google Scholar]
- Seagroves TN, Ryan HE, Lu H, Wouters BG, Knapp M, Thibault P, Laderoute K, Johnson RS. Transcription factor HIF-1 is a necessary mediator of the pasteur effect in mammalian cells. Mol Cell Biol. 2001;21:3436–3444. doi: 10.1128/MCB.21.10.3436-3444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Curr Opin Genet Dev. 2010;20:51–56. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y, Nikami H, Saito M. Sympathetic activation of glucose utilization in brown adipose tissue in rats. J Biochem. 1991;110:688–692. doi: 10.1093/oxfordjournals.jbchem.a123642. [DOI] [PubMed] [Google Scholar]
- Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, Tanokura M, Denu JM, Prolla TA. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143:802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, Gupta MP. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J Clin Invest. 2009;119:2758–2771. doi: 10.1172/JCI39162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, Jiang H, Kim HS, Flynn CR, Hill S, McDonald W. Hayes, et al. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell. 2010;40:893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennant DA, Duran RV, Gottlieb E. Targeting metabolic transformation for cancer therapy. Nat Rev Cancer. 2010;10:267–277. doi: 10.1038/nrc2817. [DOI] [PubMed] [Google Scholar]
- Tong X, Zhao F, Thompson CB. The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Curr Opin Genet Dev. 2009;19:32–37. doi: 10.1016/j.gde.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E, Hirschey MD, Finley LW, Haigis MC. Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends Biochem Sci. 2010 doi: 10.1016/j.tibs.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Xu J. Preparation, culture, and immortalization of mouse embryonic fibroblasts. Curr Protoc Mol Biol. 2005 doi: 10.1002/0471142727.mb2801s70. Chapter 28, Unit 28 21. [DOI] [PubMed] [Google Scholar]
- Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, Yu W, Li Z, Gong L, Peng Y, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324:261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.