Abstract
Respiratory syncytial virus (RSV) is a widely distributed pathogen that causes severe disease in children, the elderly, and immunocompromised individuals. Both vaccine development and drug discovery have been hampered by the inherent instability of the virus itself. Drug discovery efforts have had limited success due, at least in part, to the lack of an antiviral assay robust enough for high-throughput screening. Instability of the purified virus has long been recognized as a problem in RSV research and has been a major hurdle to producing a virus-based screening assay. Using frozen RSV-infected cells as the source of infectious material, we have overcome the problem of virus instability and validated a cell-based high-throughput screening assay to screen for inhibitors of RSV-induced cytopathic effect. The assay was validated with 1,280 compounds identified as potentially active against RSV (Long strain) in a virus-based screen. To date over 300,000 compounds have been screened over several months with minimal variability in cell or virus controls. Long-term assay stability studies are still in progress.
Introduction
Respiratory syncytial virus (RSV), from the family Paramyxoviridae, is most often associated with bronchitis and pneumonia among infants and children under 1 year of age, but the elderly or those with compromised immune systems, cardiac disease, or pulmonary disease are also at risk of severe lower respiratory disease.1 RSV infections cost more than $650 million annually due to hospitalizations and indirect medical costs.2 Available chemotherapeutic agents are limited to Ribavirin3 and the prophylactic humanized monoclonal antibody (Synagis® from MedImmune), which are used in hospitalized high-risk pediatric patients.4 Virus instability has been well documented since the early days of RSV research5 and is still problematic in both vaccine development and drug discovery today. Currently, there is no vaccine to prevent disease and the live attenuated vaccines that have been developed have a limited shelf life due to virus instability, making production and distribution of these vaccines difficult.6 A vaccine and therapeutic agents are critically needed for RSV, but drug discovery has been hampered by the lack of a robust antiviral assay for high-throughput screening (HTS). Mason et al. used a biochemical assay and identified a novel scaffold, from a small molecule screen, that inhibits the RSV viral RNA-dependent RNA polymerase.7 To identify broad classes of compounds effective against multiple targets, a cell-based antiviral assay would be highly desirable. To our knowledge, there have been no reported HTS screening efforts for identification of RSV inhibitors using cytopathic effect (CPE) as an endpoint. This is due in part to the instability of the virus and the inconsistent results produced during an HTS campaign.
We have developed a number of robust CPE-based antiviral assays that have been used for HTS campaigns.8–12 Early attempts to develop a CPE-based assay for RSV proved to be more difficult. We developed and optimized a cell-based screen for potential RSV antiviral compounds that measures the CPE induced by RSV (Long strain) infection in HEp-2 cells using the luminescent-based viability endpoint CTG® (Promega) to measure cell viability. A number of modifications to medium formulation and virus stock preparation were investigated, and a 110,000-compound screen on an internal proprietary library was run before virus instability stopped the screen.
To overcome the virus instability issue, methodologies from the fields of retrovirology and HTS were combined to produce a robust and reproducible HTS assay. Retroviruses have also been found to be unstable as purified virus stocks. As a result of this, researchers in this field have used infected cells to perpetuate the viral infection in cell culture13 and have frozen infected cells (FICs), using standard cell cryopreservation methodology, for long-term storage of infectious material. These cells remain infectious for years, if stored in liquid nitrogen, and can be used to re-establish infection by addition to uninfected cells in culture. In HTS, frozen cells are routinely used as the source of cells for screening.14 Cells are thawed, diluted, and plated in assay plates for many cell-based assays and this helps minimize the variability often observed in cell-based assays due to passage number and other variables. These two strategies have been combined to create a robust and reproducible RSV assay for HTS by using RSV FICs as the source of infectious material.
Materials and Methods
Cell Culture
HEp-2 cells (American Type Culture Collection #CCL-23) were maintained in Opti-MEM1® (Gibco) with 10% fetal bovine serum (FBS) (Gibco HI-FBS), 100 units/mL penicillin and 100 μg/mL streptomycin (Gibco) in T-125 flasks. Cells were split 1:5 twice a week and incubated at 37°C, 5% CO2, and 90% relative humidity. For passage, medium and Trypsin/EDTA (Gibco) were prewarmed. The medium was removed and the monolayer was washed with 10 mL phosphate-buffered saline, and 3 mL of 0.05% Trypsin/EDTA was added to the flask and incubated at 37°C for ∼5 min. Seven milliliters of complete Opti-MEM was added and 2 mL of the cell suspension was transferred to new T-125s containing 30 mL of the medium. Flasks were incubated at 37°C, 5% CO2, 90% relative humidity.
Frozen Infected Cells
The FIC stocks were prepared by infecting 225 cm2 tissue culture flasks at 95% confluence. The tissue culture medium was aspirated from each flask and 5 mL of the complete assay medium containing recently harvested high-titer RSV was added to each flask and distributed evenly. This provided a multiplicity of infection of ∼5. Flasks were incubated at 37°C, 5% CO2 for 1 h. After incubation, 35 mL of the complete assay medium was added to each flask and the flasks were returned to the incubator for an additional 17 h.
After 18 h total incubation, the flasks were removed from the incubator and the medium was aspirated and discarded. The cells were washed two times with phosphate-buffered saline and trypsinized in 3 mL trypsin for 5–10 min at 37°C. After the cells were harvested, they were counted, and viability was evaluated by trypan blue exclusion and determined to be at least 99% viable. Cells were then centrifuged to remove the trypsin and resuspended in 95% FBS and 5% dimethyl sulfoxide (DMSO) at a concentration of 2 × 106 viable cells/mL. The cells were dispensed in 1 mL aliquots and rate frozen at −1°C/min to −80°C and then transferred to a −150°C freezer for long-term storage. Viability was also evaluated when thawed for use and was calculated to be at least 98.5%.
Quantitation: Tissue Culture Infectious Dose 50% of FICs
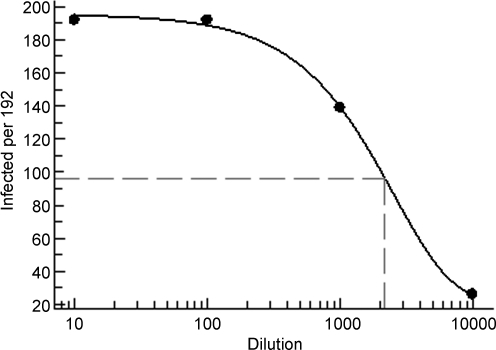
Because of the similarity of evaluating effective concentration 50% (EC50) values for compounds and tissue culture infectious dose 50% (TCID50) values for infectious materials, we used a strategy similar to the one used for evaluating dose–response data for compounds to determine TCID50 of the FICs. Activity in this case is defined as the number of RSV-positive wells and was plotted against the FIC dilution on a log scale. FICs were quantitated by diluting the FIC stock to the plating density of 80,000 cells/mL, and then log dilutions were done with uninfected cells at the same density. One hundred ninety-two wells (half of a 384-well plate) were seeded with the cell mixtures at 2,000 cells/well for dilutions from 10−1 to 10−4. Cell viability was determined 6 days postinfection using CellTiter-Glo (CTG). Infected wells showing a 50% or greater reduction in signal were scored as positive for virus. The TCID50 was determined by plotting the number of virus-positive wells versus dilution (Fig. 1). Excel XLfit formula 504 was used to calculate the dilution at which 50% infected wells (96 out of 192) occurred. TCID50 is defined as the amount of virus that produces CPE in 50% of the wells infected and is expressed as log10 of this dilution.
Fig. 1.
Plot of the number of virus-positive wells out of 192 wells tested versus dilution for TCID50 determination. TCID50 is the amount of virus that produces a CPE in 50% of the wells infected. Excel XLfit formula 504 was used to plot the data and calculate the dilution at which 96 positive wells (50% of the 192 tested) would occur. Log10 of this dilution is defined as the TCID50. TCID50 = 3.34. CPE, cytopathic effect; TCID50, tissue culture infectious dose 50%.
Assay
Assay medium
The assay medium used was Dulbecco's modified Eagle's medium/F12 (Sigma) with Penicillin-Streptomycin-Glutamine (Gibco) added to a final concentration of 1.25% (V/V). The pH was adjusted to 7.5 using 1N NaOH, and then the medium was filter sterilized. FBS was added to a final concentration of 2% (V/V) and the assay medium was stored in the dark at 4°C.
Compound Preparation
Compounds were diluted to 100 μM (10 ×) in the assay medium and then 3 μL of the diluted compound plus 2 μL of the medium was transferred to the assay plate. For cell and virus controls 5 μL of the medium containing 0.6% DMSO was dispensed to the wells. All compound handling was done on a Biomek® FXP (Beckman Coulter). Cells were added in 25 μL using a Wellmate (Matrix), for a final compound concentration of 10 μM with 0.1% DMSO and 2,000 cells/well. The control drug, Ribavirin, was diluted in the assay medium containing 0.6% DMSO to 210 μM (6 × ) and 5 μL was dispensed to the control wells for a final concentration of 35 μM and 0.1% DMSO.
Cell Preparation
HEp-2 cells were harvested as described above, centrifuged at 500 rpm for 5 min to remove the medium and trypsin, and suspended to 80,000 cells/mL using the assay medium. FICs were removed from the freezer and thawed in a room temperature water bath with constant gentle agitation. The tube was then inverted slowly 8–10 times to mix the cells. FICs were diluted 1:25 to give a final concentration of 80,000 cells/mL and added to the uninfected cells at predetermined ratio (1:100) determined empirically to produce CTG values equivalent to 3%–5% viability after 6 days, replicating the virus-based assay results that were previously obtained. TCID50s were also determined for each lot of FICs in the same way that new virus stocks were titrated before use. For this lot of FICs, the TCID50 was 3.34. This was equivalent to 4 × 104 infected cells/mL or a 50% rate of infection of the FICs. On the basis of these calculations, ∼10 RSV-infected cells were dispensed per well for the assay.
Cells were added to the assay plates (Corning 3712) in 25 μL (2,000 cells/well). Uninfected cells were added to the cell control wells (columns 1 and 2), which represents 100% inhibition. Cells mixed with FICs were added to compound wells (columns 3–22) and to virus control wells (columns 23 and 24), which represents 0% inhibition. Plates were incubated at 37°C, 5% CO2, and 95% humidity for 6 days. Syncytia were observed after 48 h. CTG was added, according to the manufacturer's protocol, on day 6 for a viability end point read on an EnVision® multilabel reader (Perkin Elmer) using a luminescent method with a 0.1 s integration time. The use of CTG as an endpoint reagent in antiviral assays has been evaluated and shown to produce results consistent with neutral red uptake and tetrazolium dye metabolism.10
Data Analysis
Data were imported into ActivityBase (IDBS) for analysis and reporting. Compound well data were reported as % Inhibition of virus-induced CPE, where percent CPE inhibition = 100 × (1 − [luminescence compound well−median luminescence virus control]/[median luminescence cell control−median luminescence virus control]). EC50 values and standard deviations were calculated based on % Inhibition at 10 concentrations for each substance using the 4 parameter Levenburg-Marquardt algorithm with the minimum and maximum parameters locked at 0 and 100, respectively.
Results and Discussion
During assay development and early HTS validation, virus stocks would retain infectivity for varying lengths of time. This could be as long as 12 weeks, which allowed completion of a 110,000-compound screen, or as short as 2 weeks, presenting a significant challenge for HTS. There were no overt indications based on assay performance that would allow prediction of a pending crash and at the end of the stable period, the virus stock would lose infectivity on a tube by tube basis rather than as a uniform drop in titer in all the aliquots. This phenomenon (Fig. 2) occurred in the relatively short period of 1 to 2 weeks and presented a major problem for conducting an HTS screening campaign. Unlike many viral-based assays, where a gradual drop in virus titer in the stock would produce a gradual reduction in the assay window and data quality, the behavior of the RSV stock presented more of a Russian Roulette outcome for a screening run. This is illustrated in Figure 2 where vials 3 and 6 still show infectivity, vial 7 is highly variable indicating a drop in titer, and vials 1, 2, 4, and 5 show minimal virus-induced CPE. The impact of this behavior on an HTS screening run would mean either a successful run with control values similar to historical controls for the assay or a complete failure, with virtually no virus infection. When considering the investments in time and resources for a single HTS run, this behavior presented an unacceptable financial risk and work on the FIC-based assay was pursued.
Fig. 2.
Seven individual vials of respiratory syncytial virus stock were evaluated for infectivity. Two rows correspond to one vial of virus stock. Cell controls with no virus are in the bottom two rows.
The purpose of developing the FIC-based assay was to produce the same assay progression and results as that seen with a virus stock, but with a more stable source of infectious material. To determine when to harvest infected cells for cryopreservation, cells were harvested at 6, 18, and 40 h postinfection and evaluated by TCID50. Cells harvested at 6 h were not very infectious and produced low TCID50s; cells harvested at 18 h produced TCID50s of 3.5 and cells harvested at 40 h had reduced viability and syncytia were observed before harvest. For assay validation and the screening campaign, 50 vials of FICs were prepared by harvesting at the 18 h time point. This will be sufficient stock to screen 1.5 million compounds. Table 1 shows the TCID50 values of the FICs in weeks postfreezing. The calculated TCID50 values have been consistent over a 42-week time period. The virus-based assay, which we were attempting to duplicate with the FICs, developed CPE sufficient to reduce cell viability by 95% in 6 days, The infectious dose of FICs required to produce this same progression was determined empirically. The low initial infectious dose of FICs allows for multiple rounds of virus replication and infection over the course of the assay. It would be expected that the source of the initial infection would have minimal impact on the classes of compounds identified in the screen. To evaluate this further, compounds were used to compare results from both assay formats.
Table 1.
Tissue Culture Infectious Dose 50% Values for Frozen Infected Cell Stock Over a 10-Month Timeframe
| Week | TCID50 |
|---|---|
| 2 |
3.27 |
| 3 |
3.34 |
| 4 |
3.59 |
| 5 |
3.47 |
| 7 |
3.42 |
| 8 |
3.38 |
| 9 |
3.39 |
| 16 |
3.66 |
| 20 |
3.67 |
| 42 | 3.34 |
TCID50, tissue culture infectious dose 50%.
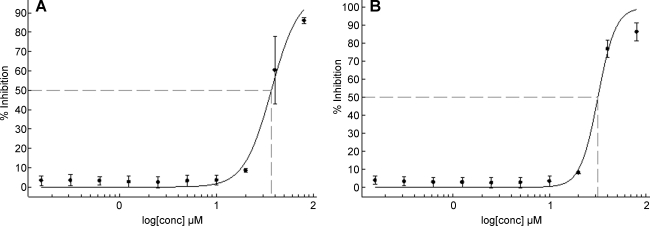
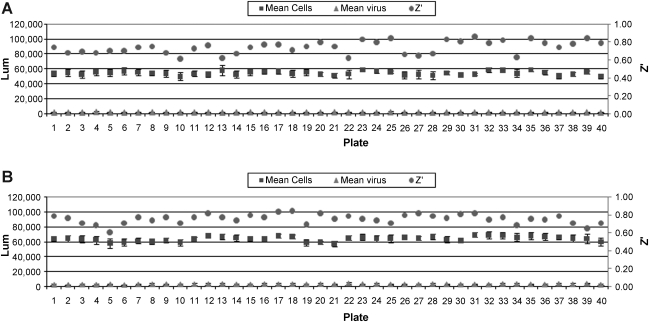
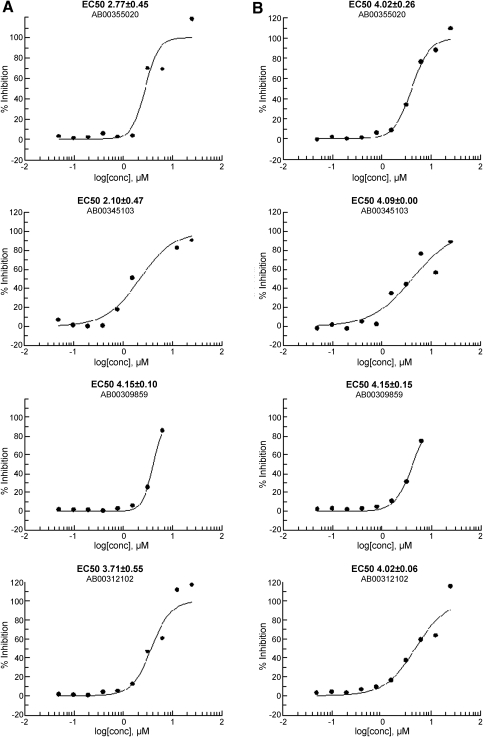
Ribavirin is the only standard reference compound available to demonstrate antiviral activity against RSV. It was used as a positive control in the HTS virus-based assay at 35 μM and produces 65%–75% inhibition of viral-induced CPE. To evaluate the FIC assay, Ribavirin dose–response assays were conducted in both the FIC and virus-based assays. Dose–response data are provided in Figure 3. FICs had an EC50 of 36.7 μM and virus-initiated infection an EC50 of 31.7 μM, panel A and B respectively. To further evaluate the use of FIC as the source of infectious material, 1,280 compounds were run in both the FIC-adapted and the original virus-based assays in dose–response format in parallel. Batch controls and Z′ values for the 40 assay plates run in duplicate are shown in Figure 4. Panel A is data from the FIC-based assay and panel B from the virus-based assay. A comparison of dose–response plots for selected hits are shown in Figure 5.
Fig. 3.
Comparison of assay formats by EC50 values derived for Ribavirin. Data are plotted as % Inhibition of CPE versus log [μM]. (A) EC50 is 36.7 ± 1.7 μM for Ribavirin using the FIC assay. (B) EC50 31.7 ± 1.8 μM for Ribavirin using the virus assay. EC50 values and standard deviations were calculated based on % Inhibition of CPE at 10 concentrations, n = 5, using the four-parameter Levenburg-Marquardt algorithm with the minimum and maximum parameters locked at 0 and 100, respectively. EC50, effective concentration 50%; FIC, frozen infected cell.
Fig. 4.
Mean of cell and virus controls from 40 compound plates run in both assay formats for comparison. Data are plotted as luminescence values/Z′ versus plate number. (A) shows the plate controls and Z′ values for the FIC assay. (B) shows plate controls and Z′ values for the virus assay. Error bars represent standard deviation calculated from n = 32 values per plate for each cell and virus controls.
Fig. 5.
Dose–response curves of selected compounds from both assay formats. Assay plates were prepared from a single compound dilution plate on the same day, using the same uninfected cells and the same medium to directly compare assay performance and minimize variables. Single replicate 10 point dose–response was done for each compound in both assay formats. Curves in (A) were derived from the FIC-based assay and curves in (B) were from the virus assay. EC50 values and standard deviations were calculated based on % Inhibition of CPE at 10 concentrations, n = 1, for each substance using the four parameter Levenburg-Marquardt algorithm with the minimum and maximum parameters locked at 0 and 100, respectively.
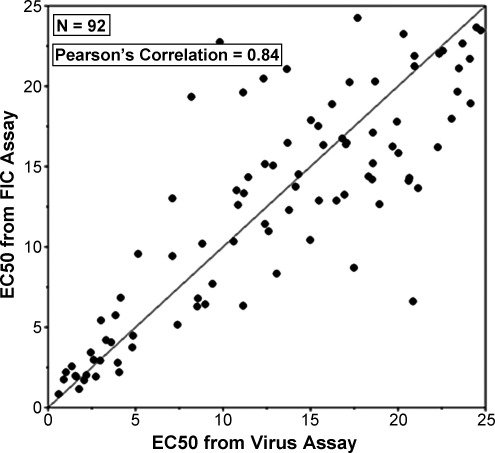
A plot of the EC50 values derived from the two assay formats for 92 compounds showing activity in either assay is shown in Figure 6. Statistical analysis of the duplicate EC50 values produced a Pearson Correlation of 0.84, which is indicative of a strong correlation between the data sets. These results indicate that the source of infectious material for the RSV antiviral assay, either FIC or high-titer infectious virus (>7), does not affect the compound EC50 data produced.
Fig. 6.
Correlation analysis of the respiratory syncytial virus assay done using FICs versus virus as the source of infectious material. A Pearson's correlation of 0.84 was calculated from this EC50 data for 92 compounds.
Since virus stability and unpredictability have been repeated obstacles to executing an HTS assay, the use of FICs was adopted for an HTS campaign. We used the RSV-FIC methodology to screen over 300,000 compounds from the Molecular Libraries Small Molecule Repository as part of a National Institute of Health Molecular Libraries Probe Production Center Network project (Severson, article in preparation). Assay performance was consistent over the course of the screen which included 995 plates screened over a 6-week period and an additional 90 plates screened in dose–response 7 weeks later using the same lot of FICs. Z′ values for the screen were between 0.5 and 0.9 with an average of 0.8. Hit rate calculated as 50% or higher inhibition was 0.2% in the FIC screen which was comparable to the hit rate of 0.3% observed in the 110K virus-based screen. Absolute values for cell and virus controls varied slightly depending on ambient conditions such as room temperature (since CTG is an enzyme-based read out) but virus controls were consistently between 2% and 5% viable for the entire screening campaign. TCID50 values for the FICs were also evaluated each week during the primary screen and periodically afterward. No significant change in infectivity was observed over a 42-week time period. The FIC methodology has been validated and used successfully for an HTS campaign, adding a novel assay development strategy for labile viruses.
We have shown that FICs can be used in place of infectious virus for an HTS antiviral screening assay. For viruses that are very stable at −80°C, such as influenza, this is unnecessary, but for viruses that are not stable as purified stocks, this is a strategy to develop an assay that is robust enough for HTS. By developing the method of using FICs in place of infectious virus, we circumvented virus instability and were able to complete the screening campaign without encountering any problems with infectivity of the stocks. However, long-term stability studies are needed to determine what the stability limits of FICs are in this application. These studies are currently in progress with TCID50 values being determined periodically. To further streamline the assay for HTS, we are evaluating the use of frozen uninfected cells in place of cells in culture. The use of all frozen cells decouples cell culture from screening and simplifies the logistics of running an HTS screen. Cost and variability associated with cell passage and cell density should both be reduced by using all frozen cells.
Abbreviations
- CPE
cytopathic effect
- CTG
CellTiter-Glo
- DMSO
dimethyl sulfoxide
- EC50
effective concentration 50%
- FBS
fetal bovine serum
- FIC
frozen infected cell
- HTS
high-throughput screening
- RSV
Respiratory Syncytial Virus
- TCID50
tissue culture infectious dose 50%
Acknowledgments
This work was supported by Southern Research Institute and the Molecular Libraries Probe Production Center Network Roadmap Initiative (U54 HG005034 awarded to Colleen B. Jonsson and NIMH 1 R03 MH082403-01A1 awarded to William Severson). The authors wish to thank Sara McKellip for compound preparation, Anna Manouvakhova for data analysis, Shuang Feng for statistical analysis, and Sue Shaddix for cell culture support.
Disclosure Statement
No competing financial interests exist.
References
- 1.Greenough A. Respiratory syncytial virus infection: clinical features, management, and prophylaxis. Curr Opin Pulm Med. 2002;8:214–217. doi: 10.1097/00063198-200205000-00011. [DOI] [PubMed] [Google Scholar]
- 2.Paramore LC. Economic impact of respiratory syncytial virus-related illness in the US: an analysis of national databases. Pharmacoeconomics. 2004;22:275–284. doi: 10.2165/00019053-200422050-00001. [DOI] [PubMed] [Google Scholar]
- 3.Crotty S. Maag D. Arnold JJ. Zhong W. Lau JY. Hong Z. Andino R. Cameron CE. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat Med. 2000;6:1375–1379. doi: 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- 4.Smith DW. Frankel LR. Mathers LH. Tang AT. Ariagno RL. Prober CG. A controlled trial of aerosolized ribavirin in infants receiving mechanical ventilation for severe respiratory syncytial virus infection. N Engl J Med. 1991;325:24–29. doi: 10.1056/NEJM199107043250105. [DOI] [PubMed] [Google Scholar]
- 5.Jordan WS., Jr. Growth characteristics of RSV. J Immunol. 1962;88:581–590. [PubMed] [Google Scholar]
- 6.Sastre P. Oomens AGP. Wertz GW. The stability of human respiratory syncytial virus is enhanced by incorporation of the baculovirus GP64 protein. Vaccine. 2007;25:5025–5033. doi: 10.1016/j.vaccine.2007.04.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mason SW, et al. Polyadenylation-dependent screening assay for respiratory syncytial virus RNA transcriptase activity and identification of an inhibitor. Nucleic Acids Res. 2004;32:4758–4767. doi: 10.1093/nar/gkh809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noah JW. Severson W. Noah DL. Rasmussen L. White EL. Jonsson CB. A cell-based luminescence assay is effective for high-throughput screening of potential influenza antivirals. Antiviral Res. 2007;73:50–59. doi: 10.1016/j.antiviral.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Chung DH. Jonsson CB. Maddox C. McKellip SN. Moore BP. Heil M. White EL. Ananthan S. Li Q. Feng S. Rasmussen L. HTS-driven discovery of new chemotypes with West Nile virus inhibitory activity. Molecules. 2010;15:1690–1704. doi: 10.3390/molecules15031690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Severson WE. Shindo N. Sosa MI. Fletcher TM., III White EL. Ananthan S. Jonsson CB. Development and validation of a high throughput screen for inhibitors of SARS CoV and its application in screening a 100,000 compound library. J Biomol Screen. 2007;12:33–40. doi: 10.1177/1087057106296688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Q. Maddox C. Rasmussen L. Hobrath JV. White EL. Assay development and high-throughput antiviral drug screening against bluetongue virus. Antiviral Res. 2009;83:267–273. doi: 10.1016/j.antiviral.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Severson WE. McDowell M. Ananthan S. Chung DH. Rasmussen L. Sosa MI. White EL. Noah J. Jonsson CB. High-throughput screening of a 100,000 compound library for inhibitors of Influenza A virus (H3N2) J Biomol Screen. 2008;13:879–887. doi: 10.1177/1087057108323123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonda MA. Braun MJ. Carter SG. Kost TA. Bess JW. Arthur LO. Van Der Maaten MJ. Characterization and molecular cloning of a bovine lentivirus related to human immunodeficiency virus. Nature. 1987;330:388–391. doi: 10.1038/330388a0. [DOI] [PubMed] [Google Scholar]
- 14.Zaman GJR. de Roos JADM. Blomenrohr J. van Koppen J. Oosterom J. Cryopreserved cells facilitate cell-based drug discovery. Drug Discov Today. 2007;12:521–526. doi: 10.1016/j.drudis.2007.05.008. [DOI] [PubMed] [Google Scholar]