Abstract
The reversible conjugation of ubiquitin and ubiquitin-like (UbL) proteins to protein substrates plays a critical role in the regulation of many cellular pathways. The removal of ubiquitin from target proteins is performed by ubiquitin proteases also known as deubiquitylases (DUBs). Owing to their substrate specificity and the central role ubiquitylation plays in cell signaling pathways, DUB are attractive targets for therapeutic development. The development of DUB inhibitors requires assays that are amenable to high-throughput screening and provide rapid assessment of inhibitor selectivity. Determination of inhibitor selectivity at an early stage of drug discovery will reduce drug failure in the clinic as well as reduce overall drug development costs. We have developed two novel assays, UbL-Enterokinase light chain and UbL-Granzyme B, for quantifying ubiquitin and UbL protease activity. In our quest to discover and characterize novel chemical entities, we have combined these assays with a previously developed assay in a multiplex format. This multiplex format allows for the detection of three distinct protease activities simultaneously, in a single well. We have demonstrated that the multiplex format is able to distinguish between selective and nonselective protease inhibitors. Specifically, we have used this assay format to characterize P022077, a selective ubiquitin-specific protease 7 inhibitor discovered at Progenra.
Introduction
Ubiquitin is a small protein that is covalently conjugated to specific lysine residues in target proteins, thereby regulating the target protein's function, localization, or stability.1–3 In addition to ubiquitin, several ubiquitin-like (UbL) proteins have been described. SUMO (small ubiquitin-like modifer) is the founding member of the UbL protein family; others include ISG15 (interferon stimulated gene 15) and NEDD8 (neural precursor cell expressed developmentally down-regulated protein 8).4 The reversible conjugation of ubiquitin and UbL proteins to target proteins regulates a plethora of cellular processes. All of these proteins utilize a similar enzymatic process for conjugation to target proteins. With respect to ubiquitin, the C-terminal glycine residue of the mature ubiquitin protein is conjugated to the ɛ-amino-group of lysine residues via a multistep, multienzyme pathway.5 This results in attachment of a single ubiquitin moiety to the target protein, which is referred to as mono-ubiquitylation. Mono-ubiquitylation of target proteins can alter their function and/or localization.6,7 Additional ubiquitins can be attached to the first ubiquitin, forming poly-ubiquitin chains. These chains can be formed via several lysine residues present in ubiquitin. Extension of poly-ubiquitin chains from all the lysines in ubiquitin has been observed in cells.8,9 The best characterized poly-ubiquitin chains are those built via lysine 48 of ubiquitin. Lysine 48-linked chains target proteins for degradation by the ubiquitin proteasome.10 The exact function of these various chains on target proteins is an area of intense interest and recent research suggests that chains that are not lysine 48-linked may also target proteins for degradation.11
Interestingly, of the UbL proteins, only SUMO has been demonstrated to form chains in vivo.12,13 SUMOylation of proteins does not target proteins for degradation, but functions primarily to regulate the activity and localization of proteins, most notably transcription factors.14,15 NEDDylation of proteins regulates the activity of substrates and is essential for the activation of the SCF family of ubiquitin E3 ligases, exemplifying the interactions among the various ubiquitin/UbL pathways.16,17
The conjugation of ubiquitin or UbL proteins can be reversed by a family of proteases collectively referred to as deubiquitylases (DUBs).18 This family of enzymes can be further subdivided into five classes: ubiquitin C-terminal Hydrolase; Ubiquitin-specific protease (USP); JAMM (JAB1/MPN/Mov34 motif metalloprotease); Machado-Josephin Domain containing protease; and Ovarian Tumor related domain containing protease proteases.18,19 Bioinformatic analysis of the human genome has identified ∼100 genes encoding DUBs.19 Dysregulation of isopeptidases has been linked to a variety of disease states.20,21 It is clear that DUB activity is responsible for regulating many vital cell functions.22 Additionally, researchers have begun to identify viral and bacterial proteins that can cleave ubiquitin from ubiquitylated proteins and many of these enzymes have a direct impact on the viability and/or virulence of the pathogenic agent.23–25 For these reasons there is a growing effort to target these enzymes for therapeutic development. The discovery and characterization of selective isopeptidase inhibitors requires robust assay systems for quantifying enzyme activity and rapidly characterizing inhibitors.
Most current assays for DUB activity employ hydrolysis of small molecule adducts such as coumarin and rhodamine from the C-terminus of ubiquitin leading to an increase in fluorescence.26,27 However, most DUBs recognize poly- or mono-ubiquitylated protein complexes and not a small molecule attached to the C-terminus of ubiquitin or UbL. To address this disparity we developed a novel assay featuring ubiquitin attached via a peptide bond to an enzyme, rendering the enzyme inactive. After specific hydrolysis of the protein-ubiquitin bond, the enzyme is activated.
Previously, we reported an assay based upon the fusion of ubiquitin to the N-terminus of phospholipase A2 (PLA2).28,29 This technology has been extended in scope, and here we describe two novel assay platforms for measuring the enzymatic activity of ubiquitin/UbL proteases. These assays involve the fusion of ubiquitin or UbL to the N-terminus of enterokinase light chain (EKL) or granzyme B (GZMB). The newly developed reporters offer a number of benefits. For example, the SUMO3-EKL fusion protein can be efficiently cleaved by sub-picomolar concentrations of SENP2core. Moreover, these assays can be combined with the ubiquitin-PLA2 reporter system in a multiplex format, which has allowed us to rapidly determine the substrate specificity of DUBs. Specifically, we have discovered that USP21 can cleave both ubiquitin and ISG15 conjugates in vitro. In addition we have used the multiplex enzyme format to characterize two novel isopeptidase inhibitors, PR-619 and P022077. The novel reporter systems will play an important role in the characterization of ubiquitin/UbL isopeptidases and the development of selective isopeptidase inhibitors as potential therapeutic agents.
Experimental Section
General Reagents
All chemicals were purchased from Sigma (St. Louis, MO) unless otherwise noted. Tetrapeptide, isolevcine-glutamic acid-proline-aspartic acid (IEPD)-7-amino-4-methylcoumarin (AMC) was purchased from EMD Biosciences (San Diego, CA) and prepared in dimethyl sulfoxide (DMSO). 2-(6-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)hexanoyl-1-hexadecanoyl-sn-glycero-3-phosphocholine (NBD C6-HPC) was purchased from Invitrogen (Carlsbad, CA) and prepared in 100% ethanol. PR-619 was purchased from LifeSensors (Malvern, PA) and prepared in DMSO. NEDD8-vinyl sulfone (VS) and NEDD8-AMC were purchased from Boston Biochem (Boston, MA). Chromatography columns and media were purchased from GE Healthsciences (Piscataway, NJ). P022077 is an analog of P5091.30 The EKL substrates I (QXL520-DDDDKGSK-FAM) and II (QXL570-DDDDKGSK-TAMRA) were procured from Anaspec (Fremont, CA).
Recombinant Protein Production
Ubiquitin-PLA2, SUMO3-PLA2, USP7, SENP1core, SENP2core (catalytic domain of SENP2), deneddylase 1 (DEN1),and PLPro were prepared as previously described.28 Bacterial expression plasmids encoding hexaHis-tagged USP21 and the fusions of ubiquitin/UbL with the active fragment of EKL were generated by standard molecular biology techniques (primers listed in Supplementary Figure S1; Supplementary Data are available online at www.liebertonline.com/adt). The ubiquitin/UbL-EKL reporter fusion proteins were expressed and purified using methods similar to those previously described for EKL.31 A fusion of SUMO3 and GZMB was generated in the pP-secSUMO3 vector (LifeSensors) using standard molecular biology techniques. Stable Pichia pustoris transformants were generated according to the manufacturer's instructions. Six isolated clones were evaluated for expression by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and western blot using anti-GZMB (AbD Serotec, Oxford, United Kingdom). Scale-up production was accomplished by inoculating 5 mL of BMGY until an OD600 of ∼10 was reached. Cultures were centrifuged, resuspended in 500 mL of BMMY, and grown at 30°C with shaking at 250 rpm for 24 h. Cultures were supplemented with methanol to 1% for 48 h. The supernatant was extensively dialyzed against phosphate-buffered saline and SUMO3-GZMB was purified by nickel chromatography using standard techniques.
Enzyme Assays
UbL-EKL assays
Unless stated otherwise, recombinant isopeptidase was mixed with UbL-EKL and EKL substrate I to final concentrations of 20, 50, and 20 nM, respectively, in a total volume of 100 μL in a well in a black-walled 96-well plate (Greiner Bio-One, Monroe, NC). All dilutions were performed in isopeptidase assay buffer (20 mM Tris-HCl, pH 8.0, 2 mM CaCl2, and 2 mM β-mercaptoethanol). The increase in fluorescence intensity over time was determined on a Perkin Elmer Envision fluorescence plate reader with excitation and emission filters corresponding to the fluorescence resonance energy transfer peptide utilized. Unless stated otherwise, net relative fluorescence units (RFUs) were determined by subtracting the blank RFU value (20 nM EKL substrate I or 100 nM EKL substrate II in isopeptidase assay buffer) from each data point. SENP2core, SUMO3-EKL sensitivity experiments were performed by mixing 0–100 fM SENP2core with 50 nM SUMO3-EKL, and 100 nM EKL substrate I in a total volume of 100 μL as above.
SUMO3-GZMB assays
Unless stated otherwise, recombinant isopeptidase was mixed with SUMO3-GZMB, and IEPD-AMC to final concentrations of 20 nM, 50 nM, and 5 μM, respectively, in a total volume of 100 μL in a well in a black-walled 96-well plate (Greiner Bio-One). All dilutions were performed in isopeptidase assay buffer (20 mM Tris-HCl, pH 8.0, 2 mM CaCl2, and 2 mM β-mercaptoethanol). The increase in fluorescence intensity over time was determined on a Perkin Elmer Envision fluorescence plate reader with excitation and emission maxima of 360 and 460 nm, respectively. Unless stated otherwise, net RFUs were determined by subtracting the blank RFU value (5 μM IEPD-AMC in isopeptidase assay buffer) from each data point. SENP2, SUMO3-GZMB concentration dependence experiments were performed as above by mixing 0–200 nM SENP2 core with 50 nM SUMO3-GZMB, and 5 μM IEPD-AMC in a total volume of 100 μL.
Multiplex Assay Format
In general, the multiplex assays included 100 nM SUMO3-GZMB, 5 μM IEPD-AMC, 30 nM ubiquitin-PLA2, 10 μM NBD C6-HPC, 100 nM NEDD8-EKL, and 1 μM EKL substrate II. In the case of USP21, NEDD8-EKL was replaced with ISG15-EKL. Fluorescence was measured at the following pairs of wavelengths: ex360/em460, ex460/em538, ex540/em590, and reported deSUMOylase, DUB, and deISGylase activities, respectively. Signal:noise ratio (SNR) was calculated with the following equation: (RFUwith enzyme − RFUminus enzyme)/(StdDevwith enzyme + StdDevminus enzyme). Z′ value is calculated as 1 − (3/SNR). Signal:background was calculated as RFUwith enzyme/RFUminus enzyme.
Isopeptidase Inhibition Assays
Dose ranges of isopeptide inhibitors were prepared in 100% DMSO and 2 μL aliquots were added to 96-well plates in triplicate. Eighty microliters of isopeptidase was added to the wells individually or as a mixture of three enzymes and incubated at room temperature for 30 min. About 20 μL of Reporter-fusion and reporter substrate was then added to all of the wells. Final concentrations of 10 mM N-ethylmaleimide (NEM) and 0.5%(v/v) DMSO were used as controls for 100% and 0% inhibition. Percent inhibition was calculated using the following equation [1 − ((RFUCompoundl − RFUNEM)/(RFUDMSO − RFUNEM))] × 100. Data were plotted in Prism and IC50 values were determined using a sigmoidal dose–response (variable slope) model.
Results and Discussion
SUMO3-EKL and SUMO3-GZMB as Novel Protease Reporters
Previously, we reported the development of a novel ubiquitin/UbL endopeptidase assay utilizing a linear fusion of ubiquitin/UbL to the N-terminus of PLA2.28 PLA2 requires a free N-terminus for activity, and fusion of ubiquitin/UbL to the N-terminus results in inactivation of the enzyme. Incubation with an ubiquitin/UbL isopeptidase results in cleavage of the α-peptide bond between the ubiquitin/UbL and PLA2 resulting in the activation of PLA2, which can be measured using a quenched fluorescent substrate of PLA2.28,29
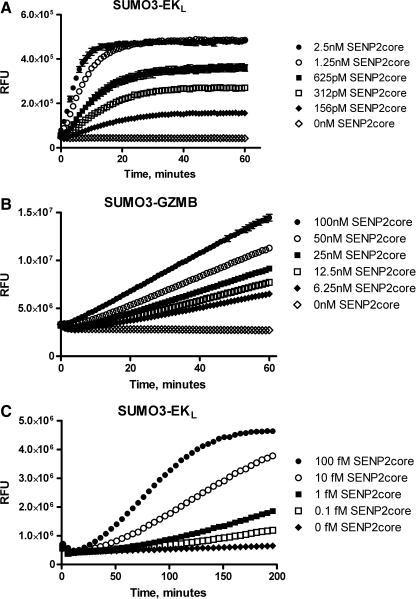
In addition to PLA2, EKL and GZMB require a free N-terminus for enzymatic activity.31–33 We generated plasmids encoding SUMO3 fused to the N-terminus of the active forms of EKL and GZMB and expressed and purified the fusion proteins. As predicted, these fusion proteins were catalytically inactive. To determine if these fusions could be cleaved by isopeptidases, we incubated them with the core catalytic domain of the deSUMOylase SENP2. Upon activation, EKL will cleave an internally quenched fluorescence resonance energy transfer peptide resulting in an increase in fluorescence. As shown in Figure 1, incubation of SENP2 with SUMO3-EKL and the EKL substrate resulted in a dramatic increase in fluorescence due to SENP2 mediated cleavage of the α-peptide bond between SUMO3 and EKL. In a similar manner, incubation of SUMO3-GZMB with SENP2 resulted in the activation of GZMB, which cleaved its substrate IEPD-AMC, leading to the release of AMC fluorophore and a concomitant increase in AMC fluorescence. In both cases the rate at which fluorescence increased depended on SENP2 concentration, demonstrating that these assays can be used to quantify deSUMOylase activity. The SUMO3-EKL system can reach a plateau within the time frame of this assay, whereas the SUMO3-GZMB signal remains linear throughout the course of the experiment. This is most likely caused by the difference in the amount of reporter substrate used in these assays. The EKL substrate is present at only 20 nM, whereas the GZMB substrate is present at 5 μM.
Fig. 1.
Detection of SENP2core protease activity with SUMO3-GZMB and SUMO3-EKL. (A) Increasing concentrations of SENP2core (0 nM ◊, 156 pM ♦, 312 pM □, 625 pM ▪, 1.25 nM ○, 2.5 nM ∙) were incubated with 100 nM SUMO3-EKL, 20 nM of EKL substrate I. The data presented are means ± SEM of triplicate wells. (B) Increasing concentrations of SENP2core (0 nM ◊, 6.25 nM ♦, 12.5 nM □, 25 nM ▪, 50 nM ○, 100 nM ∙) were incubated with 100 nM SUMO3-GZMB and 5 μM IEPD-AMC. The data presented are means ± SEM of triplicate wells. (C) Sub-picomolar concentrations of SENP2core (0 fM ♦, 0.1 fM □, 1 fM ▪, 10 fM ○, 100 fM ∙) were incubated with 500 nM SUMO3-EKL and 200 nM EKL substrate I. Data shown are representative mean data from three independent experiments. AMC, 7-amino-4-methylcoumarin; EKL, enterokinase light chain; GZMB, granzyme B.
Given the high sensitivity of the SUMO3-EKL reporter system (Fig. 1A), we further investigated the minimal concentration of SENP2 core this assay could reliably detect. We incubated sub-picomolar dose ranges of SENP2 with SUMO3-EKL and EKL substrate I and measured the change in fluorescence for 3 h. Figure 1C demonstrates that the SUMO3-EKL assay exhibited a significant increase in signal with concentrations of SENP2core as low as 0.1fM.
Subsequently, we prepared linear fusion proteins consisting of alternative ubiquitin/UbL proteins fused to the N-terminus of EKL including ubiquitin, ISG15, and NEDD8. Each of these fusions could be activated by incubation with proteases recognizing the cognate ubiquitin/UbL (Supplementary Fig. S1). Taken together, these data indicate that UbL-EKL and SUMO3-GZMB are suitable reporters for measuring protease activity.
Method for Measuring Ubiquitin/UbL Protease Activity in a Multiplex Format
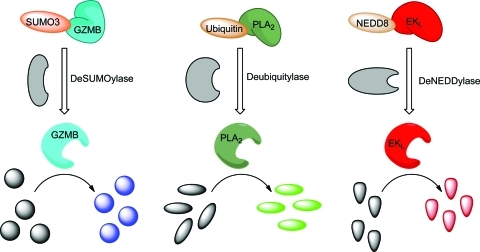
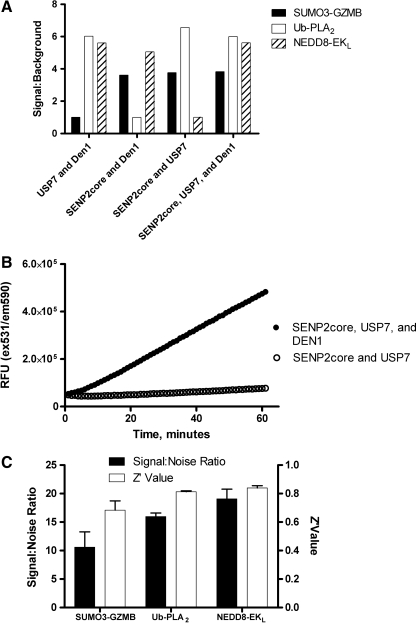
The generation of multiple ubiquitin/UbL protease reporter systems allowed us to investigate the potential for combining these assays for use in a multiplex format (Fig. 2). Due to the spectral overlap of EKL substrate I with the PLA2 substrate NBD C6-HPC, we switched to an alternative substrate for EKL (EKL substrate II) with excitation and emission maxima of 540 and 590 nm, respectively. The lack of spectral overlap of the fluorophores utilized is shown in Supplementary Figure S2. Before demonstrating that these reporter systems could be measured simultaneously in a single well, we optimized the concentrations of the assay components to yield a high SNR for each isopeptidase/reporter combination (data not shown). Subsequently, we combined 100 nM of each reporter (SUMO3-GZMB, ubiquitin-PLA2, and NEDD8-EKL) with appropriate concentrations of their substrates (Fig. 3A). When all three UbL isopeptidases were added, an increase in fluorescence intensity was observed corresponding to the activation of each reporter substrate. To verify that these signals were dependent upon the presence of the cognate isopeptidase, we repeated this experiment including, in addition to the complete reaction containing all three ubiquitin isopeptidases, controls lacking one of the isopeptidases (Fig. 3A). Figure 3B shows that the signal generated by NEDD8-EKL was completely dependent on the presence of DEN1. Similar results were obtained when omitting the other isopeptidases (Fig. 3A). To further validate this system for high-throughput screening (HTS), we calculated the SNR and Z′ value for each reporter system in this multiplex format. Each reporter yielded a Z′ >0.5 and an SNR >10, indicating that the assay format is robust and reproducible (Fig. 3C). To confirm that additional combinations of reporters were useable in a HTS multiplex format, we also validated the multiplex format utilizing SUMO3-GZMB, ubiquitin-PLA2, and ISG15-EKL (data not shown). We predict that additional combinations of reporter systems will be possible depending on the experimental design and the availability of suitable substrates.
Fig. 2.
Schematic representation of multiplexing format. To measure the activity of all three reporters in a single well, three isopeptidases are added to SUMO3-GZMB, ubiquitin-PLA2, and NEDD8-EKL. Each well contains GZMB substrate (ex340/em460), PLA2 substrate (ex460/em538) and EKL substrate II (ex540/em590). PLA2, phospholipase A2. Color images available online at www.liebertonline.com/adt
Fig. 3.
Detection of ubiquitin/ubiquitin-like protease activity in a multiplex assay format. (A) SUMO3-GZMB, ubiquitin-PLA2, and NEDD8-EKL were incubated with either the complete mixture containing SENP2core, USP7, and DEN1, or a mixture lacking one of the three isopeptidases. Fluorescence intensity was measured following a 60-minute incubation and signal:background was calculated. Representative data from one of three independent experiments are shown. Bars represent the signal:background generated by the SUMO3-GZMB reporter (solid bars), ubiquitin-PLA2 reporter (open bars), or the NEDD8-EKL reporter (crosshatched bars) (B) A cocktail of isopeptidases containing USP7 and SENP2core with (closed circles) or without (open circles) DEN1 was added to wells containing SUMO3-GZMB, IEPD-AMC, ubiquitin-PLA2, NBD C6-HPC, NEDD8-EKL, and EKL substrate II. Fluorescence intensity (ex531/em590) was measured as described in the Materials and Methods section. Error bars represent standard error of n = 3 values. (C) Utilizing the data from B, the signal:noise ratio (solid bars) and Z′ ratio (open bars) were calculated as described in Materials and Methods. The data shown are the average of three independent experiments, and the error bars represent the standard error of these three values. NBD C6-HPC, 2-(6-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)hexanoyl-1-hexadecanoyl-sn-glycero-3-phosphocholine; USP, ubiquitin-specific protease.
Characterization of DUB Activity Using Multiplexed Format
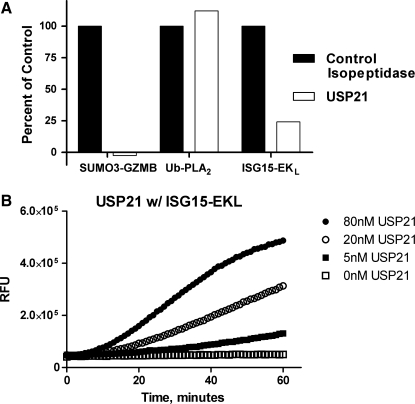
Bioinformatic analysis has successfully identified many genes that are predicted to express ubiquitin and UbL isopeptidases. Characterizing the substrate specificity of novel isopeptidases is an important first step in determining the physiological role of the proteases. We have already demonstrated that USP7, SENP2, and DEN1 generate signals only with their cognate UbLs, Ubiquitin, SUMO3, and NEDD8, respectively. In our efforts to characterize the USP class of enzymes, we expressed and purified USP21 and tested the activity of 100 nM of USP21 with SUMO3-GZMB, ubiquitin-PLA2, and ISG15-EKL and the respective substrates of these reporter enzymes in our multiplex format. For standardization purposes we included a control isopeptidase for each reporter and set the signal generated by the control isopeptidase to 100%. Control isopeptidase concentrations were selected based on similar rates of increasing fluorescent intensity, thus standardizing the assays between different UbLs. As expected, USP21 exhibited a robust signal with Ubiquitin-PLA2 and negligible activity with SUMO3-GZMB (Fig. 4A). However, in addition to the DUB activity, USP21 also exhibited a modest deISGylase activity (Fig. 4A) demonstrating the utility of the multiplex assay format to rapidly discover novel substrate specificities of isopeptidases. To confirm this result we incubated increasing concentrations of USP21 with ISG15-EKL and EKL substrate I and observed dose dependent increases in USP21 activity (Fig. 4B). Taken together, these data report a novel deISGylase activity of USP21 and demonstrate that the multiplex assay format can be used to determine the substrate selectivity of ubiquitin/UbL proteases.
Fig. 4.
USP21 is a deISGylase as well as a deubiquitylase. (A) 100 nM USP21 was incubated with 100 nM SUMO3-GZMB, 5 μM IEPD-AMC, 100 nM ubiquitin-PLA2, 20 μM NBD C6-HPC, 100 nM SUMO3-EKL, and 1 μM EKL substrate II. Fluorescence intensity was measured after a 60-minute incubation. USP21 deSUMOylase, deubiquitylase, and deISGylase activity were reported relative to control deSUMOylase (20 nM SENP2 core), deubiquitylase (20 nM USP7), and deISGylase (20 nM PLPro) activity, respectively (solid bars). The signal generated by USP21 is represented by open bars. Representative data from three independent experiments is shown. (B) Various concentrations of USP21 (0 nM □, 5 nM ▪, 20 nM ○, 80 nM ∙) were incubated with 50 nM ISG15-EKL and 20 nM EKL substrate I.
Characterization of Novel Isopeptidase Inhibitors Using Multiplexed Format
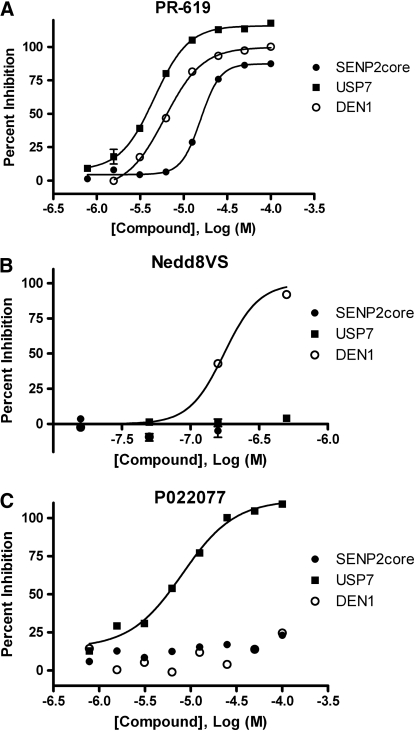
Ubiquitin isopeptidases are potential targets for therapeutic intervention. To both identify and develop isopeptidase inhibitors, it is necessary to rapidly screen novel chemical entities versus several related enzymes. The multiplex assay format described here would be predicted to accelerate screening and characterization of novel chemical entities by allowing scientists to test their effects on multiple enzymes simultaneously. To test this hypothesis we studied two small molecule isopeptidase inhibitors, PR-619 and P022077. PR-619 is a broadly active ubiquitin/UbL isopeptidase inhibitor and P022077 is an analog of the recently discovered USP7 inhibitor P5091.30 Additionally, we tested the selective deNEDDylase activity-based probe, NEDD8-VS. This probe covalently modifies the active site cysteine residue of deNEDDylases rendering the enzyme inactive. Initially, we determined the IC50 of these compounds versus SENP2, USP7, and DEN1 individually (Table 1). Subsequently, dose ranges of PR-619 were preincubated with SENP2, USP7, and DEN1 simultaneously before addition of all three reporters and their substrates and measurement of enzyme activity (Fig. 5A). From these experiments it is clear that PR-619 is a nonselective isopeptidase inhibitor with IC50 values ranging from 4 to 16 μM for the three enzymes tested. In contrast, NEDD8-VS selectively inhibited DEN1 in both the singleplex and multiplex formats (Table 1). As seen in Figure 5C, P022077 had negligible activity versus DEN1 and SENP2core over the concentration range tested, but inhibited USP7 with an IC50 of 8 μM. Comparison of the singleplexed and multiplexed data in Table 1 clearly demonstrates that the multiplex format does not dramatically alter the IC50 values or the selectivity of these inhibitors, validating the use of this multiplex format for the characterization of isopeptidase inhibitors.
Table 1.
IC50 Values for Various Isopeptidase Inhibitors
| |
IC50 (μM) |
|||||
|---|---|---|---|---|---|---|
| |
SENP2core |
USP7 |
DEN1 |
|||
| Singleplexed | Multiplexed | Singleplexed | Multiplexed | Singleplexed | Multiplexed | |
| NEM | 2100 ± 40 | 2200 ± 80 | 1300 ± 200 | 1000 ± 120 | 1580 ± 40 | 2000 ± 50 |
| PR-619 | 26 ± 8 | 23 ± 7 | 5.8 ± 2.2 | 4.3 ± 1.2 | 6.6 ± 3.4 | 9.0 ± 3.0 |
| P022077 | >50 | >50 | 8.6 ± 2.0 | 7.8 ± 3.1 | >50 | >50 |
| NEDD8VS | NT | >0.5 | NT | >0.5 | 0.17 ± 0.01 | 0.21 ± 0.06 |
IC50 values in μM for four inhibitors with three ubiquitin/UbL isopeptidases as determined in either singleplexed or multiplexed assay formats. PR-619 and NEM are broad-spectrum cysteine protease inhibitors, whereas P022077 and NEDD8-VS are selective USP7 and deNEDDylase inhibitors, respectively. Data represent mean and standard deviation of three independent experiments.
NEM, N-ethylmaleimide; UbL, ubiquitin-like; USP, ubiquitin-specific protease; VS, vinyl sulfone.
Fig. 5.
The multiplex format has the utility to identify selective isopeptidase inhibitors. (A) Various concentrations of PR-619 were incubated with 100 nM SENP2core, 20 nM USP7, and 20 nM DEN1. After 30 min, 100 nM SUMO3-GZMB, 5 μM IEPD-AMC, 100 nM ubiquitin-PLA2, 20 μM NBD C6-HPC, 100 nM NEDD8-EKL, and 1 μM EKL Substrate II were added. Fluorescence intensity was measured after 45 min at the appropriate wavelengths and the percent inhibition for each enzyme was calculated. Percent inhibition of each isopeptidase (SENP2core ∙, USP7 ▪, DEN1 ○) is shown plotted versus log of the molar concentration of compound. IC50 values were calculated using GraphPad Prism and are shown where appropriate. (B) A range of concentrations of NEDD8-vinyl sulfone were tested as described above. (C) The selective USP7 inhibitor, P022077, was tested as above.
Conclusions
The rapidly expanding field of ubiquitin research requires the development of novel technologies that will enable researchers to efficiently decipher the physiological function of the regulatory enzymes in this pathway. Ubiquitin and UbL isopeptidases are key regulators in multiple cellular signaling cascades and they represent a novel class of target for therapeutic development.20,34,35 We have described two new reporter systems, UbL-EKL and UbL-GZMB. These reporter fusions are efficiently cleaved by ubiquitin/UbL proteases and are useful for quantifying activity. Additionally, by selecting reporter enzyme substrates that generate fluorescent signals in nonoverlapping discreet wavelength ranges, one can combine them with the UbL-PLA2 reporter system in a multiplex format. This multiplexed format affords the measurement of three distinct protease activities simultaneously in a single well. We have demonstrated the utility of multiplexing enzyme assays for the characterization of enzyme specificity. In addition, we confirmed that USP21 is a DUB and simultaneously uncovered a previously unreported deISGylase activity of this enzyme. To date, only one other mammalian isopeptidase, USP18, has been shown to function as a deISGylase. The multiplex format is not able to determine absolute catalytic efficiencies of isopeptidases and additional studies are needed to fully characterize the activity of USP21. However, it is the clear that the multiplex format will facilitate the rapid initial characterization of novel ubiquitin/UbL proteases.
The therapeutic potential of targeting the ubiquitin/UbL pathways has been established with the FDA approval of the ubiquitin proteasome inhibitor, bortezomib/Velcade®, for the treatment of multiple myeloma.36 Recently, Millennium developed a selective NEDD8 E1 activating enzyme inhibitor, MLN4924, that prevents the conjugation of NEDD8 to SCF E3 ligases.37 This inhibitor is currently being developed as a novel cancer therapeutic. In addition to these targets, ubiquitin and UbL isopeptidases are considered potential drug targets. One major concern in the development of isopeptidase inhibitors is selectivity within a protease family. In the case of ubiquitin isopeptidases it will be advantageous to determine the selectivity of an inhibitor at an early stage to rapidly exclude nonselective inhibitors from the development process. To address this issue we have established a multiplex assay format that allows for the simultaneous measurement of the protease activities of three distinct isopeptidases. We have employed this multiplex format to characterize the selectivity of ubiquitin/UbL isopeptidase inhibitors. Both the singleplex and the multiplex formats report the same potencies for the inhibitors (Table 1). This multiplex assay format is amenable to HTS providing researchers with an initial determination of inhibitor selectivity during primary screens of compound libraries. It is likely that this approach will speed the discovery of selective ubiquitin/UbL isopeptidase inhibitors for further development as potential therapeutic agents as well as tool compounds for studying the biological significance of a particular protease.
Supplementary Material
Abbreviations
- AMC
7-amino-4-methylcoumarin
- DMSO
dimethyl sulfoxide
- DUB
deubiquitylase
- EKL
enterokinase light chain
- FAM
carboxyfluoroscein
- GZMB
granzyme B
- HTS
high-throughput screening
- IEPD
tetrapeptide, isolevcine-glutamic acid-proline-aspartic acid
- ISG15
interferon stimulated gene 15
- NBD C6-HPC
2-(6-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)hexanoyl-1-hexadecanoyl-sn-glycero-3-phosphocholine
- NEDD8
neural precursor cell expressed developmentally downregulated protein 8
- NEM
N-ethylmaleimide
- PLA2
phospholipase A2
- RFU
relative fluorescence unit
- SNR
signal:noise ratio
- SUMO
small ubiquitin-like modifier
- TAMRA
carboxytetramethylrhodamine
- UbL
ubiquitin-like
- USP
ubiquitin-specific protease
- VS
vinyl sulfone
Acknowledgments
We thank Dr. David Sterner for providing research materials. This work was funded in part by a grant (CA115205) awarded to Progenra Inc. by the NCI/NIH.
Disclosure Statement
No competing financial interests exist.
References
- 1.Schwartz AL. Ciechanover A. Targeting proteins for destruction by the ubiquitin system: implications for human pathobiology. Annu Rev Pharmacol Toxicol. 2009;49:73–96. doi: 10.1146/annurev.pharmtox.051208.165340. [DOI] [PubMed] [Google Scholar]
- 2.Chen BB. Mallampalli RK. Masking of a nuclear signal motif by monoubiquitination leads to mislocalization and degradation of the regulatory enzyme cytidylyltransferase. Mol Cell Biol. 2009;29:3062–3075. doi: 10.1128/MCB.01824-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ravid T. Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haas AL. Structural insights into early events in the conjugation of ubiquitin and ubiquitin-like proteins. Mol Cell. 2007;27:174–175. doi: 10.1016/j.molcel.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Chen ZJ. Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 7.Yin H. Gui Y. Du G. Frohman MA. Zheng XL. Dependence of phospholipase D1 multi-monoubiquitination on its enzymatic activity and palmitoylation. J Biol Chem. 2010;285:13580–13588. doi: 10.1074/jbc.M109.046359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li W. Ye Y. Polyubiquitin chains: functions, structures, and mechanisms. Cell Mol Life Sci. 2008;65:2397–2406. doi: 10.1007/s00018-008-8090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pickart CM. Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 10.Pickart CM. Targeting of substrates to the 26S proteasome. FASEB J. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- 11.Xu P. Duong DM. Seyfried NT. Cheng D. Xie Y. Robert J. Rush J. Hochstrasser M. Finley D. Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vertegaal AC. Small ubiquitin-related modifiers in chains. Biochem Soc Trans. 2007;35:1422–1423. doi: 10.1042/BST0351422. [DOI] [PubMed] [Google Scholar]
- 13.Tatham MH. Jaffray E. Vaughan OA. Desterro JM. Botting CH. Naismith JH. Hay RT. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem. 2001;276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- 14.Cho G. Lim Y. Golden JA. SUMO interaction motifs in Sizn1 are required for promyelocytic leukemia protein nuclear body localization and for transcriptional activation. J Biol Chem. 2009;284:19592–19600. doi: 10.1074/jbc.M109.010181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du JX. Bialkowska AB. McConnell BB. Yang VW. SUMOylation regulates nuclear localization of Kruppel-like factor 5. J Biol Chem. 2008;283:31991–32002. doi: 10.1074/jbc.M803612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Read MA. Brownell JE. Gladysheva TB. Hottelet M. Parent LA. Coggins MB. Pierce JW. Podust VN. Luo RS. Chau V. Palombella VJ. Nedd8 modification of cul-1 activates SCF(beta(TrCP))-dependent ubiquitination of IkappaBalpha. Mol Cell Biol. 2000;20:2326–2333. doi: 10.1128/mcb.20.7.2326-2333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morimoto M. Nishida T. Honda R. Yasuda H. Modification of cullin-1 by ubiquitin-like protein Nedd8 enhances the activity of SCF(skp2) toward p27(kip1) Biochem Biophys Res Commun. 2000;270:1093–1096. doi: 10.1006/bbrc.2000.2576. [DOI] [PubMed] [Google Scholar]
- 18.Amerik AY. Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 19.Nijman SM. Luna-Vargas MP. Velds A. Brummelkamp TR. Dirac AM. Sixma TK. Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Nicholson B. Marblestone JG. Butt TR. Mattern MR. Deubiquitinating enzymes as novel anticancer targets. Future Oncol. 2007;3:191–199. doi: 10.2217/14796694.3.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shanmugham A. Ovaa H. DUBs and disease: activity assays for inhibitor development. Curr Opin Drug Discov Devel. 2008;11:688–696. [PubMed] [Google Scholar]
- 22.Reyes-Turcu FE. Ventii KH. Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rytkonen A. Poh J. Garmendia J. Boyle C. Thompson A. Liu M. Freemont P. Hinton JC. Holden DW. SseL, a Salmonella deubiquitinase required for macrophage killing and virulence. Proc Natl Acad Sci USA. 2007;104:3502–3507. doi: 10.1073/pnas.0610095104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Catic A. Misaghi S. Korbel GA. Ploegh HL. ElaD, a Deubiquitinating protease expressed by E. coli. PLoS ONE. 2007;2:e381. doi: 10.1371/journal.pone.0000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Z. Wang Y. Ratia K. Mesecar AD. Wilkinson KD. Baker SC. Proteolytic processing and deubiquitinating activity of papain-like proteases of human coronavirus NL63. J Virol. 2007;81:6007–6018. doi: 10.1128/JVI.02747-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tirat A. Schilb A. Riou V. Leder L. Gerhartz B. Zimmermann J. Worpenberg S. Eidhoff U. Freuler F. Stettler T. Mayr L. Ottl J. Leuenberger B. Filipuzzi I. Synthesis and characterization of fluorescent ubiquitin derivatives as highly sensitive substrates for the deubiquitinating enzymes UCH-L3 and USP-2. Anal Biochem. 2005;343:244–255. doi: 10.1016/j.ab.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 27.Dang LC. Melandri FD. Stein RL. Kinetic and mechanistic studies on the hydrolysis of ubiquitin C-terminal 7-amido-4-methylcoumarin by deubiquitinating enzymes. Biochemistry. 1998;37:1868–1879. doi: 10.1021/bi9723360. [DOI] [PubMed] [Google Scholar]
- 28.Nicholson B. Leach CA. Goldenberg SJ. Francis DM. Kodrasov MP. Tian X. Shanks J. Sterner DE. Bernal A. Mattern MR. Wilkinson KD. Butt TR. Characterization of ubiquitin and ubiquitin-like-protein isopeptidase activities. Protein Sci. 2008;17:1035–1043. doi: 10.1110/ps.083450408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leach CA. Tian X. Mattern MR. Nicholson B. Detection and characterization of SUMO protease activity using a sensitive enzyme-based reporter assay. Methods Mol Biol. 2009;497:269–281. doi: 10.1007/978-1-59745-566-4_18. [DOI] [PubMed] [Google Scholar]
- 30.Cao P. Nicholson B. Weinstock J. Kingsbury W. Mattern MR. Goldenberg SJ. McDermott J. Leach CA. Kizhakkethil-George SK. Butt TR. Antineoplastic methods, compounds and compositions, Patent Application, PCT/US2010/029358. 2009.
- 31.Gasparian ME. Ostapchenko VG. Schulga AA. Dolgikh DA. Kirpichnikov MP. Expression, purification, and characterization of human enteropeptidase catalytic subunit in Escherichia coli. Protein Expr Purif. 2003;31:133–139. doi: 10.1016/s1046-5928(03)00159-1. [DOI] [PubMed] [Google Scholar]
- 32.Song HW. Choi SI. Seong BL. Engineered recombinant enteropeptidase catalytic subunit: effect of N-terminal modification. Arch Biochem Biophys. 2002;400:1–6. doi: 10.1006/abbi.2001.2737. [DOI] [PubMed] [Google Scholar]
- 33.McGuire MJ. Lipsky PE. Thiele DL. Generation of active myeloid and lymphoid granule serine proteases requires processing by the granule thiol protease dipeptidyl peptidase I. J Biol Chem. 1993;268:2458–2467. [PubMed] [Google Scholar]
- 34.Cheon KW. Baek KH. HAUSP as a therapeutic target for hematopoietic tumors (review) Int J Oncol. 2006;28:1209–1215. [PubMed] [Google Scholar]
- 35.Ponts N. Yang J. Chung DW. Prudhomme J. Girke T. Horrocks P. Le Roch KG. Deciphering the ubiquitin-mediated pathway in apicomplexan parasites: a potential strategy to interfere with parasite virulence. PLoS ONE. 2008;3:e2386. doi: 10.1371/journal.pone.0002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bross PF. Kane R. Farrell AT. Abraham S. Benson K. Brower ME. Bradley S. Gobburu JV. Goheer A. Lee SL. Leighton J. Liang CY. Lostritto RT. McGuinn WD. Morse DE. Rahman A. Rosario LA. Verbois SL. Williams G. Wang YC. Pazdur R. Approval summary for bortezomib for injection in the treatment of multiple myeloma. Clin Cancer Res. 2004;10:3954–3964. doi: 10.1158/1078-0432.CCR-03-0781. [DOI] [PubMed] [Google Scholar]
- 37.Brownell JE. Sintchak MD. Gavin JM. Liao H. Bruzzese FJ. Bump NJ. Soucy TA. Milhollen MA. Yang X. Burkhardt AL. Ma J. Loke HK. Lingaraj T. Wu D. Hamman KB. Spelman JJ. Cullis CA. Langston SP. Vyskocil S. Sells TB. Mallender WD. Visiers I. Li P. Claiborne CF. Rolfe M. Bolen JB. Dick LR. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol Cell. 2010;37:102–111. doi: 10.1016/j.molcel.2009.12.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.