Abstract
Decellularized skeletal muscle is a promising model that can be used to study cell–matrix interactions and changes that occur in muscle extracellular matrix (ECM) in myopathies and muscle wasting diseases. The goal of this study is to develop a novel method to decellularize skeletal muscle that maintains the native biochemical composition and structure of the ECM. This method consists of sequential incubation of mouse tibialis anterior muscles in latrunculin B, high ionic strength salt solution, and DNase I and avoids use of proteases or detergents that degrade the ECM. Characterization of the decellularized muscles using hematoxylin and eosin staining along with DNA quantification suggested complete removal of DNA, whereas biochemical analyses indicated no loss of collagens and only a slight reduction in glycosaminoglycans. Western blot analysis of decellularized tissues showed removal of the vast majority of the contractile proteins actin and myosin, and morphological analysis using scanning electron microscopy suggested removal of myofibers from decellularized muscle tissues. Passive mechanical testing of decellularized muscle bundles revealed the typical nonlinear behavior, similar to that of intact muscle. Together, these results suggest that the protocol developed successfully decellularizes skeletal muscle without altering its composition and mechanical function.
Introduction
Decellularized tissues are useful for a number of biomedical applications such as understanding the physicochemical properties of extracellular matrix (ECM) and providing a tissue specific scaffold for engineering functional tissues. For instance, by harnessing the cell–matrix interactions that are crucial for cell growth and differentiation,1 several studies have promoted tissue formation by combining cells with tissue-derived ECM from decellularized heart, urinary bladder, skeletal muscle, and small intestinal submucosa.2–6 A number of different decellularization methods have been developed,7 but the native biochemical, mechanical, and structural properties of the decellularized ECM are altered depending upon the method used.
This study describes the development of a decellularization method that maintains the mechanical and structural integrity of skeletal muscle connective tissue with minimal disruption to the ECM. Skeletal muscle ECM plays an important role in tissue maintenance and regeneration of skeletal muscle,8 and modulates cell adhesion and migration, growth factor storage and release, and satellite cell activation and differentiation.9–11 The composition, structure, and mechanical properties of skeletal muscle ECM may change with age and various muscle pathologies, and these ECM associated changes could play an important role in determining the success of therapeutic interventions that rely on multiple biological and biomechanical cues from the ECM.
Previous attempts to decellularize skeletal muscles have incorporated relatively harsh physical methods such as freeze–thawing12,13 and detergent and enzymatic treatment with Triton X-100, sodium deoxycholate, sodium dodecyl sulfate, and trypsin.5,14–22 These methods inherently result in ECM degradation and/or compromised mechanical and structural properties, which is not desirable for studies that require maintenance of skeletal muscle ECM biochemical and mechanical properties. To this end, we developed a decellularization method that utilizes only osmotic shock and actin and myosin depolymerization, and specifically does not use either proteases or detergents.
Materials and Methods
Decellularization of muscle tissue
All animal handling and experimental procedures were in accordance with the protocol approved by the UCSD Institutional Animal Care and Use Committee and NIH guidelines for animal welfare. After euthanasia, tibialis anterior (TA) muscles were removed from 2-month-old female C57BL/6 mice (Harlan Sprague Dawley, Indianapolis, IN). Upon removal, TA muscles were incubated in 50 nM latrunculin B (Cayman Chemical, Ann Arbor, Michigan) in high-glucose Dulbecco's modified Eagle's medium (DMEM; Gibco, Carlsbad, CA) for 2 h at 37°C with agitation. All further steps were performed with agitation at room temperature unless otherwise stated. Muscle tissues were washed with distilled water twice for 15 min between incubation steps. After incubation in latrunculin B, muscles were incubated in 0.6 M potassium chloride (Fluka Chemicals, Milwaukee, WI) for 2 h followed by 1.0 M potassium iodide (Fisher Scientific, Waltham, MA) for 2 h.23 After the salt solution incubations, muscles were washed in distilled water overnight and then the potassium chloride and potassium iodide incubations were repeated, followed by incubation in DNase I (1 kU/mL; Sigma, St. Louis, MO) for 2 h. Finally, treated muscles were washed in distilled water for a minimum of 2 days with daily water changes to remove remaining reagents. Control muscles were analyzed immediately after harvesting.
For comparative purposes, the decellularization method of Stern et al.21 was performed on intact TA muscles from 2-month-old female C57BL/6 mice. Briefly, the following steps were performed at 4°C with continuous agitation: ultrapure water for 2 days, 1 h in 0.05% trypsin with EDTA (Mediatech, Manassas, VA), neutralized in DMEM with 10% fetal bovine serum (Gemini Bio-Products, West Sacramento, CA) overnight, 5 days of exposure to 1% Triton X-100 (Sigma), rinsed in ultrapure water for 2 days, and rinsed in phosphate-buffered saline (PBS) for 1 day. Biochemical assays for DNA, glycosaminoglycan (GAG), and collagen quantification were performed on these tissues. Note that this method was originally optimized for slices of muscle tissue (<500 μm). Here, we have performed this method on intact muscle tissues to make a valid direct comparison.
DNA quantification
Total DNA from lyophilized and papain-digested (4 μg/mg tissue; Worthington Biochemical, Lakewood, NJ) muscles (16–18 h at 65°C) was quantified using the fluorescent Quant-iT Picogreen reagent (Life Technologies, Carlsbad, CA) according to the manufacturer's instructions. Fluorescence was measured using a Beckman Coulter DTX 880 Multimode Detector (excitation: 480 nm, emission: 520 nm) and values were compared with a lambda DNA standard.
Sulfated GAG content
Sulfated GAG content of the samples was quantified using the dimethylmethylene blue dye assay (Sigma).24 Muscles were lyophilized and papain-digested followed by incubation with dimethylmethylene blue dye. Absorbance of the samples at 525 nm was measured with a Beckman Coulter DU730 UV/Vis Spectrophotometer using chondroitin sulfate (Sigma) as the standard.
Collagen content
The proximal halves of TA muscles were hydrolyzed in 6 N HCl for 18 h at 115°C. Hydroxyproline content was then measured after reaction with chloramine T (EMD Chemicals, Gibbstown, NJ) and p-dimethylaminobenzaldehyde (Fluka Chemicals).25 Absorbance of the samples was measured at 550 nm using a Bio-Tek Synergy HT1 microplate reader, and values were compared with a L-4-hydroxyproline (Fluka Chemicals) standard. Collagen content was calculated using a 1:7.46 hydroxyproline:collagen ratio.26
Histology and immunohistochemistry
Muscles were allowed to equilibrate in 1:1 PBS:optimal cutting temperature compound (OCT; Sakura Finetek, Torrance, CA) for 30 min before being frozen in OCT in liquid nitrogen-cooled isopentane. Embedded muscles were cryosectioned at 10 μm thickness perpendicular to the fiber direction as evidenced by tightly packed polygonal fibers with no preferred long axis. Hematoxylin and eosin staining (Statlab Medical Products, McKinney, TX) was performed and sections were mounted with Permount mounting medium (Fisher Scientific). Sections for immunohistochemistry were permeabilized in 3% bovine serum albumin (Sigma) and 0.5% Triton X-100 (Fisher Scientific) in PBS for 30 min and incubated in primary antibody (Pax7 diluted 1:250; Developmental Studies Hybridoma Bank, Iowa City, IA) for 1 h. The secondary antibody (Alexa Fluor 568 goat anti-mouse IgG diluted 1:250; Life Technologies) was applied for 1 h and slides were coverslipped in mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA). Slides were viewed using a Zeiss Axio Observer A1 fluorescence microscope and images were recorded using AxioView software.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and western blot analysis
Muscles were homogenized in the presence of protease and phosphatase inhibitors. Total protein content was measured using the Bradford dye-binding method. Equal amounts of protein from each sample were loaded into wells of 4%–15% gradient polyacrylamide gels (Bio-Rad, Hercules, CA). Proteins were transferred to polyvinylidene fluoride membranes that were blocked in tris buffered saline with 0.1% Tween-20 and 5% nonfat dry milk for 1 h at room temperature. The primary antibody (actin [JLA20] diluted 1:1000 or myosin [A4.1519] diluted 1:20,000; Developmental Studies Hybridoma Bank) was applied for 1 h at room temperature. Membranes were washed and incubated with the secondary antibody (goat anti-mouse IgG horseradish peroxidase-conjugated diluted 1:10,000; Santa Cruz Biotechnology, Santa Cruz, CA) for 30 min at room temperature. Membranes were washed followed by incubation with chemiluminescent substrate (ECL Plus; GE Healthcare, Waukesha, WI) and imaged using a Storm Imager 840. Precision Plus Protein Standard (Bio-Rad) was used for molecular weight indication.
Scanning electron microscopy analysis
Muscles were lightly fixed in 4% paraformaldehyde for 45 min at room temperature and subsequently dehydrated in graded ethanol. Muscles were then snap-frozen in liquid nitrogen and broken transversely to the fiber direction into three slices (∼2 mm thickness) and lyophilized. Lyophilized muscle slices were sputter coated with chromium using an Emitech K575 × Sputter Coater and observed using a Philips XL30 Environmental Scanning Electron Microscope.
Passive mechanics testing on decellularized muscles
Tissue was placed in a relaxing solution composed of ethylene–glycol tetraacetic acid (7.5 mM), potassium propionate (170 mM), magnesium acetate (2 mM), imidazole (5 mM), creatine phosphate (10 mM), adenosine triphosphate (4 mM), leupeptin (a protease inhibitor, 17 mg/mL), and E64 (a protease inhibitor, 4 mg/mL).27 This relaxing solution has been shown to prevent proteolytic degradation and has therefore been used for mechanical characterization of intact muscle tissues.28 Small bundles of ∼20 endomysial sheaths were dissected free from the tissue for mechanical testing.
Dissected bundles were secured on either side to 125 μm titanium wires using 10-0 silk suture loops. One wire was secured to an ultrasensitive force transducer (Model 405A, sensitivity 10 V/g; Aurora Scientific, Ontario, Canada) and the other was secured to a servomotor (Model 318B; Aurora Scientific) on a micromanipulator as previously described for muscle fibers.28 Bundle length was set to the minimum length that produced measurable forces. Baseline sample diameters were measured optically with a cross-hair reticule mounted on a dissecting microscope and micromanipulators on an x–y mobile stage. Force–strain data were generated for each mounted bundle in ∼10% strain increments with force recorded for 3-min to allow stress-relaxation. Bundles were elongated to ∼100% strain. Force data were converted to stress by dividing force by the baseline cross-sectional area. Data were fit to a viscoelastic model that contained a spring in parallel with a series of a spring and dashpot. Relaxed stress was fit with a strain dependent parallel spring creating a quadratic stress–strain equation. Quadratic curve fits were averaged for untreated and decellularized samples and plotted at 10% strain increments.
Culture of C2C12 cells on decellularized muscles
To evaluate cytocompatibility of the decellularized muscles, C2C12 cells were cultured on decellularized tissues. C2C12 cells (4 × 105 cells in 40 μL culture medium) were pipetted onto intact decellularized muscles in low adhesion Petri dishes. The seeded muscles were maintained at 37°C for 30 min before adding medium to allow adhesion of cells onto the decellularized tissues. Growth medium (DMEM [Gibco], 10% fetal bovine serum [Atlanta Biologicals, Atlanta, GA], 2 mM L-glutamine [Gibco], and 1% penicillin/streptomycin [Gibco]) was then added and cell seeded muscles were maintained at 37°C and 5% CO2 for 4 days. The C2C12-seeded muscles were then fixed in 4% paraformaldehyde for 1.5 h, embedded in OCT, frozen, and cryosectioned at 10 μm. Sections were mounted with DAPI and viewed as described above.
Statistical analysis
Biochemical data were obtained from four animals with one TA muscle from each animal being decellularized and the remaining TA muscle serving as the untreated control, and from three animals for TA muscles decellularized using the Stern et al. method.21 Experiments involving mechanical testing were performed on untreated (n = 9) and decellularized (n = 8) TA bundles. Data were calculated as mean ±standard error and significance was determined by performing one-way analysis of variance with Tukey-Kramer multiple comparison post tests or two-tailed Student's t-tests (Prism 5; GraphPad Software, La Jolla, CA).
Results
Removal of cellular components
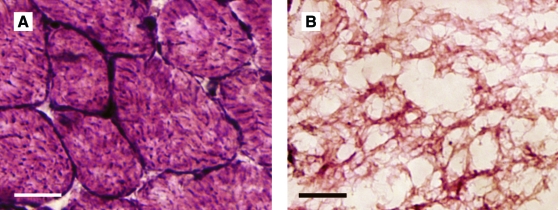
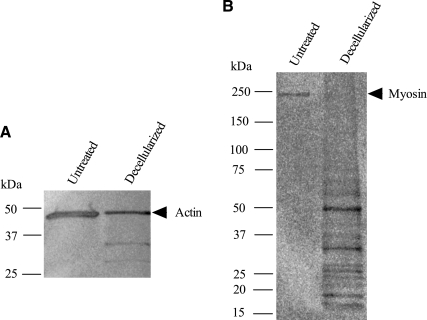
DNA content was significantly reduced from 2.92 ± 0.14 μg/mg dry weight in untreated muscle to 0.12 ± 0.01 μg/mg dry weight in decellularized muscle (p < 0.001). Cell nuclei were absent in decellularized muscles stained with hematoxylin and eosin compared with untreated muscles (Fig. 1). Similar findings were observed with DAPI and Pax7 staining, where the latter is a marker for satellite cells (data not shown). Evidence for removal of sarcomeric components was based on actin and myosin protein analysis. Western blotting for actin revealed bands at 43, 35, and 30 kDa with the 43 kDa band corresponding to full size actin (Fig. 2A). The amount of intact actin in decellularized muscles was greatly decreased compared with that of untreated muscles. Western blotting for myosin detected no full size myosin (200 kDa) in decellularized muscles, but several bands corresponding to low-molecular-weight fragments (<50 kDa size) were observed, which indicates degradation of myosin (Fig. 2B).
FIG. 1.
Hematoxylin and eosin staining of untreated (A) and decellularized (B) mouse tibialis anterior muscles. Absence of dark stained peripheral nuclei was confirmed in decellularized muscle. Scale bars represent 20 μm. Color images available online at www.liebertonline.com/ten.
FIG. 2.
Western blots of untreated and decellularized mouse tibialis anterior muscles for actin (A) and myosin (B). Both actin and myosin content were greatly reduced in decellularized muscles; however, a small 43 kDa band for actin was still present. A 200 kDa band for myosin was not observed in decellularized muscles.
Decellularized muscle composition and structure
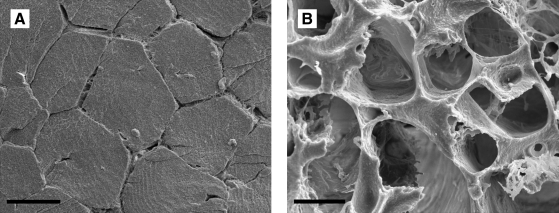
Major ECM components were quantified to determine the effect of decellularization on the biochemical composition of the ECM. Total GAG content was decreased from 4.93 ± 0.18 μg/mg dry weight in untreated muscles to 2.93 ± 0.18 μg/mg dry weight in decellularized muscles (p < 0.001). Collagen content remained constant between the untreated and decellularized muscles (3.01 ± 0.11 and 3.49 ± 0.24 μg/mg wet weight respectively; p = 0.10). We next examined the structure of the muscles to ensure that the decellularization process did not alter the gross structure of the ECM. Scanning electron microscopy (SEM) images obtained from untreated and decellularized muscles showed that, unlike untreated muscles, decellularized muscles showed a hollow, tubular structure (Fig. 3). This observation implied that fibers were still present in the untreated muscle, but had been removed from the decellularized muscle.
FIG. 3.
Scanning electron micrographs of untreated (A) and decellularized (B) mouse tibialis anterior muscles cut transversely to fiber direction. Unlike untreated muscles, decellularized muscles had a hollow tubular structure, suggesting the removal of muscle fibers. Scale bar represents 20 μm.
Mechanical properties of decellularized muscle
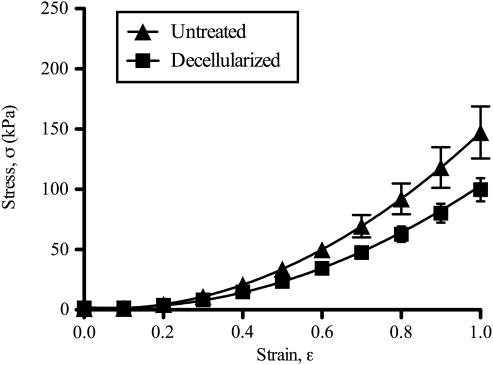
Passive mechanical testing of untreated and decellularized muscle bundles yielded the nonlinear stress–strain behavior as reported previously for muscle fiber bundles (Fig. 4).29 Relaxed stress values were not significantly different between untreated and decellularized samples at any strain ranging from 10% to 100% strain (p = 0.06–0.56). Similarly, the tangent modulus value at 100% strain was not significantly different between untreated and decellularized samples (308 ± 51.1 kPa and 218 ± 24.5 kPa, respectively; p = 0.14).
FIG. 4.
Mechanical properties of untreated and decellularized mouse tibialis anterior muscle bundles. Stress (σ) vs. strain (ɛ) relationship represented as average stress (untreated: n = 9; decellularized: n = 8) ± standard error. Relationships were approximated by quadratic fits with σ = 162ɛ2 − 15.7ɛ + 1.18 (r2 = 0.979) for untreated bundles and σ = 117ɛ2 − 16.0ɛ+ 2.09 (r2 = 0.993) for decellularized bundles.
Decellularized muscles support cell survival
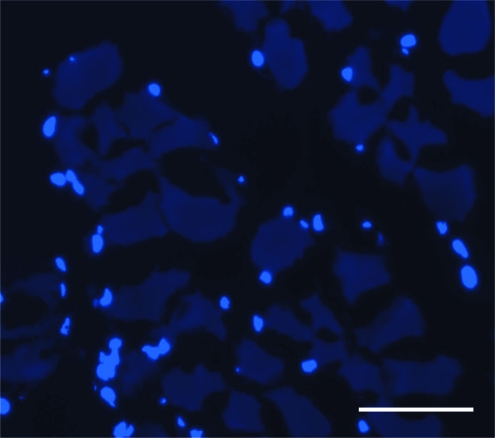
The C2C12 cells cultured on decellularized muscles were viable after 4 days in culture as evidenced by positive DAPI staining (Fig. 5). Cells were seeded on the surface of the muscle; whereas the majority of cells remained on the surface of the muscle, some cells had begun to migrate into the decellularized tissue after 4 days.
FIG. 5.
Cryosection of C2C12 cells cultured on decellularized mouse tibialis anterior muscle for 4 days and stained with 4′,6-diamidino-2-phenylindole. The fact that the cells survived 4 days and the presence of the 4′,6-diamidino-2-phenylindole-stained nuclei indicated the cytocompatibility of decellularized muscles and confirmed the removal of any potentially cytotoxic decellularization reagents. Scale bar represents 50 μm. Color images available online at www.liebertonline.com/tec.
Comparison with an existing decellularization method
Finally, we compared the decellularization method described in this article with the method of Stern et al.21 When performed on whole muscles, the latter method was unable to remove the nuclear components completely as characterized by the DNA content. Specifically, the Stern et al. method resulted in a DNA content of 1.71 ± 0.17 μg/mg dry weight compared with 0.12 ± 0.01 μg/mg dry weight for our method (p < 0.001 compared with our method and untreated).
We also compared the biochemical composition of the decellularized tissues. A significant reduction in the GAG content of the decellularized tissues was found using the Stern et al. method where the GAG content decreased from 4.93 ± 0.18 μg/mg dry weight in untreated muscles to 1.23 ± 0.04 μg/mg dry weight (p < 0.001; p < 0.001 compared with our method). This corresponds to a 75% of reduction in GAG compared with a 40% reduction observed with our method. In contrast to GAG, the collagen content remained relatively constant between the untreated and decellularized muscles for both methods (3.80 ± 0.14 μg/mg wet weight for Stern et al.; p < 0.05 compared with untreated).
Discussion
The present study describes a decellularization method for intact skeletal muscle without the use of detergents or proteolytic enzymes. The method utilized actin disruption by treatment with latrunculin B, cell lysis by osmotic shock, myosin depolymerization by exposure to high ionic strength salt solution, and DNase I treatment to remove residual DNA from skeletal muscle. This method successfully removed muscle fibers and degraded sarcomeric components in the muscle tissue without altering the ECM structure or mechanical properties.
Since DNA quantification indicated the presence of a small amount of DNA in the decellularized tissue, tissue sections were stained for satellite cells that may be more resistant to the decellularization protocol. Negative Pax7 (satellite cells) and DAPI staining indicated that any DNA remaining in the decellularized muscles was degraded and not contained within intact cell nuclei.
An approximately 40% reduction in the GAG content of the decellularized tissue was observed, which could be due to removal of GAGs associated with the cell membrane; almost 30% of GAGs are associated with the cell membrane,30 and these GAGs may have been removed with the sarcolemma during the decellularization process. Decellularization methods using trypsin and Triton X-100 remove GAGs from skeletal muscle,21 but this effect appears to be tissue dependent since the use of trypsin and/or Triton X-100 on other tissues has yielded disparate results.31–33 Collagen content was unchanged in decellularized muscle, showing that this method did not remove the most abundant ECM structural component. Together, these results provide evidence that the decellularization method described here is minimally disruptive to native ECM. Further, comparison with the method of Stern et al. showed that our decellularization method was more efficient at removing DNA and retaining GAGs in intact muscle compared with a method that uses a proteolytic enzyme (trypsin) and a detergent (Triton X-100).21
SEM images from decellularized muscles (Fig. 3) suggested that muscle fibers were removed from the tissue, but that the overall architecture of the ECM was maintained when compared with the untreated tissue. SEM images of skeletal muscle endomysium obtained by Trotter and Purslow show a thinner, more fibrous matrix than we obtained in this study.34,35 The differences in the endomysial thickness could be attributed to the different sample preparation methods. Totter et al. treated muscles with NaOH, which removed the sarcolemma, basement membrane, and any GAGs associated with the endomysium, leaving only collagen fibrils within the ECM.34 The thicker ECM observed in our decellularized tissues is likely due to the presence of GAGs and the basement membrane.
Mechanical analysis of the decellularized muscles confirmed the mechanical integrity of the decellularized tissue since no difference was observed between the untreated and decellularized muscle bundle stress–strain relationship (Fig. 4). These results provide support for the concept that the ECM is the primary passive load bearing structure in skeletal muscle, not the fibers themselves. This result must be tempered by the fact that there was no unambiguous reference length available for determination of the stress–strain relation for decellularized muscle bundles. However, future studies can potentially overcome this limitation by referencing strain to sarcomere length before decellularization or by measuring the relationship between collagen crimp pattern and tissue strain.
The ability of the decellularized muscles to support adhesion and survival of C2C12 cells indicates their cytocompatibility, which is in agreement with other studies.4,5,21 The use of decellularized muscle has clinical applications in repair of severe skeletal muscle injuries where the muscles cannot undergo complete regeneration. Detergent-decellularized muscle has been used as a scaffold to repair muscle defects in animal models4,5,18; however, complete functional regeneration has not yet been shown. Biochemical and mechanical signals of the ECM play a key role in the activation of muscle progenitor cells and their differentiation,11,21,36 and thus the use of muscle ECM that contains these signals may be more effective at achieving functional regeneration of impaired muscles. This decellularization method may be used to study the changes that occur in muscle ECM with various myopathies and may serve as a diagnostic tool by identifying characteristic patterns in muscle ECM composition or architecture. In addition, tissues decellularized by this method may be used to measure directly the passive mechanical properties of skeletal muscle ECM. Since the decellularization method described here relies on diffusion of reagents into the tissue, extending this protocol to other tissues may require optimization depending upon the size and composition of the targeted tissue. The development of this method is a first step toward understanding not only the biochemical cues that exist within skeletal muscle ECM, but also the structural and mechanical signals for tissue maintenance.
Acknowledgments
The authors thank Dr. Ramsés Ayala, Dr. Chien-Wen Chang, Phil Kyriakakis, Gretchen Meyer, YongSung Hwang, Evie Lin, Ryan Anderson, and Soo Yeon Kim for discussion and technical support. The Pax7 monoclonal antibody developed by Atsushi Kawakami, actin monoclonal antibody developed by Jim Jung-Ching Lin, and myosin monoclonal antibody developed by Helen M. Blau were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA 52242. This work was supported by California Institute of Regenerative Medicine grant (#RN2-00945-1), NIH Grant R01AR057393, and a National Science Foundation Graduate Research Fellowship (A.R.G.).
Disclosure Statement
No competing financial interests exist.
References
- 1.Juliano R.L. Haskill S. Signal transduction from the extracellular matrix. J Cell Biol. 1993;120:577. doi: 10.1083/jcb.120.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ott H.C. Matthiesen T.S. Goh S.K. Black L.D. Kren S.M. Netoff T.I. Taylor D.A. Perfusion-decellularized matrix: using nature's platform to engineer a bioartificial heart. Nat Med. 2008;14:213. doi: 10.1038/nm1684. [DOI] [PubMed] [Google Scholar]
- 3.Yoo J.J. Meng J. Oberpenning F. Atala A. Bladder augmentation using allogenic bladder submucosa seeded with cells. Urology. 1998;51:221. doi: 10.1016/s0090-4295(97)00644-4. [DOI] [PubMed] [Google Scholar]
- 4.De Coppi P. Bellini S. Conconi M.T. Sabatti M. Simonato E. Gamba P.G. Nussdorfer G.G. Parnigotto P.P. Myoblast-acellular skeletal muscle matrix constructs guarantee a long-term repair of experimental full-thickness abdominal wall defects. Tissue Eng. 2006;12:1929. doi: 10.1089/ten.2006.12.1929. [DOI] [PubMed] [Google Scholar]
- 5.Conconi M.T. De Coppi P. Bellini S. Zara G. Sabatti M. Marzaro M. Zanon G.F. Gamba P.G. Parnigotto P.P. Nussdorfer G.G. Homologous muscle acellular matrix seeded with autologous myoblasts as a tissue-engineering approach to abdominal wall-defect repair. Biomaterials. 2005;26:2567. doi: 10.1016/j.biomaterials.2004.07.035. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y. Kropp B.P. Lin H.K. Cowan R. Cheng E.Y. Bladder regeneration with cell-seeded small intestinal submucosa. Tissue Eng. 2004;10:181. doi: 10.1089/107632704322791835. [DOI] [PubMed] [Google Scholar]
- 7.Gilbert T.W. Sellaro T.L. Badylak S.F. Decellularization of tissues and organs. Biomaterials. 2006;27:3675. doi: 10.1016/j.biomaterials.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 8.Grounds M.D. Towards understanding skeletal muscle regeneration. Pathol Res Pract. 1991;187:1. doi: 10.1016/S0344-0338(11)81039-3. [DOI] [PubMed] [Google Scholar]
- 9.Crawley S. Farrell E.M. Wang W. Gu M. Huang H.Y. Huynh V. Hodges B.L. Cooper D.N. Kaufman S.J. The α7β1 integrin mediates adhesion and migration of skeletal myoblasts on laminin. Exp Cell Res. 1997;235:274. doi: 10.1006/excr.1997.3671. [DOI] [PubMed] [Google Scholar]
- 10.Flaumenhaft R. Moscatelli D. Saksela O. Rifkin D.B. Role of extracellular matrix in the action of basic fibroblast growth factor: matrix as a source of growth factor for long-term stimulation of plasminogen activator production and DNA synthesis. J Cell Physiol. 1989;140:75. doi: 10.1002/jcp.1041400110. [DOI] [PubMed] [Google Scholar]
- 11.Cornelison D.D. Filla M.S. Stanley H.M. Rapraeger A.C. Olwin B.B. Syndecan-3 and syndecan-4 specifically mark skeletal muscle satellite cells and are implicated in satellite cell maintenance and muscle regeneration. Dev Biol. 2001;239:79. doi: 10.1006/dbio.2001.0416. [DOI] [PubMed] [Google Scholar]
- 12.Ide C. Nerve regeneration through the basal lamina scaffold of the skeletal muscle. Neurosci Res. 1984;1:379. [Google Scholar]
- 13.Di Benedetto G. Zura G. Mazzucchelli R. Santinelli A. Scarpelli M. Bertani A. Nerve regeneration through a combined autologous conduit (vein plus acellular muscle grafts) Biomaterials. 1998;19:173. doi: 10.1016/s0142-9612(97)00200-7. [DOI] [PubMed] [Google Scholar]
- 14.Borschel G.H. Dennis R.G. Kuzon W.M., Jr. Contractile skeletal muscle tissue-engineered on an acellular scaffold. Plast Reconstr Surg. 2004;113:595. doi: 10.1097/01.PRS.0000101064.62289.2F. [DOI] [PubMed] [Google Scholar]
- 15.Qing Q. Qin T. Optimal method for rat skeletal muscle decellularization. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2009;23:836. [PubMed] [Google Scholar]
- 16.Gamba P.G. Conconi M.T. Lo Piccolo R. Zara G. Spinazzi R. Parnigotto P.P. Experimental abdominal wall defect repaired with acellular matrix. Pediatr Surg Int. 2002;18:327. doi: 10.1007/s00383-002-0849-5. [DOI] [PubMed] [Google Scholar]
- 17.Fawcett J.W. Keynes R.J. Muscle basal lamina: a new graft material for peripheral nerve repair. J Neurosurg. 1986;65:354. doi: 10.3171/jns.1986.65.3.0354. [DOI] [PubMed] [Google Scholar]
- 18.Merritt E.K. Hammers D.W. Tierney M. Suggs L.J. Walters T.J. Farrar R.P. Functional assessment of skeletal muscle regeneration utilizing homologous extracellular matrix as scaffolding. Tissue Eng Part A. 2010;16:1395. doi: 10.1089/ten.TEA.2009.0226. [DOI] [PubMed] [Google Scholar]
- 19.Mligiliche N. Tabata Y. Endoh K. Ide C. Peripheral nerve regeneration through a long detergent-denatured muscle autografts in rabbits. Neuroreport. 2001;13:1719. doi: 10.1097/00001756-200106130-00040. [DOI] [PubMed] [Google Scholar]
- 20.Liu X.L. Arai T. Sondell M. Lundborg G. Kanje M. Dahlin L.B. Use of chemically extracted muscle grafts to repair extended nerve defects in rats. Scand J Plast Reconstr Surg Hand Surg. 2001;35:337. doi: 10.1080/028443101317149291. [DOI] [PubMed] [Google Scholar]
- 21.Stern M.M. Myers R.L. Hammam N. Stern K.A. Eberli D. Kritchevsky S.B. Soker S. Van Dyke M. The influence of extracellular matrix derived from skeletal muscle tissue on the proliferation and differentiation of myogenic progenitor cells ex vivo. Biomaterials. 2009;30:2393. doi: 10.1016/j.biomaterials.2008.12.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y. He Y. Bharadwaj S. Hammam N. Carnagey K. Myers R. Atala A. Van Dyke M. Tissue-specific extracellular matrix coatings for the promotion of cell proliferation and maintenance of cell phenotype. Biomaterials. 2009;30:4021. doi: 10.1016/j.biomaterials.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Granzier H.L. Irving T.C. Passive tension in cardiac muscle: contribution of collagen, titin, microtubules, and intermediate filaments. Biophys J. 1995;68:1027. doi: 10.1016/S0006-3495(95)80278-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farndale R.W. Buttle D.J. Barrett A.J. Improved quantitation and discrimination of sulphated glycosaminoglycans by use of dimethylmethylene blue. Biochim Biophys Acta. 1986;883:173. doi: 10.1016/0304-4165(86)90306-5. [DOI] [PubMed] [Google Scholar]
- 25.Stegemann H. Stalder K. Determination of hydroxyproline. Clin Chim Acta. 1967;18:267. doi: 10.1016/0009-8981(67)90167-2. [DOI] [PubMed] [Google Scholar]
- 26.Neuman R.E. Logan M.A. The determination of collagen and elastin in tissues. J Biol Chem. 1950;186:549. [PubMed] [Google Scholar]
- 27.Wood D.S. Zollman J. Reuben J.P. Brandt P.W. Human skeletal muscle: properties of the “chemically skinned” fiber. Science. 1975;187:1075. doi: 10.1126/science.187.4181.1075. [DOI] [PubMed] [Google Scholar]
- 28.Fridén J. Lieber R.L. Spastic muscle cells are shorter and stiffer than normal cells. Muscle Nerve. 2003;26:157. doi: 10.1002/mus.10247. [DOI] [PubMed] [Google Scholar]
- 29.Lieber R.L. Runesson E. Einarsson F. Fridén J. Inferior mechanical properties of spastic muscle bundles due to hypertrophic but compromised extracellular matrix material. Muscle Nerve. 2003;28:464. doi: 10.1002/mus.10446. [DOI] [PubMed] [Google Scholar]
- 30.Mertens G. Cassiman J.J. Van den Berghe H. Vermylen J. David G. Cell surface heparan sulfate proteoglycans from human vascular endothelial cells. Core protein characterization and antithrombin III binding properties. J Biol Chem. 1992;267:20435. [PubMed] [Google Scholar]
- 31.Yang B. Zhang Y. Zhou L.H. Sun Z.Y. Zheng J.H. Chen Y. Dai Y.T. Development of a porcine bladder acellular matrix with well preserved extracellular bioactive factors for tissue engineering. Tissue Eng Part C Methods. 2010;16:1201. doi: 10.1089/ten.TEC.2009.0311. [DOI] [PubMed] [Google Scholar]
- 32.Wainwright J.M. Czajka C.A. Patel U.B. Freytes D.O. Tobita K. Gilbert T.W. Badylak S.F. Preparation of cardiac extracellular matrix from an intact porcine heart. Tissue Eng Part C Methods. 2010;16:525. doi: 10.1089/ten.tec.2009.0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gratzer P.F. Harrison R.D. Woods T. Matrix alteration and not residual sodium dodecyl sulfate cytotoxicity affects the cellular repopulation of a decellularized matrix. Tissue Eng. 2006;12:2975. doi: 10.1089/ten.2006.12.2975. [DOI] [PubMed] [Google Scholar]
- 34.Trotter J.A. Purslow P.P. Functional morphology of the endomysium in series fibered muscles. J Morphol. 1992;212:109. doi: 10.1002/jmor.1052120203. [DOI] [PubMed] [Google Scholar]
- 35.Purslow P.P. Trotter J.A. The morphology and mechanical properties of endomysium in series-fibred muscles: variations with muscle length. J Muscle Res Cell Motil. 1994;15:299. doi: 10.1007/BF00123482. [DOI] [PubMed] [Google Scholar]
- 36.Engler A.J. Griffin M.A. Sen S. Bönnemann C.G. Sweeney H.L. Discher D.E. Myotubes differentiate optimally on substrates with tissue-like stiffness: pathological implications for soft or stiff microenvironments. J Cell Biol. 2004;166:877. doi: 10.1083/jcb.200405004. [DOI] [PMC free article] [PubMed] [Google Scholar]