Abstract
An outstanding aspect of pharmaceutical nanotechnology lies in the characterization of nanocarriers for targeting of drugs and other bioactive agents. The development of microscopic techniques has made the study of the surface and systems architecture more attractive. In the field of pharmaceutical nanosystems, researchers have collected vital information on size, stability, and bilayer organization through the microscopic characterization of liposomes. This paper aims to compare the results obtained by atomic force microscopy, environmental scanning electron microscopy, transmission electron microscopy, and confocal laser scanning microscopy to point out the limits and advantages of these applications in the evaluation of vesicular systems. Besides this comparative aim, our work proposes a simple confocal laser scanning microscopy procedure to rapidly and easily detect the liposomal membrane.
Keywords: atomic force microscopy, transmission electron microscopy, environmental scanning electron microscopy, confocal laser scanning microscopy
Introduction
Colloidal carriers and particularly liposomes have become widely used as pharmaceutical devices in numerous clinical applications.1–3 The increase in their therapeutic applications has developed analytical and technological approaches to characterize the carriers in terms of morphology, size, polydispersity index, number of lamellae, charge, bilayer fluidity, lipidic composition, and encapsulation efficiency.4 The application of dynamic light scattering (DLS, also known as photon correlation spectroscopy, PCS), nuclear magnetic resonance (NMR), X-ray photoelectron spectroscopy (XPS), electron paramagnetic resonance (EPR), differential scanning calorimetry (DSC) has improved the evaluation of physicochemical and technological properties of these drug delivery systems.5–7 In particular, the size and the unilamellar or multilamellar structure of the liposomes can be also provided by small-angle X-ray scattering (SAXS).8,9
During the 1970s and 1980s, freeze-fracture electron microscopy was the first tool able to provide detail on liposome organization that was impossible to achieve by conventional thin-section electron microscopy.10,11 Modern microscopical techniques have increased our ability to characterize nanopharmaceutical systems.
This rapid communication aims to determine the details of the morphology and structure of a conventional liposomal formulation obtained by well-known microscopy techniques. The results of the analysis obtained by atomic force microscopy (AFM), environmental scanning electron microscopy (ESEM), transmission electron microscopy (TEM) and confocal laser scanning microscopy (CLSM; labeling using a fluorochrome marker) were compared in order to emphasize advantages and bias of these techniques. We propose a simple procedure to analyze the liposome membrane by CLSM.
Methods
Preparation of liposomes
A typical and commonly used liposomal formulation was prepared by a thin-layer evaporation method using phosphatidylcholine (PC) and cholesterol (CHOL) at 1:0.1 (mol:mol) ratio.12 The total lipid amount was fixed at 1 mg/mL. Briefly, lipids (5 mg) were dissolved in 2 mL of chloroform. The solvent was removed by rotary evaporation (B-480; Büchi, Büchs, Switzerland) for 1 hour under vacuum (10 mbar) at 20 ± 1°C. Then, the dried lipidic film was vacuum-dried for 3 hours (0.15 mbar) and then hydrated at room temperature with 5 mL of MilliQ water (Millipore, Billerica, MA). The preparation was alternatively vortexed for 3 minutes (Zx3; Velp Scientifica, Usmate, Italy) and warmed in a water bath at room temperature for 3 minutes. The cycle was repeated three times. The sample, prepared in three lots, was stored at 4°C, protected from light and analyzed within 15 days. Liposomes were analyzed for particle size and zeta potential by PCS and laser Doppler anemometry using a Zetasizer Nano ZS (laser 4 mW He-Ne, 633 nm, automatic laser attenuator, transmission 100% to 0.0003%, detector avalanche photodiode, QE > 50% at 633 nm, T = 25°C; Malvern Instruments, Malvern, UK) without any dilution of the samples.
Tapping mode atomic force microscopy
The AFM experiments were performed with a Nanoscope IIIa (Digital Instruments, Santa Barbara, CA) operating in tapping mode at room temperature. Both the height and the phase imaging data were simultaneously acquired using a commercial silicon tip-cantilever (high resolution noncontact “GOLDEN” Silicon Cantilevers NSG-11, NT-MDT, tip diameter ≅5–10 nm; Zelenograd, Moscow, Russia) with stiffness about 40 Nm−1 and a resonance frequency around 170 KHz. All the AFM images were obtained without any dilution of the samples, with a scan rate of 0.7 or 1 Hz over a selected area in the dimension of 5 μm × 5 μm or 0.65 μm × 0.65 μm using freshly cleaved mica as the substrate. The force applied to the surface was roughly adjusted by the ratio of the engaged or set-point amplitude Asp to the free air amplitude A0.
According to the literature, the set-point amplitude has to be adjusted to 10%–25% of the free air amplitude for “high force” and to 40%–70% or 75%–90% of the amplitude for the “moderate- and low-force” imaging, respectively.13 In this communication, the experiments were carried in “moderate force” mode and the set point was adjusted to 50%–60%. Images were processed and analyzed using a program obtained from Digital Instruments (version V5–31; Veeco Group, Santa Barbara, CA). The height and diameter of liposomes were measured from the profile section of AFM line scans analyzing the height images.
Environmental scanning electron microscopy
A Quanta 200 ESEM (FEI Company, Hillsboro, OR) was used in the ESEM investigations. The sample was put on stage and observed in real time (hydration/dehydration step) into the ESEM chamber adjusting the temperature with the Peltier stage (Emott AG, Zurich, Switzerland). At the beginning of the experimental procedure, the chamber pressure and sample temperature were respectively set at about 6.45 Torr and 4°C. In these conditions, relative humidity reached 100%. The sample was maintained under these “initial conditions” for about 2 minutes and an image of interest was chosen after this time (initial state–wet state). The chamber pressure is then slowly taken back at the dehydration condition (P ≅4–2.65 Torr) and the sample temperature stabilized at 9°C. The images taken describe the final dehydration state.
Negative staining transmission electron microscopy
Briefly, a drop of a water-diluted suspension of the liposomes (about 0.05 mg/mL) was placed on a 200-mesh formvar copper grid (TABB Laboratories Equipment, Berks, UK), allowed to adsorb and the surplus was removed by filter paper. A drop of 2% (w/v) aqueous solution of uranyl acetate was added and left in contact with the sample for 5 minutes (initially, we tested different concentrations of uranyl acetate solution; the condition applied in this study was the most suitable to maintain the integrity of the sample during the preparation). The surplus water was removed and the sample was dried at room conditions before the vesicles were imaged with a TEM operating at an acceleration voltage of 200 KV (model JEM 2010; JEOL, Peabody, MA).
Confocal microscopy
CLSM analysis was performed with a DM IRE2 microscope (Leica, Mannheim, Germany) and a Leica confocal system equipped with a 3-channel multiband Leica scanner TCS SP2 with AOBS, laser diode blu COH (405 nm/25 mW), laser Ar (458 nm/5 mW, 476 nm/5 mW, 488 nm/20 mW, 496 nm/5 mW, 514 nm/20 mW), laser HeNe (543 nm/1.2 mW, 594 nm Orange, 633 nm/102 mW) and with a 63× λ-blu corrected oil immersion objective. Before the analysis, the dried lipidic film of the liposomes was hydrated with a water solution of rhodamine 123 (0.015 mg/mL; Sigma-Aldrich Company, Milan Italy). The excess fluorochrome solution was removed by dialysis technique using a dialysis tube (CelluSep MWCO 3500; Membrane Filtration Products, Seguin, TX). A small aliquot of the dialyzed sample (usually 10 μL) was transferred to a coverslip (slide) and directly observed.
Results and discussions
Preliminary results: liposome properties
The formulation was heterogeneous (D(10) = 120 ± 34 nm; D(50) = 550 ± 123 nm; D(90) = 2540 ± 342 nm) with a polydispersity index (PDI; an estimate of the width of distribution) about 0.45 ± 0.1 and the zeta potential (z-p) of about −6.5 ± 2.4 mV.
AFM studies
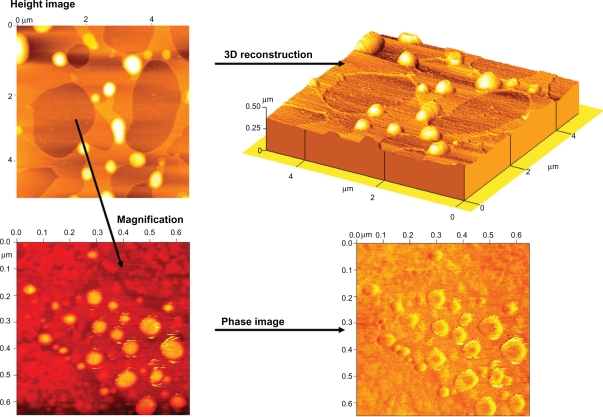
The AFM images of liposomes on mica are shown in Figure 1. As described previously,14 AFM in the tapping and noncontact mode approaches allows the observation of the liposomal morphology without any sample manipulation such as staining, labeling, or fixation (see height images in Figure 1). Particularly the intermittent contact motion of the tip (tapping) eliminates lateral or shear forces which would deform or scrape the sample.15,16 The main advantage of the technique is the possibility to operate with high resolution in air or in fluid in real-time and at the nanometer scale. But, liposomes can change their shape once deposited on mica support also using tapping mode and operating in aqueous solution (within 10–15 minutes from deposition when liposomes were still hydrated and plunged in water). In fact, the interaction between the sample and the substrate, as well as the continuous movement of the tip, can induce deformation17 depending chiefly on the vesicle composition.18
Figure 1.
TM-AFM analysis (images acquired within 10–15 minutes from deposition on mica support).
Abbreviations: TM-AFM, tapping mode atomic force microscopy.
Our experimental results agree with the hypotheses in the literature. The comparison between the diameter and the height values of our liposomes emphasized the flattening of vesicles on the support just few minutes after deposition. This indicated only a moderate stability of the liposomes on a mica substrate. In fact the liposomes maintained a spherical, well-defined shape, although the diameters were higher than the related heights (Figure 1, 3D reconstruction). Twenty minutes after deposition, liposomes showed a progressive tendency to turn into an asymmetrical and defeat structure described as planar vesicles (data not shown). This behavior can identify dried or partially-dried liposomes as others have reported previously.19,20
Along with this application, AFM was able to identify the important details of the surface properties.21 The AFM phase images display the variation of the phase during the cantilever oscillation, especially considering the oligo- and unilamellar vesicles, which are well studied by small scansions. After having fixed the operation conditions (as Asp, A0, etc), the phase shift observed during the image acquisition could be affected by the bilayer wetting and hydration and consequently by the local surface properties of liposomes.22 The phase images confirmed the instability of our liposomal formulation on mica surface. The dark area (negative phase shift-attractive force between tip and sample) can be related to a collapsed structure, characterized by a flattened layer of lipids with high viscosity, while the bright contour (positive phase shift- repulsive force between tip and sample) suggested that the lipids were still well hydrated with a relative low viscosity environment.
In conclusion, the possibility of obtaining local information about the surface of liposomes using TM-AFM strengths the versatility and the applicability of this technique in the pharmaceutical and medical field.
ESEM studies
Generally, liposomes may suffer structural perturbations as a result of the high-vacuum conditions and the staining process required by some electron microscopy techniques. Therefore, scanning electron microscopy (SEM) is a less frequently used imaging technique because analysis requires the sample to be dried or fixed before imaging.
However, ESEM images wet systems without previous sample preparation (conductive coating). ESEM is based on the use of a multiple aperture, graduated vacuum system, which allows the chamber to be maintained at pressures of up to 40 Torr, which allows specimens to be imaged under water vapor or other auxiliary gases. Moreover, by using a correct pump-down procedure and by controlling the temperature of the specimen, which in the ESEM is usually performed using a Peltier stage, dehydration of wet samples can be inhibited and hence samples can be imaged in their ‘natural state’.23
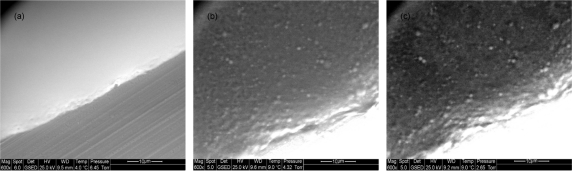
In our case, we tried to study both the formation and morphology of liposomes in real time producing a progressive dehydration of the sample by change of the pressure and temperature into the ESEM chamber. Figure 2 shows the ESEM micrographs of liposomes both at the initial condition (wet sample) and at the reduced pressure adopted to evaluate the morphology (until 2.65 Torr) maintaining a sufficient hydration of sample. Under these conditions, spherical and separated lipidic structures were clearly observed. Further reduction of pressure to 2 Torr resulted in the aggregation of the liposomes which give rise to an undefined and flattened structure (data not shown). Unfortunately, the resolution of ESEM analysis don’t provide detailed information related to the surface and the architecture of the nanoscale structures, as already pointed out by Mohammed et al.24
Figure 2.
ESEM micrographs of liposomes: a) 4.0°C, 6.45 Torr; b) 9.0°C, 4.32 Torr; c) 9.0°C, 2.65 Torr.
TEM analysis
Several methods can be used to apply TEM in the evaluation of morphology and architecture of liposomes. The freeze-fracture electron microscopy is an optimal technique for examining the ultrastructure of rapidly frozen biological samples by TEM, but the preparation of the samples (cryofixation, fracturation and the following operation of shading with evaporated platinum or gold) required caution and long time.25,26 In our opinion, negative staining is an easier and faster procedure. According to this procedure, liposomes are surrounded or embedded in a suitable electron dense material providing high contrast and good reproducibility. We used a cationic negative stain (uranyl cation) that binds the phosphate group of phospholipids, poorly penetrating the lipidic bilayer; nevertheless, it allows the indicative evaluation of the liposomal internal structure without discriminating on the fine details.27
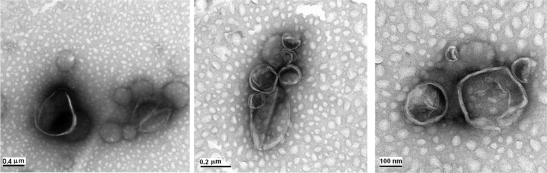
The negative staining of our liposomes (Figure 3) confirms the results obtained by the AFM analysis. The images describe a population of heterogeneous vesicles in which it is possible to emphasize the presence of close bilayer structures spaced by free internal structure. Nevertheless, the shape of liposomes appeared distorted, although this electronic microscopical technique ensures the complete structural analysis of the thin transparent samples. The possible artefacts are due both to the staining process (the interaction between the sample and the negative stain) and the distortion/alteration induced during the drying steps are caused by the exposition of the samples to a vacuum.
Figure 3.
NS-TEM images of liposomes.
CLSM analysis
To study the structure of liposomes by confocal microscopy (CLSM), we loaded the liposomes with a fluorochrome marker which localizes in the lipidic bilayer. This approach could be considered innovative with reference to the potential for high-resolution imaging, as the nondestructive technique allows a 3D reconstruction using a sophistical and sensible range of fluorochromes (commercially available fluoroprobes are designed sensible to the environment in order to operate within different physiological ranges of pH, ionic strength, and water content with high sensitivity, selectivity and versatility).28,29 The literature describing the physical characterization of pharmaceutical systems using this technique is far less extensive and between whiles it provides examples that illustrate the potential and scope.
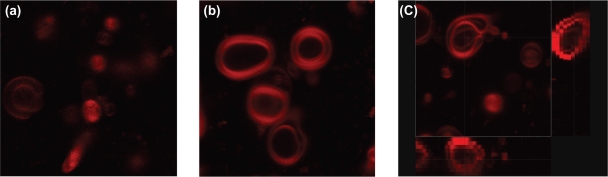
Chiefly in the case of large multilamellar vesicles, this technique allows us to easily appreciate and evaluate the internal structure of the lipidic systems which is not possible to investigate directly with the other techniques described previously (Figure 4). In fact, the layers of the multilamellar vesicle structure appeared well identified by a close bilayer (colored in red) and separated by aqueous phase. The ability to reject light from outside the focal plane obtaining a good contrast and clarity allows the acquisition of images of planes at various depths. The projection of each plane joined with both vertical and horizontal sections and the related 3D reconstruction show the liposomes in their tridimensional architecture, bypassing the necessity of any procedure of sample fixation (as in TEM approach by negative staining or freeze fracture), staining by 31P-NMR (as in the nuclear magnetic resonance technique)30 or using complicated techniques that related to neutrons and X-ray applications.31 Unfortunately, the acquisition of high definition detailed images of small unilamellar or oligolamellar liposomes appears limited by the native resolution of this technology, which cannot resolve structures sized under 200 nm. The recent development in microscopic instrumentation as the introduction of multiphoton and stimulated emission depletion microscopy (STED) with a lateral resolution less than 50 nm could solve the problem.
Figure 4.
Confocal images illustrating the architecture of liposomes (a and b). Rhodamine 123 was localized into the bilayer structures. c) Three-dimensional projection of liposomes identifying lamellae of multilamellar liposomes.
Conclusion
In summary, microscopic studies improve the characterization of nanoscale structures of liposomes and provide information about shape and morphology (AFM, TEM), dimensions (AFM, ESEM, TEM, and CLSM), surface properties (AFM), and internal structure (CLSM). More critical aspects regarding the sample preparation and the observation should be considered and carefully evaluated. A great potential for completing the physico-chemical characterization of liposomes by using CLSM is to exploit the rhodamine labeling proposed in this study. In fact, by using this method, one important advantage is that the preparation of sample is easy to operate avoiding any possible sample alterative process and obtaining the detailed evaluation of the liposomal architecture.
Acknowledgments
The authors thank Dott. Massimo Tonelli (Centro Interdipartimentale Grandi Strumenti, University of Modena and Reggio Emilia) for the technical supports on AFM and ESEM measurements. The authors report no conflicts of interest in this work.
References
- 1.Juliano RL, Layton D. Liposomes as a drug delivery system. In: Juliano RL, editor. Drug Delivery Systems: Characteristics and biomedical applications. New York, NY: Oxford University Press; 1980. pp. 189–236. [Google Scholar]
- 2.El Aneed A. An overview of current delivery systems in cancer gene therapy. J Control Rel. 2004;94:1–14. doi: 10.1016/j.jconrel.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 3.Torchilin VP. Multifunctional nanocarriers. Adv Drug Deliv Rev. 2006;58:1532–1555. doi: 10.1016/j.addr.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Edwards KA, Baeumner AJ. Analysis of liposomes. Talanta. 2006;68:1432–1441. doi: 10.1016/j.talanta.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 5.Egelhaaf SU, Wehrli E, Müller M, Adrian M, Schurtenberger P. Determination of the size distribution of lecithin liposomes: a comparative study using freeze fracture, cryoelectron microscopy and dynamic light scattering. J Microsc. 1996;184:214–228. [Google Scholar]
- 6.Gabrijelčicč V, Šentjurc M, Schara M. The measurement of liposome entrapped molecules’ penetration into the skin: A 1D-EPR and EPR kinetic imaging study. Int J Pharm. 1994;102:151–158. [Google Scholar]
- 7.Demetzos C. Differential Scanning Calorimetry (DSC): a tool to study the thermal behavior of lipid bilayers and liposomal stability. J Liposome Res. 2008;18:159–173. doi: 10.1080/08982100802310261. [DOI] [PubMed] [Google Scholar]
- 8.Skalko N, Bouwstra J, Spies F, Stuart M, Frederik PM, Gregoriadis G. Morphological observations on liposomes bearing covalently bound protein: Studies with freeze-fracture and cryo electron microscopy and small angle X-ray scattering techniques. Biochim Biophys Acta. 1998;1370:151–160. doi: 10.1016/s0005-2736(97)00256-3. [DOI] [PubMed] [Google Scholar]
- 9.Stark B, Pabst G, Prass R. Long-term stability of sterically stabilized liposomes by freezing and freeze-drying: Effects of cryoprotectants on structure. Eur J Pharm Sci. 2010;41:546–555. doi: 10.1016/j.ejps.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 10.Elias PM, Goeke J, Friend DS, Brown BE. Freeze-fracture identification of sterol-digitoxin complexes in cell and liposome membranes. J Cell Biol. 1978;78:577–596. doi: 10.1083/jcb.78.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guiot P, Baudhuin P, Gotfredsen C. Morphological characterization of liposome suspensions by stereological analysis of freeze-fracture replicas from spray-frozen samples. J Microsc. 1980;120:159–174. doi: 10.1111/j.1365-2818.1980.tb04132.x. [DOI] [PubMed] [Google Scholar]
- 12.Ruozi B, Battini R, Tosi G, Forni F, Vandelli MA. Liposome-oligonucleotides interaction for in vitro uptake by COS I and HaCaT cells. J Drug Target. 2005;13:295–304. doi: 10.1080/10611860500215014. [DOI] [PubMed] [Google Scholar]
- 13.McLean RS, Sauer BB. Tapping-mode AFM studies using phase detection for resolution of nanophases in segmented polyurethanes and other block copolymers. Macromolecules. 1997;30:8314–8317. [Google Scholar]
- 14.Ruozi B, Tosi G, Leo E, Vandelli MA. Application of atomic force microscopy to characterize liposomes as drug and gene carriers. Talanta. 2007;73:12–22. doi: 10.1016/j.talanta.2007.03.031. [DOI] [PubMed] [Google Scholar]
- 15.Albrecht TR, Grütter P, Horne D, Rugard D. Frequency modulation detection using high-Q cantilevers for enhanced force microscope sensitivity. J Appl Phys. 1991;69:668–673. [Google Scholar]
- 16.Zhong Q, Inniss D, Kjoller K, Elings VB. Fractured polymer/silica fiber surface studied by tapping mode atomic force microscopy. Surface Sci Lett. 1993;290:688–692. [Google Scholar]
- 17.Jass J, Tjärnhage T, Puu G. From liposomes to supported, planar bilayer structures on hydrophilic and hydrophobic surfaces: An atomic force microscopy study. Biophys J. 2000;79:3153–3163. doi: 10.1016/S0006-3495(00)76549-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruozi B, Tosi G, Forni F, Fresta M, Vandelli MA. Atomic force microscopy and photon correlation spectroscopy: two techniques for rapid characterization of liposomes. Eur J Pharm Sci. 2005;25:81–89. doi: 10.1016/j.ejps.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 19.Liang X, Mao G, Ng KYS. Probing small unilamellar EggPC vesicles on mica surface by atomic force microscopy. Colloids Surf B Biointerfaces. 2004;34:42–51. doi: 10.1016/j.colsurfb.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 20.Colas JC, Shi W, Rao VS, Omri A, Mozafari MR, Singh H. Microscopical investigations of nisin-loaded nanoliposomes prepared by Mozafari method and their bacterial targeting. Micron. 2007;38(8):841–847. doi: 10.1016/j.micron.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 21.Koop-Marsaudon S, Leclère Ph, Dubourg F, Lazzaroni R, Aimé JP. Quantitative measurement of the mechanical contribution to tapping-mode atomic force microscopy images of soft materials. Langmuir. 2000;16:8432–8437. [Google Scholar]
- 22.Ruozi B, Tosi G, Costantino L, et al. AFM phase imaging of soft-hydrated samples: a versatile tool to complete the chemical-physical study of liposomes. J Liposome Res. 2009;19(1):59–67. doi: 10.1080/08982100802584071. [DOI] [PubMed] [Google Scholar]
- 23.Dragnevski KD, Donald AM. Applications of environmental scanning electron microscopy (ESEM) in the study of novel drying latex films. J Phys Conf Ser. 2008;126:012077. doi: 10.1088/1742-6596/126/1/012077. [DOI] [Google Scholar]
- 24.Mohammed AR, Weston N, Coombes AGA, Fitzgerald M, Perrie Y. Liposome formulation of poorly water soluble drugs: optimisation of drug loading and ESEM analysis of stability. Int J Pharm. 2004;285:23–34. doi: 10.1016/j.ijpharm.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 25.Robenek H, Severs NJ. Recent advances in freeze-fracture electron microscopy: the replica immunolabeling technique. Biol Proced Online. 2008;10:9–19. doi: 10.1251/bpo138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frederik PM, Stuart MCA, Bomans PHH, Lasic DD. Cryo-electron microscopy of liposomes. In: Lasic DD, Barenholz Y, editors. Nonmedical Applications of Liposomes. Boca Raton, FL: CRC Press; 1996. pp. 309–322. [Google Scholar]
- 27.Harris JR. A comparative negative staining study of aqueous suspensions of sphingomyelin. Micron Microsc Acta. 1986;17:175–200. [Google Scholar]
- 28.Pygall SR, Whetstone J, Timmins P, Melia CD. Pharmaceutical applications of confocal laser scanning microscopy: the physical characterization of pharmaceutical systems. Adv Drug Deliv Rev. 2007;59:1434–1452. doi: 10.1016/j.addr.2007.06.018. [DOI] [PubMed] [Google Scholar]
- 29.Haugland RP. Handbook of Fluorescent Probes and Research Products. 9th ed. Carlsbad, CA: Molecular Probes; 2000. [Google Scholar]
- 30.Jousma H, Talsma H, Spies F, Joosten JGH, Junginger HE, Crommelin DJA. Characterization of liposomes. The influence of extrusion of multilamellar vesicles through polycarbonate membranes on particle size, particle size distribution and number of bilayers. Int J Pharm. 1987;35:263–274. [Google Scholar]
- 31.Kučerka N, Nieh MP, Pencer J, Harroun T, Katsaras J. The study of liposomes, lamellae and membranes using neutrons and X-rays. Curr Opin Colloid Interface Sci. 2007;12:17–22. [Google Scholar]