Abstract
The rapidly accumulating genome sequence data allow researchers to address fundamental biological questions that were not even asked just a few years ago. A major problem in genomics is the widening gap between the rapid progress in genome sequencing and the comparatively slow progress in the functional characterization of sequenced genomes. Here we discuss two key questions of genome biology: whether we need more genomes, and how deep is our understanding of biology based on genomic analysis. We argue that overly specific annotations of gene functions are often less useful than the more generic, but also more robust, functional assignments based on protein family classification. We also discuss problems in understanding the functions of the remaining “conserved hypothetical” genes.
Introduction
The year 2010 marks the 15th anniversary of the publication of the 1,830,138-base genome of the bacterium Haemophilus influenzae Rd Kw20 - the first cellular life form to have its entire genome sequenced [1]. Aided by the tremendous progress in sequencing technology, genome sequencing is advancing at an ever-increasing pace. By the end of 2009, 1052 genomes representing 720 individual species (636 bacteria, 61 archaea, and 23 eukaryotes) were completely sequenced, deposited in the public nucleotide sequence databases (GenBank\EMBL\DDBJ) and made freely available over the internet. Many more genomes were at various stages of sequencing and assembly, including almost 100 eukaryotic genomes whose preliminary descriptions have been published [2]. Thanks to the advent of the new generation of sequencing technologies, the costs of genome sequencing have dropped so much that the projects to sequence the entire human microbiome (http://nihroadmap.nih.gov/hmp/, [3]) and to generate ~5,000 reference genomes for every major prokaryotic lineage (the Genomic Encyclopedia of Bacteria and Archaea: http://www.jgi.doe.gov/programs/GEBA/, [4]) have become realistic. Given these remarkable advances, it seems timely to address two lingering questions: ‘How many more genomes do we need?’ and ‘How deep is our understanding of biology derived from genome analysis?’
Don’t we already have enough genomes?
An interesting, perhaps provocative question is whether a sufficient number of genomes have already been sequenced. Most biologists subscribe to “the more the merrier“ view [4], but others have argued that microbial genomics has already reached the stage of diminishing returns, such that each new genome yields information of progressively decreasing utility [5, 6]. There seems to be some substance to this claim; for example, it is unlikely that we ever see a single bacterial chromosome that is much longer than 13,033,779 nucleotides (as in the myxobacterium Sorangium cellulosum). On the other end of the spectrum, intracellular cycada symbiont Candidatus Hodgkinia cicadicola, with its 143,795-bp genome, could be considered a cellular organelle rather than an independent organism or, at best, a bacterium far on its way to become an organelle [7], so there is hardly any room for further genome reduction of cellular life forms. With respect to other common parameters, such as G+C content, the number of encoded proteins, and metabolic and signaling complexity [8], the extremes might already have been reached, or will be in the near future. Perhaps more importantly, the set of highly conserved genes (that is, those represented in the majority of genomes) is clearly approaching saturation [9]. Similarly, in structural genomics projects, the chances of discovering a new protein fold or even a new superfamily are progressively dropping.
Nevertheless, genome sequencing is here to stay, and there are several compelling reasons for that. First of all, the value of the sequence information is in the eye of the beholder. Many biologists still passionately argue for sequencing their own favorite organism, strain or isolate, no matter how many close relatives already have been sequenced. Indeed, not having a genome sequence for an experimental model is increasingly - and for good reasons - perceived as being stuck in the "dark ages". The availability of the genome sequence allows researchers to easily clone and express any gene, create microarrays to analyze gene expression, and reconstruct the metabolic and signaling networks. Having genomic sequences from closely related organisms opens the door to the quantitative study of mutational patterns, selective regimes, adaptations to ecological factors and, in the case of microbial pathogens, virulence determinants. Potentially even more important is the possibility to identify genes and traits that are not present in the given genome - a task that clearly requires a complete genome sequence.
Secondly, the available genome collection, despite its rapid expansion, still barely scratches the surface of the real biological diversity. The availability of genomic data already led to a revolution in systematics, especially with regard to bacteria and archaea, having put this field on a solid evolutionary footing and giving rise to the new discipline of phylogenomics [10, 11]. Still, judging from the metagenomic data, as many as 90% of the microbial species on Earth remain uncultivated [12, 13], which complicates reconstruction of the global carbon and nitrogen cycles. Genome analysis has already led to several important advances in these areas. Thus, the genome of the marine α-proteobacterium SAR11 (now renamed Candidatus Pelagibacter ubique), apparently the most abundant organism on this planet, opened our eyes to a peculiar role of bacteriorhodopsin-mediated photosynthesis as an auxiliary energy source in the extremely streamlined metabolism of this bacterium [14]. The genome sequence of the deep-sea proteobacterium Idiomarina loihiensis revealed mostly proteolytic, in contrast to the expected saccharolytic, metabolism [15], indicating that the marine habitat of this bacterium contains enough dissolved protein to support a peptide-based diet. The genomes of recently discovered anammox bacteria have yielded valuable insights into the evolution of the global nitrogen cycle and the biochemical reactions that convert nitrate and nitrite into nitrogen gas [16]. This list of unexpected discoveries with biogeochemical implications could be easily extended.
Thirdly, hidden sampling biases in genome sequencing are becoming apparent. For example, starting with Mycoplasma genitalium in 1995, more than 20 mollicute genomes have been sequenced, none of which encoded a single environmental sensor [17]. However, the perception that mollicutes have no signal transduction systems was shattered upon the completion of the (slightly larger) genome of the soil mollicute Acholeplasma laidlawii, which encodes two sensory histidine kinases, three response regulators, an adenylate cyclase, and at least 15 proteins involved in c-di-GMP-mediated regulation (http://www.ncbi.nlm.nih.gov/Complete_Genomes/SignalCensus.html).
Fourthly, although obtaining complete genome sequences from every major lineage [4] would certainly be a dramatic step forward, a single representative genome is by no means sufficient to assess the true biological diversity of a taxon. As a case in point, the sequencing of several genomes from the cyanobacterium Prochlorococcus marinus - a widespread inhabitant of ocean surface waters - was originally aimed at establishing the principal differences between “high-light” and “low-light” ecotypes [18]. However, different strains of P. marinus proved to have vastly different gene repertoires, indicative of high rates of gene acquisition and loss by these organisms. These findings have shown that: (i) the core set of genes shared by all P. marinus isolates is very limited – and shrinking; and (ii) the P. marinus pan-genome, that is the sum total of genes represented in at least one P. marinus strain, is extremely large – and expanding [19]. This crucial yet unexpected development puts into question the very rationale for assigning organisms with dramatically different genome contents – but (nearly) identical 16S rRNA sequences – to the same “species” (such as P. marinus or Escherichia coli) and puts the study of pan-genomes to the forefront of genomic research.
Finally, there remains the crucial issue of using genome sequencing to improve human health. For obvious reasons, the first sequenced genomes were mostly those of common bacterial pathogens. Then the human genome and representative genomes from popular model organisms emerged. As sequencing costs continue to decrease, the use of genomic data for fighting disease becomes more and more attractive. For many bacterial pathogens, multiple strains have been sequenced, often providing clues to the virulence factors, host specificity and drug resistance. Some biologists advocate developing a system of constant genome-based monitoring of various points on the globe, hoping to catch new emerging pathogens before they cause a new epidemic. Such an effort is already well underway for influenza viruses [20, 21]. The human cancer genome projects aims at sequencing thousands of tissue samples from various tumors, in hopes of delineating the whole spectrum of mutations that could contribute to cancer [22]. Although this approach has been criticized [6], the perspective of obtaining the full list of potentially oncogenic mutations – thereby achieving a “complete understanding” of the causes of cancer – is certainly too attractive to pass.
What part of the genome do you not understand?
With sequenced genomes being released almost every day, how well do we understand the functions of the genes in each new one? The answer to this question will depend on the exact meaning of the word “understanding” (as well as “function”). Modern dictionaries associate “understanding” with such terms as “appreciation”, “comprehension”, “explanation”, “insight”, “interpretation”, “knowledge”, and “mastery”. Accordingly, understanding a genome starts from the “knowledge” of the nucleotide sequence and the sequences of encoded proteins and RNAs, and includes “interpretation” of their functions, “insight” into their complex interactions, and “explanation” of the evolutionary history that shaped each particular genome. This leads to the “comprehension” of the potential activity of each component of the cell, which must be tempered by the “appreciation” that proteins often have additional (e.g. moonlighting [23, 24]) function. Finally, this understanding can be extended into “mastery” – the ability to modify the genome for certain (e.g. biotechnological) applications. Therefore, the problem of understanding the genome can be rephrased as follows: how good is the “parts list” that is compiled for each genome in the form of functional annotation of the predicted protein-coding and RNA-coding genes?
Obviously, this list is never complete. Almost 10 years ago, Peer Bork described the “70% hurdle”: on average, for approximately one-third of the genes in any given genome, the functions could not be predicted through traditional methods of genome analysis; perhaps even worse, the accuracy of functional prediction was only ~70% for the remaining genes [25]. Bork warned that hopes to cross this 70% barrier and achieve a better understanding of the functional content of genomes with the help of high-throughput analytical methods would be tempered by the fact that these methods themselves have high error rates and are most effective when used in concert [25]. Looking back, Bork’s sobering prediction was right on target. High-throughput analyses of gene and protein expression, protein-protein interactions, and ligand binding led to a dramatic increase in the amount of data pertaining to any given gene in model genomes [26]. However, as illustrated in Box 1, accumulation of such data does not necessarily translate into clarity regarding gene function, at least not immediately, and not without much work.
Box 1. The long and circuitous path from experiment to “understanding”.
Many “conserved hypothetical” genes remain without an assigned function simply because they have never been studied experimentally. In other cases, experimental studies brought contradictory results that could not be easily reconciled. To illustrate the difficulty of, in the words of Sydney Brenner, "converting data into knowledge and knowledge into understanding", let us consider the history of functional characterization of three widespread genes.
YgjD/Gcp/Kae1/QRI1/OSGEPL family
The E. coli ygjD (gcp) gene has orthologs in almost every bacterial, archaeal and eukaryotic genome. In many eukaryotes it is found in two paralogous copies, such as QRI7 and Kae1 in yeast, At4g22720 and At2g45270 in Arabidopsis thaliana, and OSGEPL and OSGEPL1 in human. In addition, there is a family of more distant bacterial paralogs, represented by E. coli YeaZ and B. subtilis YdiC. We have previously discussed the potential functions of this protein family (which contains an actin/HSP70 superfamily ATPase domain), and expressed doubts about its annotation as "O-sialoglycoprotease", which was based on a single experimental observation, and further suggested an association of this protein with translation (e.g. co-translational degradation of misfolded proteins) [39]. In the past several years, proteins of this family have been studied in several model organisms, and the crystal structures of several family members have been solved [46, 59]. An archaeal YgjD family member showed no protease activity, but has been reported to bind DNA and possess an apurinic endonuclease activity [62]. In yeast, Kae1 is a subunit of the KEOPS complex which regulates transcription, telomere uncapping and telomere length, and is required for cell growth; this protein is targeted to mitochondria and appears to be essential for genome maintenance [46]. Despite all these observations, the actual function of the YgjD family proteins remains enigmatic [46, 60]. A recent study suggested their involvement in biosynthesis of threonylcarbamoyladenosine (t6A), a universal tRNA base modification occurring at position 37 in a subset of tRNAs decoding the ANN codons [63]. If so, translational defects resulting from impaired t6A biosynthesis could explain at least some properties of the ygjD mutants.
YebC/YrbC/DUF28/UPF0082 family
The E. coli yebC is another widespread gene with orthologs in most bacterial and eukaryotic genomes. In some organisms, there are two paralogs, such as yebC and yeeN in E. coli, or yeeI and yrbC in B. subtilis. Products of these genes are listed as “domain of unknown function” (DUF28, PF01709) in the Pfam database [29] and as uncharacterized protein family UPF0082 in UniProt [40]. Crystal structures of three members of this family have been solved [64], but those structures have provided no clear indication of protein function. Analysis of the genome neighborhoods of the yebC-like genes has revealed potential association with the Holliday junction resolvase RuvABC and suggested that YebC might be involved in DNA recombination and repair in bacteria and mitochondria [39]. Indeed, a recent study demonstrated DNA binding by a Pseudomonas aeruginosa YebC family protein, and suggested a role in transcription regulation [65]. However, another study [48] mapped the cytochrome c oxidase deficiency in humans (late-onset Leigh syndrome) to a mutation in the human YebC ortholog CCDC44 (renamed TACO1) and concluded that this protein was required for the proper translation of the mitochondrial COX1 protein. Three possible mechanisms of YebC action to ensure translation of the full-length COX1 polypeptide were considered: (i) securing an accurate start of translation, (ii) stabilizing the elongating polypeptide, and (iii) interacting with the peptide release factor [48]. While involvement in translation appears very likely for such a widespread protein family, its apparent capacity to bind DNA remains to be confirmed and/or explained.
YjgF/YabJ/YER057c/UK114 family
The E. coli yjgF gene has highly conserved homologs in bacteria, archaea and eukaryotes, often with multiple paralogs in the same genome. Representatives of the YjgF protein family are known as "purine regulatory protein YabJ" in B. subtilis and as "tumour-associated antigen UK114" in human and other mammals. Members of this family have been reported to possess ribonuclease activity, to function as a molecular chaperone, calpain activator, transcriptional regulator, and translational inhibitor, and also to affect photosynthesis, isoleucine biosynthesis and mitochondrial genome maintenance (reviewed in [66, 67]). Crystal structures of bacterial, archaeal and eukaryotic members of this family have been solved, revealing an inter-subunit cleft that is capable of binding a variety of small molecule ligands [58]. Despite all these efforts, the cellular functions of the members of the YjgF family remain unclear.
Another important issue here is the definition of “function”, particularly as it applies to (semi)-automated genome annotations. For a limited set of essential genes, the notion of function seems quite straightforward: the function is what the gene product needs to do to allow cell growth. Operationally, if a gene is knocked-out, the cell dies, and the cause of death can be assumed to be the function of the gene in question. For non-essential genes the picture is more complicated as the functions of many, if not most, proteins are inherently multifaceted and complicated. For example, a single oxidoreductase would use a range of substrates and a variety of electron acceptors, making a precise functional assignment difficult, if not impossible. Should the function be assigned on the basis of the substrate with the lowest KM or the highest Vmax, or the one that is most likely to be physiologically relevant? This problem becomes particularly severe for high-throughput enzyme assays, which helped assign general biochemical functions to products of previously uncharacterized genes, but were often unable to pinpoint the natural substrates for the respective enzymes [27, 28]. Furthermore, many proteins, particularly in eukaryotes, lack any (known) enzymatic activity and appear to function exclusively in protein-protein interactions. It could be argued that the “understanding” of a protein function should, at the very least, include knowledge of (i) biochemical activity, if any (i.e. the nature of the catalyzed reaction and the range of utilized substrates and products); (ii) the biological process (pathway, stress response, cell cycle) for which this activity is (most) important; and (iii) the evolutionary aspects, such as phyletic distribution, level of sequence conservation, and frequency of mutation, gene loss and/or non-orthologous gene displacement.
Owing to the paucity of experimental data, this information is rarely available in its entirety, and functional assignments for the majority of the genes are based solely on the sequence similarity of their products to experimentally characterized proteins in a handful of model organisms such as E. coli, Bacillus subtilis, yeast, Dictyostelium, Drosophila, Caenorhabditis elegans, zebrafish, or mouse. Automatic transfer of functional annotation often leads to confusion when, for example, the product of a widespread prokaryotic gene (ytaB in B. subtilis) is often annotated as “mitochondrial benzodiazepine receptor” (it is hard to imagine why B. subtilis and hundreds of other bacteria and archaea would need a receptor for Valium, even apart from the fact that they have no mitochondria). Alternative annotations for the products of this gene family in various organisms include “tryptophan-rich sensory protein TspO”, “carotenoid biosynthetic protein CrtK” and “18 kD translocator protein”. However, despite these discordant annotations, there is little doubt that all members of the TspO/MBR protein family (Pfam family PF03073 [29]) are very similar and perform closely related – and important – functions, which remain to be uncovered [30]. In our opinion, a more productive route towards sensible functional annotation is to replace annotation of individual proteins (particularly, those from poorly studied organisms) with annotation of protein families. In essence, this approach substitutes protein classification (something that we generally know how to do) for specific protein annotation, which except possibly for a handful of obvious cases, will remain questionable until each protein is experimentally characterized, even when predictions appear entirely plausible and supported by high similarity to experimentally characterized homologs and/or operon structure. As experimental assays increasingly lag behind the avalanche of genomic data, such experimental validation of predicted protein function becomes progressively less likely. By contrast, protein family classification is becoming increasingly robust. As an example, recognition of a Fis-type or a winged helix-turn-helix domain allows the recognition of a protein as a DNA-binding transcriptional regulator although the regulated genes (operons) may be difficult to predict [31]. Likewise, numerous membrane proteins are reliably recognized as transporters, whereas the range of their substrates often remains uncertain. The notable success of protein family databases, such as Pfam, InterPro, COGs and CDD [29, 32–34], is probably due not only to the fact that these were – and still are – comprehensive collections with many useful features. It could be argued that another key to their success lies in the abandonment of the elusive goal of annotating every single sequence and instead concentrating on the common traits of protein families. In doing so, these databases provided a reasonably robust common framework for annotating the entire protein sets encoded in newly sequenced genomes. Thus, annotation pipelines used at most genome sequencing projects now include comparison against at least one of the available protein family databases.
It is important to note that family assignment is only the first step towards understanding, which, as discussed above, requires knowledge of both the biochemical activity of the protein and the cellular process in which the protein is involved. As an example, the sequence-based prediction that the conserved bacterial protein Era is a GTPase was a good first step in its characterization, and recognition of its involvement in translation was another step forward. However, “true understanding” of the role of this GTPase in the translation process – and its proper functional annotation – came only after an experimental study that revealed the participation of Era in processing and maturation of 16S rRNA [35].
“Conserved hypothetical” and “putative uncharacterized” genes: when and how will their functions become known?
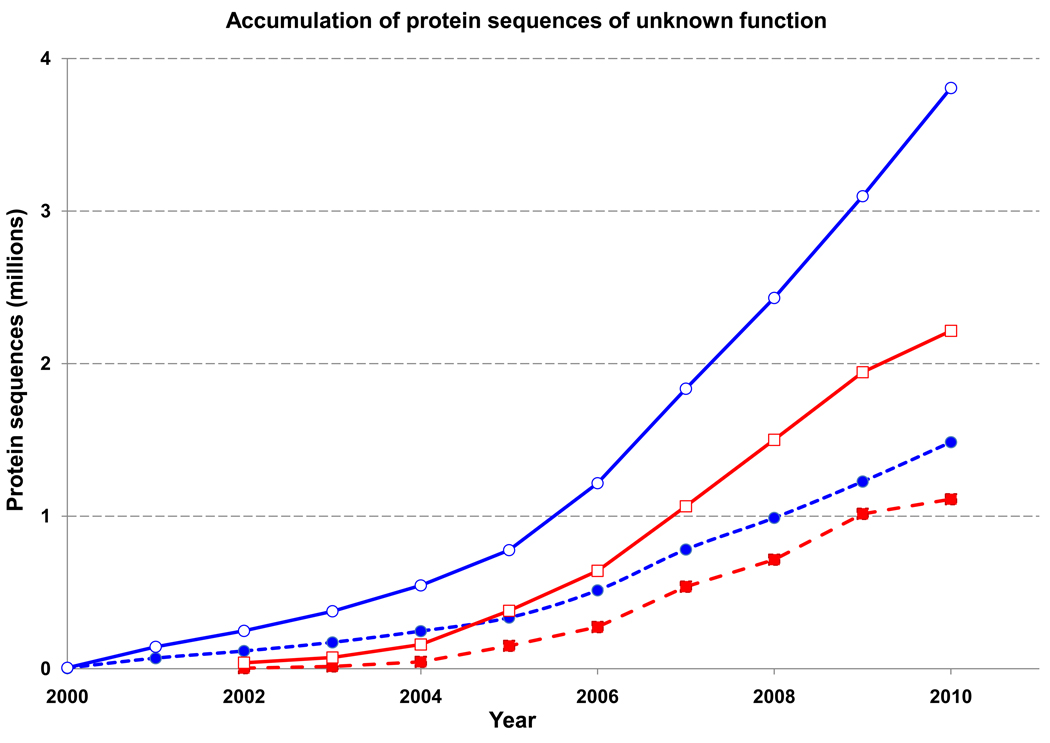
Even in the relatively well-studied model organisms, the great majority of genes have never been experimentally characterized. E. coli K-12 and yeast Saccharomyces cerevisiae appear to be the only organisms for which at least 50% of the genes have been studied experimentally [36–38]. Despite the best efforts of experimental and computational biologists, a substantial – and constantly growing, given the acceleration of genome sequencing – number of deduced proteins have no known function (Figure 1). This is hardly surprising in case of lineage-specific genes that are found, for example, only in Vibrio or Burkholderia - bacterial lineages that are extensively sampled by genome sequencing, but do not include well-characterized model organisms. However, some genes that are widespread among bacteria, archaea and/or eukaryotes still remain without functional annotation [39]. The protein products of these genes have been variously referred to as “hypothetical”, “conserved hypothetical”, “uncharacterized” or even “putative uncharacterized” (as of May 1, 2010, 3,118,564 proteins in UniProt were annotated this way [40]). Several lists of “conserved hypothetical” proteins have been compiled, including Domains of Unknown Function (DUFs) in Pfam, R- and S-COGs in the COG database, and Uncharacterized Protein Families (UPFs) in UniProtKB\Swiss-Prot [29, 33, 40]. These lists have been extensively used to guide structural genomics efforts, which resulted in structural (albeit usually not functional) characterization of many such proteins [41, 42].
Figure 1.
Accumulation of protein sequences of unknown function in the genome databases. Open symbols indicate the total number of protein sequences encoded in prokaryotic (blue) and eukaryotic (red) genomes; filled symbols indicate the number of “hypothetical” or “uncharacterized” proteins. The data are taken from the NCBI’s RefSeq database [68]; the numbers for 2010 are extrapolated from the first 4 months.
To highlight the distinction between the “hypothetical” genes whose functions remained completely unknown and those that could be assigned a general biochemical function (e.g. a methyltransferase, an oxidoreductase, a transcriptional regulator or a membrane transporter), we denoted the former category of genes “unknown unknown” and the latter category “known unknown” [39]. The “known unknown” category includes also genes of unknown biochemical function that have (partially) known cellular function, such as a “cell division protein” or a “stress response protein”. In purely operational terms, there are more or less clear ways of establishing function for “known unknown” genes, but not for “unknown unknowns”.
Six years ago we analyzed widely conserved “hypothetical” genes and compiled the “top 10” lists of “known unknown” and “unknown unknown” genes [39]. A re-examination of these lists shows that, despite mounting observations, nearly half of those genes still remain without an assigned function (Tables 1 and 2). Some of the genes in the two lists have been experimentally characterized, and in a few cases the function has been established [43]. In eukaryotes, products of some, albeit not all, of these widely conserved genes appear to be targeted to mitochondria [44–48]. In two instances, mutations in these genes were linked to mitochondrial diseases, such as hereditary paraganglioma [44] and the late-onset Leigh syndrome [48]. In other cases, however, experimental results were contradictory (Box 1). Apparently, the problem was not in the lack of effort to characterize these genes, but in the pleiotropic phenotypes of their mutations, which made it difficult to pinpoint the primary function.
Table 1.
Updated “top 10” list of widespread “known unknown” genes
| Gene name | Protein family | PDB entry |
Initial predictions (2004) | Updated functional annotation, reference | |||
|---|---|---|---|---|---|---|---|
| E. coli | Yeast | Human | Pfam | COG | |||
| ygjD | QRI7 | OSGEP | PF00814 | 0533 | 2VWB | Putative metal- and ATP-dependent protease. Fused to a Ser/Thr protein kinase domain in some archaea. Gene neighborhoods suggest association with translation | DNA binding protein with apurinic endonuclease activity [62]; threonylcarbamoyladenosine biosynthesis in tRNA [63] |
| ychF | YBR025c | PTD004 | PF06071 | 0012 | 1JAL | Predicted GTPase; binds double-stranded RNA; coexpressed with peptidyl-tRNA hydrolase; predicted to be a translation factor | An ATPase in the GTPase family [69] |
| yrdC | SUA5 | YRDC | PF01300 PF03481 | 0009 | 1HRU | Double-stranded RNA binding protein, predicted translation initiation factor; induced by ischemia in humans | Ribosome maturation factor RimN [70]; threonylcarbamoyladenosine biosynthesis in tRNA [43] |
| ybeM | NIT2 | NIT1 | PF00795 | 0388 | 1EMS | A member of nitrilase superfamily, predicted amidase. Some members might function as glutaminase subunits of NAD synthetase. In worm and fly fused to Fhit domain (diadenosine triphosphatase), a potential tumor suppressor | Omega-amidodicarboxylate amidohydrolase activity [71] |
| yihA | YDR336w | HSPC135 | PF01926 | 0218 | 1PUI | A GTP-binding protein required for E. coli cell division; co-occurrence with the gene for ClpP protease in several genomes suggests involvement in regulated (perhaps co-translational) protein degradation | Crucial for ribosome assembly or stability in Staphylococcus aureus [72] |
| yigB | YMR130c | C9orf158 | PF00702 | 1011 | 1NRW | A phosphatase of haloacid dehalogenase (HAD) superfamily; adjacency to the XerC DNA recombinase gene suggests a role in DNA recombination and/or repair | Flavin mononucleotide (FMN) phosphatase activity [28] |
| yfhR | YNL320w | BEM46 | PF00561 | 1073 | 2WTM | Predicted enzyme of the alpha/beta hydrolase fold, most likely an esterase; possible role in yeast budding | Important for cell polarity [73] |
| yfcE | VPS29 | PEP11 | PF00149 | 0622 | 1SU1 | A phosphoesterase of the calcineurin-like superfamily; vacuolar sorting protein in yeast. Gene neighborhood is compatible with a role in RNA metabolism | A phosphodiesterase with variable activity against 2',3'-cAMP [74, 75] |
| - | NUG1 | GNL3 | PF01926 | 1161 | 1PUJ | Predicted GTPase; genome context suggests possible involvement in translation. In yeast, required for nuclear export of 60S pre-ribosomal particles. In humans, nucleolar protein, important for cell proliferation | No news [76] |
| yhcM | AFG1 | LACE1 | PF03969 | 1485 | n/a | Predicted ATPase, in eukaryotes localized to the mitochondria | Promotes degradation of cytochrome c oxidase mitochondrially encoded subunits [45] |
Modified from Table 2 from Ref [39] with permission from Oxford University Press. Additional information on the listed gene products is available from the respective online resources: for Pfam [29], in the http://pfam.sanger.ac.uk/family?PF00814 format; for COGs [33], in the http://www.ncbi.nlm.nih.gov/COG/grace/wiew.cgi?COG0533 format; for Protein DataBank (PDB), in the http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=2VWB format.
Abbreviations: n/a, not available; COG, Clusters of Orthologous Groups of proteins database.
Table 2.
Updated “top 10” list of widespread “unknown unknown” genes
| Gene name | Protein family | PDB entry |
Initial predictions (2004) | Updated annotation | |||
|---|---|---|---|---|---|---|---|
| E. coli | Yeast | Human | Pfam | COG | |||
| yebC | YGR021w | PRO0477 | PF01709 | 0217 | 1KON | Often encoded in the same operon with Holliday junction resolvasome (RuvABC) subunits. However, also found in eukaryotes (mitochondrial protein) whose resolvases are unrelated to RuvABC. Potential role in DNA repair and/or recombination | DNA-binding transcriptional regulator [65]; translational activator of COX I; mutation causes cytochrome c oxidase deficiency and late-onset Leigh syndrome [48] |
| ybgI | NIF3 | NIF3L1 | PF01784 | 0327 | 1NMO | In yeast, interacts with transcriptional coactivator NGG1p. Could be a transcriptional regulator | Mitochondrial localization |
| ybeB | - | C7orf30 | PF02410 | 0799 | 2ID1 | Homologs of plant protein Iojap, required for normal function of chloroplast ribosomes. In most bacteria, adjacent to the gene for nicotinic acid mononucleotide adenylyltransferase, suggesting a role in NAD metabolism and/or bacterial cell division | Co-migrates with the 50S subunit [77]; NAD-dependent nucleic acid AMP ligase, releases NMN from NAD (V. de Crecy-Lagard, pers. commun). |
| yjeF | YNL200c | AIBP | PF03853 | 0062 | 1JZT | In many prokaryotes, fused to a sugar kinase domain. In plants, fused to a pyridoxamine 5-phosphate oxidase-like domain. In humans, is secreted by kidney cells; binds apolipoprotein A-I. Domain fusions suggest a role in RNA processing | Mitochondrial localization, role in spermiogenesis and oogenesis [47] |
| yigZ | YDL177c | IMPACT | PF01205 | 1739 | 1VI7 | Imprinted gene in mouse, but not in human, a candidate gene for bipolar affective disorder. In fungi, fused with UDP-glucose 4-epimerase, suggesting that it could be an enzyme of sugar metabolism | Binds to the translational activator GCN1, inhibits protein kinase GCN2 [78] |
| rtcB | - | HSPC117 | PF01139 | 1690 | 1UC2 | In several bacteria, encoded in the same operon with RNA 3'-terminal phosphate cyclase RtcA, suggesting that RtcB could be an RNA modification enzyme | Binds vinculin, potential role in cell adhesion [79] |
| ydjX | YKR088c | TMEM64 | PF00597 | 0398 | n/a | Predicted membrane protein, may be involved in utilization of 4-hydroxybutyrate; moderate growth defect in yeast mutants | No news |
| ygfY | SDH5 | PGL2 | PF03937 | 2938 | 1X6I | In yeast, required for sporulation and for growth on respiratory substrates; possible transcriptional regulator | Required for insertion of flavin into the succinate dehydrogenase complex, mutation leads to paraganglioma [44] |
| - | YOR289w | AMMECR1 | PF01871 | 2078 | n/a | In humans, absent in the Alport syndrome, mental retardation, midface hypoplasia, and elliptocytosis | Predicted RNA-modifying enzyme [80] |
| ydiU | YPL222w | SELO | PF02696 | 0397 | n/a | Selenoprotein O, in yeast localized to mitochondria; not found in archaea | No news |
Modified from Table 3 from Ref [39] with permission from Oxford University Press. Additional information on the listed gene products is available from the respective online resources: for Pfam [29], in the http://pfam.sanger.ac.uk/family?PF00814 format; for COGs [33], in the http://www.ncbi.nlm.nih.gov/COG/grace/wiew.cgi?COG0533 format; for Protein DataBank (PDB), in the http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=2VWB format.
What could be the functions of all those “hypothetical” genes?
Given that universally conserved genes are typically involved in translation, transcription or ribosome biogenesis [9], widespread genes are likely to function in these or related processes as well. Indeed, several recently characterized “conserved hypothetical” genes are involved in post-transcriptional modification of tRNA [43, 49]. Besides, a significant number of “orphan” enzyme activities have not been associated with any protein sequence [50], suggesting that some “hypothetical” genes might have well-known functions. Characterization of the “missing” genes in various metabolic pathways allowed assigning functions to a number of formerly “conserved hypothetical” genes [51, 52]}.
Less common “hypothetical” genes are far more abundant in the genomes of free-living organisms than in the relatively streamlined genomes of parasites, symbionts and saprophytes [53]. Based on the observation that the fraction of metabolic and particularly regulatory genes increases with the genome size [17, 54, 55], sophisticated regulation of gene expression and complex (secondary) metabolism, including various post-transcriptional and post-translational modifications, appear to be plausible roles for a fair number of the remaining uncharacterized genes.
Recent studies have highlighted an additional class of functions that might account for the abundance of uncharacterized genes in free-living organisms, namely, detoxification (usually hydrolysis) of potentially hazardous side-products of various metabolic reactions [56]. These activities, commonly referred to as “house-cleaning”, are particularly important for aerobic organisms that have to cope with spontaneous oxidation of nucleotides, amino acids, lipids, and other cellular components. For example, the recently characterized “conserved hypothetical” gene yebR (renamed msrC) has been shown to encode an enzyme that hydrolyzes methionine-(R)-sulfoxide, a product of methionine oxidation [57]. Other cellular reactions that might require house-cleaning include methylation, acetylation and adenylation, among potentially many others. It is probably no coincidence that many poorly characterized proteins appear to function as hydrolases [27, 28].
Finally, it has to be kept in mind that a considerable fraction of genes in many genomes might not have definable cellular functions, but rather originate from viruses and mobile elements and only transiently pass through microbial genomes. Genomes are highly dynamic entities, and each sequence is a temporal snapshot that is likely to include many short-lived elements that are not maintained by selection. The very notion of annotation for such “selfish” genes is different from that applied to “regular” genes with distinct cellular functions [9].
Concluding remarks
In conclusion, it might be worthwhile to make several basic generalizations regarding genomes and the understanding of gene functions:
Functions of many widespread genes are known; all universal genes are involved in translation [9]
Widespread genes with unknown functions remain uncharacterized for a reason: they often affect multiple processes and their mutations typically are pleiotropic (Box 1)
The functions of a substantial fraction of genes in each sequenced genome remain unknown
Not every experiment on an unknown gene yields useful clues regarding function.
Structural characterization of a protein rarely gives direct clues to its function [41, 42, 58]).
Analysis of gene expression rarely gives direct clues to gene functions
Delineation of a protein interaction network involving the gene of interest rarely gives direct clues to its function [26, 59, 60]
Functional assignments for previously uncharacterized, widely conserved genes are just like any biological discoveries: they require a lot of hard work and a bit of luck
So far there is no single high-throughput approach that would finally reveal the functions of all “hypothetical” genes encoded in the sequenced genomes. This goal may be reachable only through sustained efforts of numerous experimental, computational and structural biologists [61]. At the end of 2009, NIH awarded a grant to the COMputational BRidge to EXperiments (COMBREX, http://www.combrex.org/, formerly SciBay) consortium project that aims to coordinate collaborative efforts of various research groups towards computational identification of the most interesting families of “conserved hypothetical” proteins and their experimental characterization (http://www.nigms.nih.gov/News/Results/gogrant_112309.htm). This project seems to have considerable potential to accelerate functional characterization of the remaining “hypothetical” genes, thereby bringing us closer to the “complete understanding” of genomes – and the organisms themselves.
Acknowledgements
This study was supported by the Intramural Research Program of the National Library of Medicine at the U.S. National Institutes of Health.
References
- 1.Fleischmann RD, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 2.Liolios K, et al. The Genomes On Line Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2010;38:D346–D354. doi: 10.1093/nar/gkp848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ley RE, et al. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu D, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature. 2009;462:1056–1060. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitworth DE. Genomes and knowledge - a questionable relationship? Trends Microbiol. 2008;16:512–519. doi: 10.1016/j.tim.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Kaiser J. A skeptic questions cancer genome projects. ScienceInsider, 23 April 2010. 2010 ( http://news.sciencemag.org/scienceinsider/2010/04/a-skeptic-questions-cancer-genom.html)
- 7.McCutcheon JP, et al. Origin of an alternative genetic code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000565. e1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galperin MY, Kolker E. New metrics for comparative genomics. Curr. Opin. Biotechnol. 2006;17:440–447. doi: 10.1016/j.copbio.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koonin EV, Wolf YI. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 2008;36:6688–6719. doi: 10.1093/nar/gkn668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisen JA, Fraser CM. Phylogenomics: intersection of evolution and genomics. Science. 2003;300:1706–1707. doi: 10.1126/science.1086292. [DOI] [PubMed] [Google Scholar]
- 11.Koonin EV. The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol. 2010;11:209. doi: 10.1186/gb-2010-11-5-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hugenholtz P. Exploring prokaryotic diversity in the genomic era. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-2-reviews0003. REVIEWS0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeLong EF. The microbial ocean from genomes to biomes. Nature. 2009;459:200–206. doi: 10.1038/nature08059. [DOI] [PubMed] [Google Scholar]
- 14.Giovannoni SJ, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- 15.Hou S, et al. Genome sequence of the deep-sea gamma-proteobacterium Idiomarina loihiensis reveals amino acid fermentation as a source of carbon and energy. Proc. Natl. Acad. Sci. USA. 2004;101:18036–18041. doi: 10.1073/pnas.0407638102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klotz MG, Stein LY. Nitrifier genomics and evolution of the nitrogen cycle. FEMS Microbiol. Lett. 2008;278:146–156. doi: 10.1111/j.1574-6968.2007.00970.x. [DOI] [PubMed] [Google Scholar]
- 17.Galperin MY. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 2005;5:35. doi: 10.1186/1471-2180-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rocap G, et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424:1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- 19.Scanlan DJ, et al. Ecological genomics of marine picocyanobacteria. Microbiol. Mol. Biol. Rev. 2009;73:249–299. doi: 10.1128/MMBR.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McHardy AC, Adams B. The role of genomics in tracking the evolution of influenza A virus. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000566. e1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee CW, et al. Large-scale evolutionary surveillance of the 2009 H1N1 influenza A virus using resequencing arrays. Nucleic Acids Res. 2010;38:e111. doi: 10.1093/nar/gkq089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeffery CJ. Moonlighting proteins. Trends Biochem. Sci. 1999;24:8–11. doi: 10.1016/s0968-0004(98)01335-8. [DOI] [PubMed] [Google Scholar]
- 24.Sriram G, et al. Single-gene disorders: what role could moonlighting enzymes play? Am. J. Hum. Genet. 2005;76:911–924. doi: 10.1086/430799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bork P. Powers and pitfalls in sequence analysis: the 70% hurdle. Genome Res. 2000;10:398–400. doi: 10.1101/gr.10.4.398. [DOI] [PubMed] [Google Scholar]
- 26.Jensen LJ, et al. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuznetsova E, et al. Enzyme genomics: Application of general enzymatic screens to discover new enzymes. FEMS Microbiol. Rev. 2005;29:263–279. doi: 10.1016/j.femsre.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 28.Kuznetsova E, et al. Genome-wide analysis of substrate specificities of the Escherichia coli haloacid dehalogenase-like phosphatase family. J. Biol. Chem. 2006;281:36149–36161. doi: 10.1074/jbc.M605449200. [DOI] [PubMed] [Google Scholar]
- 29.Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chapalain A, et al. Bacterial ortholog of mammalian translocator protein (TSPO) with virulence regulating activity. PLoS One. 2009;4:e6096. doi: 10.1371/journal.pone.0006096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galperin MY. Diversity of structure and function of response regulator output domains. Curr. Opin. Microbiol. 2010;13:150–159. doi: 10.1016/j.mib.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hunter S, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tatusov RL, et al. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marchler-Bauer A, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tu C, et al. Structure of ERA in complex with the 3' end of 16S rRNA: implications for ribosome biogenesis. Proc. Natl. Acad. Sci. USA. 2009;106:14843–14848. doi: 10.1073/pnas.0904032106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riley M, et al. Escherichia coli K-12: a cooperatively developed annotation snapshot--2005. Nucleic Acids Res. 2006;34:1–9. doi: 10.1093/nar/gkj405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keseler IM, et al. EcoCyc: a comprehensive view of Escherichia coli biology. Nucleic Acids Res. 2009;37:D464–D470. doi: 10.1093/nar/gkn751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christie KR, et al. Functional annotations for the Saccharomyces cerevisiae genome: the knowns and the known unknowns. Trends Microbiol. 2009;17:286–294. doi: 10.1016/j.tim.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Galperin MY, Koonin EV. 'Conserved hypothetical' proteins: prioritization of targets for experimental study. Nucleic Acids Res. 2004;32:5452–5463. doi: 10.1093/nar/gkh885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.The UniProt Consortium. The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2009;37:D169–D174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rigden DJ. Understanding the cell in terms of structure and function: Insights from structural genomics. Curr. Opin. Biotech. 2006;17:457–464. doi: 10.1016/j.copbio.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 42.Lee D, et al. Predicting protein function from sequence and structure. Nat. Rev. Mol. Cell Biol. 2007;8:995–1005. doi: 10.1038/nrm2281. [DOI] [PubMed] [Google Scholar]
- 43.El Yacoubi B, et al. The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Res. 2009;37:2894–2909. doi: 10.1093/nar/gkp152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hao HX, et al. SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science. 2009;325:1139–1142. doi: 10.1126/science.1175689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khalimonchuk O, et al. Evidence for a pro-oxidant intermediate in the assembly of cytochrome oxidase. J. Biol. Chem. 2007;282:17442–17449. doi: 10.1074/jbc.M702379200. [DOI] [PubMed] [Google Scholar]
- 46.Oberto J, et al. Qri7/OSGEPL, the mitochondrial version of the universal Kae1/YgjD protein, is essential for mitochondrial genome maintenance. Nucleic Acids Res. 2009;37:5343–5352. doi: 10.1093/nar/gkp557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rudolph C, et al. ApoA-I-binding protein (AI-BP) and its homologues hYjeF_N2 and hYjeF_N3 comprise the YjeF_N domain protein family in humans with a role in spermiogenesis and oogenesis. Horm. Metab. Res. 2007;39:322–335. doi: 10.1055/s-2007-977699. [DOI] [PubMed] [Google Scholar]
- 48.Weraarpachai W, et al. Mutation in TACO1, encoding a translational activator of COX I, results in cytochrome c oxidase deficiency and late-onset Leigh syndrome. Nat. Genet. 2009;41:833–837. doi: 10.1038/ng.390. [DOI] [PubMed] [Google Scholar]
- 49.Phillips G, et al. Discovery and characterization of an amidotransferase involved in the modification of archaeal tRNA. J. Biol. Chem. 2010;285:12706–12713. doi: 10.1074/jbc.M110.102236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pouliot Y, Karp PD. A survey of orphan enzyme activities. BMC Bioinformatics. 2007;8:244. doi: 10.1186/1471-2105-8-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Osterman A, Overbeek R. Missing genes in metabolic pathways: a comparative genomics approach. Curr. Opin. Chem. Biol. 2003;7:238–251. doi: 10.1016/s1367-5931(03)00027-9. [DOI] [PubMed] [Google Scholar]
- 52.Hanson AD, et al. 'Unknown' proteins and 'orphan' enzymes: the missing half of the engineering parts list--and how to find it. Biochem. J. 2010;425:1–11. doi: 10.1042/BJ20091328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kolker E, et al. Global profiling of Shewanella oneidensis MR-1: Expression of hypothetical genes and improved functional annotations. Proc. Natl. Acad. Sci. USA. 2005;102:2099–2104. doi: 10.1073/pnas.0409111102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Nimwegen E. Scaling laws in the functional content of genomes. Trends Genet. 2003;19:479–484. doi: 10.1016/S0168-9525(03)00203-8. [DOI] [PubMed] [Google Scholar]
- 55.Konstantinidis KT, Tiedje JM. Trends between gene content and genome size in prokaryotic species with larger genomes. Proc. Natl. Acad. Sci. USA. 2004;101:3160–3165. doi: 10.1073/pnas.0308653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Galperin MY, et al. House cleaning, a part of good housekeeping. Mol. Microbiol. 2006;59:5–19. doi: 10.1111/j.1365-2958.2005.04950.x. [DOI] [PubMed] [Google Scholar]
- 57.Lin Z, et al. Free methionine-(R)-sulfoxide reductase from Escherichia coli reveals a new GAF domain function. Proc. Natl. Acad. Sci. USA. 2007;104:9597–9602. doi: 10.1073/pnas.0703774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burman JD, et al. The crystal structure of Escherichia coli TdcF, a member of the highly conserved YjgF/YER057c/UK114 family. BMC Struct. Biol. 2007;7:30. doi: 10.1186/1472-6807-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Handford JI, et al. Conserved network of proteins essential for bacterial viability. J. Bacteriol. 2009;191:4732–4749. doi: 10.1128/JB.00136-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Msadek T. Grasping at shadows: revealing the elusive nature of essential genes. J. Bacteriol. 2009;191:4701–4704. doi: 10.1128/JB.00572-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roberts RJ, et al. An experimental approach to genome annotation. The American Academy of Microbiology colloquium report. 2004 American Society for Microbiology http://academy.asm.org/images/stories/documents/experimentalapproachgenomeannotationcolor.pdf. [Google Scholar]
- 62.Hecker A, et al. An archaeal orthologue of the universal protein Kae1 is an iron metalloprotein which exhibits atypical DNA-binding properties and apurinic-endonuclease activity in vitro. Nucleic Acids Res. 2007;35:6042–6051. doi: 10.1093/nar/gkm554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.El Yacoubi B, et al. Function of the YrdC/YgjD conserved protein network: the t6A lead. In: Weil T, Santos M, editors. 23rd tRNA workshop: From the origin of life to biomedicine. 2010. p. 7. ( http://bioinformatics.ua.pt/trna2010/book.html) [Google Scholar]
- 64.Shin DH, et al. Crystal structure of conserved hypothetical protein Aq1575 from Aquifex aeolicus. Proc. Natl. Acad. Sci. USA. 2002;99:7980–7985. doi: 10.1073/pnas.132241399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liang H, et al. The YebC family protein PA0964 negatively regulates the Pseudomonas aeruginosa quinolone signal system and pyocyanin production. J. Bacteriol. 2008;190:6217–6227. doi: 10.1128/JB.00428-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Christopherson MR, et al. YjgF is required for isoleucine biosynthesis when Salmonella enterica is grown on pyruvate medium. J. Bacteriol. 2008;190:3057–3062. doi: 10.1128/JB.01700-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thakur KG, et al. Mycobacterium tuberculosis Rv2704 is a member of the YjgF/YER057c/UK114 family. Proteins. 2010;78:773–778. doi: 10.1002/prot.22623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sayers EW, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koller-Eichhorn R, et al. Human OLA1 defines an ATPase subfamily in the Obg family of GTP-binding proteins. J. Biol. Chem. 2007;282:19928–19937. doi: 10.1074/jbc.M700541200. [DOI] [PubMed] [Google Scholar]
- 70.Kaczanowska M, Ryden-Aulin M. The YrdC protein--a putative ribosome maturation factor. Biochim. Biophys. Acta. 2005;1727:87–96. doi: 10.1016/j.bbaexp.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 71.Krasnikov BF, et al. Identification of the putative tumor suppressor Nit2 as omega-amidase, an enzyme metabolically linked to glutamine and asparagine transamination. Biochimie. 2009;91:1072–1080. doi: 10.1016/j.biochi.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cooper EL, et al. YsxC, an essential protein in Staphylococcus aureus crucial for ribosome assembly/stability. BMC Microbiol. 2009;9:266. doi: 10.1186/1471-2180-9-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mercker M, et al. The BEM46-like protein appears to be essential for hyphal development upon ascospore germination in Neurospora crassa and is targeted to the endoplasmic reticulum. Curr. Genet. 2009;55:151–161. doi: 10.1007/s00294-009-0232-3. [DOI] [PubMed] [Google Scholar]
- 74.Miller DJ, et al. Structural and biochemical characterization of a novel Mn2+-dependent phosphodiesterase encoded by the yfcE gene. Protein Sci. 2007;16:1338–1348. doi: 10.1110/ps.072764907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Keppetipola N, Shuman S. A phosphate-binding histidine of binuclear metallophosphodiesterase enzymes is a determinant of 2',3'-cyclic nucleotide phosphodiesterase activity. J. Biol. Chem. 2008;283:30942–30949. doi: 10.1074/jbc.M805064200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rosby R, et al. Knockdown of the Drosophila GTPase nucleostemin 1 impairs large ribosomal subunit biogenesis, cell growth, and midgut precursor cell maintenance. Mol. Biol. Cell. 2009;20:4424–4434. doi: 10.1091/mbc.E08-06-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jiang M, et al. The Escherichia coli GTPase CgtAE is involved in late steps of large ribosome assembly. J. Bacteriol. 2006;188:6757–6770. doi: 10.1128/JB.00444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pereira CM, et al. IMPACT, a protein preferentially expressed in the mouse brain, binds GCN1 and inhibits GCN2 activation. J. Biol. Chem. 2005;280:28316–28323. doi: 10.1074/jbc.M408571200. [DOI] [PubMed] [Google Scholar]
- 79.de Hoog CL, et al. RNA and RNA binding proteins participate in early stages of cell spreading through spreading initiation centers. Cell. 2004;117:649–662. doi: 10.1016/s0092-8674(04)00456-8. [DOI] [PubMed] [Google Scholar]
- 80.Balaji S, Aravind L. The RAGNYA fold: a novel fold with multiple topological variants found in functionally diverse nucleic acid, nucleotide and peptide-binding proteins. Nucleic Acids Res. 2007;35:5658–5671. doi: 10.1093/nar/gkm558. [DOI] [PMC free article] [PubMed] [Google Scholar]