Abstract
Genetic studies are rapidly identifying variants that shape risk for disorders of human cognition, but the question of how such variants predispose to neuropsychiatric disease remains. Noninvasive human brain imaging allows assessment of the brain in vivo, and the combination of genetics and imaging phenotypes remains one of the only ways to explore functional genotype-phenotype associations in human brain. Common variants in contactin-associated protein-like 2 (CNTNAP2), a neurexin superfamily member, have been associated with several allied neurodevelopmental disorders, including autism and specific language impairment, and CNTNAP2 is highly expressed in frontal lobe circuits in the developing human brain. Using functional neuroimaging, we have demonstrated a relationship between frontal lobar connectivity and common genetic variants in CNTNAP2. These data provide a mechanistic link between specific genetic risk for neurodevelopmental disorders and empirical data implicating dysfunction of long-range connections within the frontal lobe in autism. The convergence between genetic findings and cognitive-behavioral models of autism provides evidence that genetic variation at CNTNAP2 predisposes to diseases such asautism in part through modulation of frontal lobe connectivity.
INTRODUCTION
The mechanisms by which common genetic variation can predispose individuals to neuropsychiatric disorders are largely unknown. Recent work suggests a relationship between common genetic variation in contactin-associated protein-like 2 (CNTNAP2) and language ability across a range of developmental neuropsychiatric disorders, including autism (1). Mutations in CNTNAP2 were first identified in a family with Tourette syndrome and obsessive compulsive disorder (2)and in Old Order Amish children with cortical dysplasia, epilepsy, and microcephaly, 70% of whom also had an autism spectrum disorder (3). Furthermore, a highly circumscribed region of ~10 kb within this 2.3 Mb gene has previously shown replicated association with quantitative language endophenotypes in autism [rs2710102; P = 0.0006 (4, 5)] and specific language impairment (SLI) [P = 0.002 to 5 × 10−5 (6)].
CNTNAP2 messenger RNA (mRNA) is significantly enriched in the developing human brain in the frontal and temporal lobes, as well as in striatal circuits and in the frontal cortex of adult brain, but shows no such anterior cortical enrichment in rodents (7). In humans, these regions are posited to support speech and language learning (8), as well as other forms of implicit learning (9, 10), further supporting a role for CNTNAP2 in neural circuits underlying language and cognition. CNTNAP2 clusters voltage-gated channels (Kv1.1) at the nodes of Ranvier (11) and is likely also involved in interactions between neuronal axons and glia (12). During development, Caspr2, the protein encoded by CNTNAP2, is thought to assist in interactions important for cellular migration and subsequent laminar organization (3), indicating a role for CNTNAP2 in the construction of neural circuits. Additionally, a recent structural imaging investigation identified reduced frontal gray matter and abnormal white matter connectivity in carriers of an alternative autism risk allele of CNTNAP2 (13).
The frontal lobe is responsible for a number of higher-order cognitive functions, including planning, decision making, and abstraction, and thusisaprimarycandidate fordysfunction in many neurodevelopmental and neuropsychiatric disorders. One function of the frontal lobe is cognitive control, or the coordination of goal-directed thoughts and actions. Recently, a rostro-caudal, or anterior-posterior, information-processing gradient has been proposed to underlie frontal lobe organization (14). In this model, the anterior prefrontal cortex (PFC) directs information processing by posterior regions during the execution of abstract goals. There is growing evidence for at least two rostro-caudal gradients in the frontal lobe: dorsolateral and ventro-lateral. It has been hypothesized that the dorsal path could be more directly involved in the planning and execution of goal-directed motor actions, whereas the ventral path is involved in contextual processing. Human neuroimaging studies provide support for these anterior-to-posterior gradients (15), and structural equation models provided the first preliminary evidence in support of anterior influence on posterior regions (16). The frontal lobe is also important for a number of social and communication functions. Specifically, the inferior frontal gyrus (IFG) is a major player in speech and language and is involved in both semantic and syntactic processing of linguistic inputs as well as the motor production of speech (17). The IFG is also implicated in the mirror neuron system, a mechanism by which primates are able to understand the actions of others through the firing of mirror neurons when an action is observed as well as when it is executed (18). There is evidence for both language [see (19) for review] and mirror neuron system dysfunction (20-23) in autism. The focal expression pattern of CNTNAP2 in anterior frontal circuits in humans, together with a growing body of evidence implicating disruption of frontal cortical circuit connectivity in disorders such as autism (24, 25), attention deficit–hyperactivity disorder (ADHD) (26), and schizophrenia (27), prompted us to test the hypothesis that variation at the CNTNAP2 risk locus might predispose to cognitive dysfunction in humans through modulation of neural activity in the frontal lobe and its functional connectivity.
RESULTS
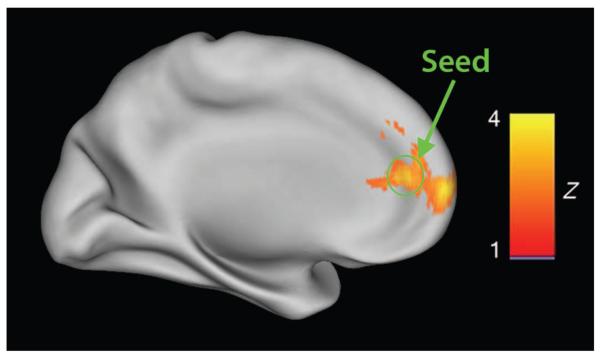
We first scanned 32 children (16 typically developing and 16 with autism) (table S1) using functional magnetic resonance imaging (fMRI) during a variation of a reward-guided implicit learning task known to engage frontostriatal circuits (28), regions that overlap areas of CNTNAP2 gene expression during development (7). Genotype frequency was similar between the typically developing and the autism groups, and data were collapsed across groups to investigate the specific effects of the risk variant on brain function. Comparisons between risk allele carriers (n = 9) and nonrisk allele carriers (n = 23) showed significantly reduced activity in the medial PFC (mPFC) during reward feedback processing in the nonrisk group (P =0.001) (Fig. 1). This finding was robust when examined within diagnostic groups (fig. S1), supporting a dominant effect of the rs2710102 risk allele on brain function. This finding is consistent with previous studies that have demonstrated a dominant effect of the autism and SLI CNTNAP2 risk allele on language endophenotypes, such that heterozygous individuals perform similarly to homozygous risk carriers (1, 6). The mPFC is part of the default mode network (29), which is typically more active during resting baseline than during externally directed attention. Thus, the risk group shows less of the normally observed decrease in medial prefrontal activity than does the nonrisk group during a task requiring externally directed attention (fig. S2).
Fig. 1.
Comparison between risk and nonrisk carriers of the autism-associated CNTNAP2 allele (rs2710102) during reward processing. (Top) Significantly reduced activation in mPFC in the nonrisk individuals compared to risk carriers (Z > 2.3, P < 0.05, after correction for multiple comparisons). The location of the seed region for subsequent analysis in the replication cohort is circled in green.
On the basis of the known expression patterns for CNTNAP2 in the frontal cortex and its role in synaptic transmission, we hypothesized that frontal functional connectivity may serve as a biologically relevant intermediate phenotype for an association study with CNTNAP2. Recently, it has been established that functional connectivity within the default mode is under genetic control, with a family-based estimated heritability of 0.42 (30). Abnormal patterns of functional connectivity have also been observed in a number of neurodevelopmental disorders with large genetic determinants, including both autism and schizophrenia [see (31) for review]. Therefore, we tested functional connectivity networks for association with our risk allele. To investigate whether this genetic risk allele modulates functional interactions between frontal systems and more posterior cortical regions, irrespective of the functional demands of the task, we performed functional connectivity analysis on residual time series after accounting for the task effects (functional connectivity MRI; see Supplementary Material). This analysis revealed a left-lateralized network composed of the left IFG, insula, anterior temporal pole, superior temporal gyrus, and angular gyrus in the nonrisk group (Fig. 2A, first row, and table S2). Conversely, the risk carriers had more widespread and bilateral connectivity throughout the frontal cortex and anterior temporal poles (Fig. 2A, second row, and table S2). Both groups show some evidence of connectivity with medial posterior regions such as the posterior cingulate. In contrast to the diffuse bilateral network in the risk carriers, the discrete left-lateralized frontotemporal network in the nonrisk group overlaps with regions known to be important in language processing, such as the IFG and superior temporal gyrus, and is particularly interesting given the previous association of CNTNAP2 with language abilities and the expression pattern of CNTNAP2 mRNA.
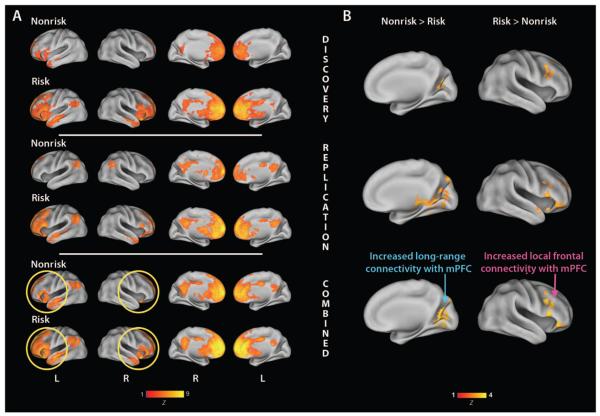
Fig. 2.
Functional connectivity with the mPFC is associated with CNTNAP2. (A) Yellow circles highlight the discrete left-lateralized mPFC functional connectivity network observed in the nonrisk group, in contrast to the more distributed, bilateral network observed in the risk group. Lateral (columns 1 and 2) and medial (columns 3 and 4) views are displayed with left (L) and right (R) hemispheres indicated. Maps are corrected for multiple comparisons at the cluster level (Z >2.3, P <0.05).(B) A direct contrast between risk and nonrisk groups shows relatively greater long-range anterior-posterior connectivity in nonrisk versus risk carriers (blue arrow) and relatively greater right frontal connectivity in risk carriers compared to the nonrisk group (pink arrow). Discovery (row 1), replication (row 2), and combined cohort (row 3) analyses are illustrated separately. Maps are corrected for multiple comparisons at the cluster level (Z > 2.3, P <0.05).
To confirm the diagnosis-independent relationship between CNTNAP2 genotype and functional connectivity, we examined this genotype-phenotype relationship in a separate cohort restricted to 39 typically developing children (16 female/23 male; 10 nonrisk/29 risk) scanned during a different language-learning paradigm (see Supplementary Material for details). Supporting a role for CNTNAP2 in structuring frontal connectivity, the same networks were observed (Fig. 2A, third and fourth rows; see rows 5 and 6 for combined analyses), replicating findings obtained in our discovery cohort. In both the discovery and the replication samples, the differences in connectivity patterns between risk and nonrisk allele carriers were statistically significant, such that nonrisk showed stronger long-range anterior-posterior connectivity between the mPFC and the medial occipital and ventral temporal cortices (Pdiscovery = 2.44 × 10−6; Preplication = 5.47 × 10−9; Fig. 2B, left), whereas the risk carriers had stronger local connectivity between the mPFC and the right frontal cortex (Pdiscovery = 4.21 × 10−4; Preplication = 1.01 × 10−4; Fig. 2B, right). These results were consistent when examined in males only, indicating that the results were not confounded by gender (table S4). Both risk and nonrisk subjects from the discovery and replication samples demonstrated similar negative correlations with mPFC activity in ventral visual cortices, superior parietal lobe, and cerebellum (table S5). To confirm replication in the exact regions identified as significantly different between risk and nonrisk carriers in the discovery sample, we next conducted a region-of-interest (ROI) analysis based on these clusters. We were able to replicate the CNTNAP2 risk allele effects in both the right middle frontal gyrus for risk > nonrisk (P = 0.0081) and the left intracalcarine cortex for the contrasting nonrisk > risk (P = 0.0003) (table S6).
To verify that our replication and discovery samples were not significantly different, we ran between-group comparisons, restricting our investigation to all significant voxels identified in the analysis of the discovery sample. We found no significant differences between the discovery and the replication samples for risk carriers. In the nonrisk carriers, we detected a small cluster in the right orbitofrontal cortex (3 voxels; x, y, z = 28, 28, −16; Z =4.94, P < 0.05 corrected) as significantly different between discovery > replication samples and a small cluster in the left paracingulate cortex (23 voxels; x, y, z = −8, 54, 12; Z = 5.84, P <0.05 corrected) for the contrast replication > discovery. These clusters did not overlap with regions of significantly different functional connectivity observed in between-group comparisons for risk and nonrisk carriers in either discovery or replication samples. Thus, these minor differences did not influence our main results of increased local frontal connectivity and reduced long-range connectivity in risk allele carriers.
DISCUSSION
Here, we demonstrated a significant and replicated association between variation in CNTNAP2 and a functional connectivity phenotype in the human brain. The general paradigm used here—in which functional neuroanatomy, supported by gene expression data, is used to explore the functional consequences of a known disease-associated variant in a highly focused manner—is likely to be of widespread utility in the elucidation of mechanisms underlying disorders of human cognition. Furthermore, we replicated the initial finding in an independent cohort of healthy adolescents. We found that functional connectivity with the mPFC, a region showing differential activity as a function of genotype, identified a network that overlaps with regions of known expression of CNTNAP2 during development. Specifically, we found that the nonrisk groups showed a circumscribed pattern of left lateralized mPFC connectivity, restricted to the inferior frontal cortices and anterior and superior temporal cortices, whereas risk carriers demonstrated a more diffuse pattern of dorsal and ventral frontal lobe connectivity. This abnormal, less lateralized pattern in the risk allele carriers is interesting in light of both the association between CNTNAP2 and language ability (6) and the evidence of aberrant cortical asymmetry in autism and SLI (32). Comparisons between risk allele carriers and nonrisk individuals revealed a pattern of greater long-range anterior-posterior connectivity in the nonrisk individuals and increased local frontal connectivity in the risk allele carriers. Recent work has demonstrated that brain maturation is reflected in a weakening of short-range and a strengthening of long-range cortical-cortical connectivity (33). From this perspective, risk variant carriers' brains connectivity may reflect a more immature connectivity pattern. Thus, we provide the first link between development of normal functional brain connectivity and a specific genetic factor. That the association is independent of disease status indicates that this variant mediates risk by modulating the continuum of normal brain function, as would be expected for intermediate phenotypes related to cognition or behavior (34).
Our demonstration of an association between a CNTNAP2 risk allele and increased local and reduced long-range frontal connectivity is also consistent with the abnormalities observed in both autism and SLI. Converging cognitive-behavioral and neurological evidence supports an overabundance of local connections, particularly within the frontal lobe, and deficient long-range connections along the anterior-posterior aspect as a possible unifying mechanism underlying core cognitive and behavioral deficits in autism [see (35) for review]. This “developmental disconnection” model (25) posits that key disconnections between multiple frontal and temporal lobe association cortices disrupt experience-dependent processes during development that are important for creating and maintaining neural connections. These early disconnections hinder the development of appropriate behaviors (for example, joint attention and language skills) that have consequences for both later development and experience-dependent neuroplasticity. Moreover, the anterior-posterior functional connectivity pattern is consistent with patterns of reduced gray and white matter volume in healthy carriers of a distinct autism-related CNTNAP2 allele (36), separated by over 1 Mb and showing no linkage disequilibrium with the variant studied here. Specifically, Tan and colleagues (13) found that healthy homozygous carriers of the CNTNAP2 autism risk allele showed reduced volumes in the right frontal cortex, ventral temporal cortex, and occipital pole-regions consistent with our functional connectivity measures. These data further implicate CNTNAP2 in modulation of frontal and temporal lobe development and function and suggest a mechanism whereby CNTNAP2 is involved in maturation of cortical networks from local to long-range networks (37).
Here, we leveraged gene expression patterns and neurocognitive hypotheses to identify a strong neuroimaging-based intermediate phenotype to test for association with an a priori candidate gene. The utility of functional brain imaging to reveal stable endophenotypes for genetic association studies is in the early days of exploration. A true endophenotype should be state-independent, associated with the disease, and present in nonaffected family members at a higher rate than in the general population (38), and thus, it will be important to validate putative functional endophenotypes in longitudinal and family-based studies. By limiting our analysis to a known genetic association and a carefully focused biological hypothesis, we mitigate power problems associated with multiple testing and small sample sizes used in functional neuroimaging studies. Our findings are consistent with the hypothesis that genetic variation within CNTNAP2 predisposes to language and cognitive difficulties through functional reorganization of frontal circuits during development (24). CNTNAP2 has been associated with risk for neuropsychiatric disorders other than autism and SLI, including ADHD (39), Tourette syndrome (2), and schizophrenia (40). Therefore, these findings are likely to have broad relevance.
MATERIALS AND METHODS
Participants
Participants for the discovery sample were recruited through referrals from the University of California, Los Angeles (UCLA) Autism Evaluation Clinic and through flyers posted around the UCLA campus and the greater Los Angeles area. Participants for the replication sample were recruited from the greater Los Angeles area via summer camps, posted flyers, and mass mailings. Written informed child assent and parental consent were obtained from all children and their parents, respectively, according to the guidelines set forth by the UCLA Institutional Review Board.
Scan acquisition
Scans were acquired on a Siemens Allegra 3 Tesla head-only MRI scanner at the Ahmanson-Lovelace Brain Mapping Center at UCLA. For each participant, a high-resolution structural T2-weighted echo-planar imaging (EPI) volume [spin-echo, repetition time (TR) = 5000 ms, echo time (TE) = 33 ms, matrix size = 128 by 128, field of view (FOV) = 20 cm, 36 slices, 1.56-mm in-plane resolution, 3 mm thick] was acquired coplanar with the functional scans to allow for spatial registration of each participant's data into a standard coordinate system. For the reward-learning run, 180 functional images were acquired with an EPI gradient-echo acquisition covering the whole cerebral volume (TR = 2000 ms, TE = 30 ms, flip angle = 90°, matrix size = 64 by 64, FOV = 20 cm, 33 slices, 3.125-mm in-plane resolution, 4 mm thick). For the language-learning run, 174 images were acquired with a similar EPI acquisition (TR = 3000 ms, TE = 25 ms, flip angle = 90°, matrix size = 64 by 64, FOV = 20 cm, 36 slices, 3.125-mm in-plane resolution, 4 mm thick). Two volumes at the beginning of each functional run were used to allow equilibration to steady state and were subsequently excluded from the analysis.
Statistical analysis
fMRI analysis was carried out with FEAT (FMRI Expert Analysis Tool) version 5.63, part of FSL (FMRIB Software Library) versions 3.3 and 4.0. Time-series statistical analysis was carried out with FILM with local autocorrelation correction. Regressors of interest were created by convolving a delta function representing trial onset times with an optimal set of four basis functions generated with FMRIB's Linear Optimal Basis Sets (FLOBS), along with their temporal derivative. Motion parameters for each subject were entered as covariates of no interest. Functional images were aligned with FMRIB's Linear Image Registration Tool (FLIRT) to high-resolution coplanar images via an affine transformation with six degrees of freedom. The high-resolution coplanar images were then aligned to the standard Montreal Neurological Institute (MNI) average of 152 brains by using an affine transformation with 12 degrees of freedom. Functional connectivity analyses, controlling for task-related activity, were conducted within and between genotype groups. Within the replication cohort analysis, the seed ROI was a 6-mm sphere centered at the peak voxel coordinate in the mPFC cluster used as the seed region for the discovery cohort (MNIx,y,z = 12, 26, 14). First, a nuisance model including pre-whitening with task as an explanatory variable, and white matter average time series, cerebrospinal fluid average time series, and motion included as covariates, was run and residuals were computed. The average time series from the seed ROI was extracted from the residual image. Both the extracted time series and the residual image were then normalized. Next, subject-wise correlations were computed between the normalized time series and the residual image. Contrast images from single subjects were transformed with Fisher's Z transformation [1/2 log (1 + r)/(1 − r)] before being entered into higher-level ordinary least-squares analyses to create more normally distributed values and decrease bias in reverse transformation of second-level correlations. Group contrasts were cluster corrected at i > 2.3 (P < 0.05). To confirm the qualitative replication of the between-group comparisons observed in the replication sample, we ran a secondary ROI analysis based on significant voxels from the between-group comparisons in the discovery cohort. We used a mask of voxels surviving correction for multiple comparisons from the risk > nonrisk and nonrisk > risk discovery sample comparisons to prethreshold our between-group comparisons in the replication sample, restricting our search to a priori voxels of interest. On the basis of the results from the discovery sample, we were able to estimate the direction of the effect and used a one-tailed P < 0.01.
Acknowledgments
We thank the Brain Mapping Medical Research Organization, Brain Mapping Support Foundation, Pierson-Lovelace Foundation, Ahmanson Foundation, William M. and Linda R. Dietel Philanthropic Fund at the Northern Piedmont Community Foundation, Tamkin Foundation, Jennifer Jones-Simon Foundation, Capital Group Companies Charitable Foundation, Dickinson Family Foundation, Robson Family, and Northstar Fund. We thank K. McNealy for providing replication data.
Funding: This work was in part supported by grants from the National Institute of Child Health and Human Development (P01 HD035470, P50 HD055784, and R01 HD065280-01), the National Alliance for Autism Research, Autism Speaks, and Whitehall Foundation, as well as by the Training Program in Neurobehavioral Genetics (T32 MH073526), and a National Research Service Award predoctoral fellowship (F31 MH079645). The project described was in part also supported by grants (RR12169, RR13642, and RR00865, UL1 RR025774) from the National Center for Research Resources (NCRR), a component of the NIH, and R01-hd065280 and P50-hd055784-03s1 from the NIH (to S.Y.B.); its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH.
Footnotes
SUPPLEMENTARY MATERIAL
www.sciencetranslationalmedicine.org/cgi/content/full/2/56/56ra80/DC1
Materials and Methods
Fig. S1. Percent signal change in mPFC during reward processing within diagnostic groups.
Fig. S2. Average time course in mPFC during monetary reward feedback for risk and nonrisk groups.
Fig. S3. Functional connectivity maps for risk and nonrisk in discovery sample.
Fig. S4. Functional connectivity maps for risk and nonrisk in replication sample.
Fig. S5. Functional connectivity maps for risk and nonrisk in combined samples.
Fig. S6. Functional connectivity maps for risk versus nonrisk groups.
Table S1. Sample demographics.
Table S2. Differences in mPFC connectivity between risk and nonrisk carriers in discovery and replication samples.
Table S3. Cluster peaks for combined between-group analysis.
Table S4. Cluster peaks for combined male-only between-group analysis.
Table S5. Negative correlations with mPFC in discovery and replication samples.
Table S6. Significant between-group region-of-interest comparisons in replication sample.
References
Author contributions: A.A.S.-V.Z., S.Y.B., M.D., and D.H.G. developed the initial idea and design of the study. A.A.S.-V.Z., D.H.G., and R.A.P. developed the fMRI task. J.A.M. contributed to statistical analysis of the data. B.S.A., A.I.A.-R., A.A.S.-V.Z., J.D.R., and L.I.S. performed genotyping and genetic analyses. B.S.A., S.Y.B., M.D., D.H.G., and A.A.S.-V.Z. interpreted the findings and contributed to the manuscript. All authors revised and commented on drafts of the manuscript.
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Abrahams BS, Geschwind DH. Genetics of autism. In: Speicher MR, Antonarakis SE, Motulsky AG, editors. Human Genetics: Problems and Approaches. Springer-Verlag; Heidelberg: 2010. [Google Scholar]
- 2.Verkerk AJ, Mathews CA, Joosse M, Eussen BH, Heutink P, Oostra BA. Tourette Syndrome Association International Consortium for Genetics, Cntnap2 is disrupted in a family with Gilles de la Tourette syndrome and obsessive compulsive disorder. Genomics. 2003;82:1–9. doi: 10.1016/s0888-7543(03)00097-1. [DOI] [PubMed] [Google Scholar]
- 3.Strauss KA, Puffenberger EG, Huentelman MJ, Gottlieb S, Dobrin SE, Parod JM, Stephan DA, Morton DH. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N. Engl. J. Med. 2006;354:1370–1377. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- 4.Alarcón M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bomar JM, Sebat J, Wigler M, Martin CL, Ledbetter DH, Nelson SF, Cantor RM, Geschwind DH. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am. J. Hum. Genet. 2008;82:150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arking DE, Cutler DJ, Brune CW, Teslovich TM, West K, Ikeda M, Rea A, Guy M, Lin S, Cook EH, Jr., Chakravarti A. A common genetic variant in the neurexin super-family member CNTNAP2 increases familial risk of autism. Am. J. Hum. Genet. 2008;82:160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vernes SC, Newbury DF, Abrahams BS, Winchester L, Nicod J, Groszer M, Alarcón M, Oliver PL, Davies KE, Geschwind DH, Monaco AP, Fisher SE. A functional genetic link between distinct developmental language disorders. N. Engl. J. Med. 2008;359:2337–2345. doi: 10.1056/NEJMoa0802828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abrahams BS, Tentler D, Perederiy JV, Oldham MC, Coppola G, Geschwind DH. Genome-wide analyses of human perisylvian cerebral cortical patterning. Proc. Natl. Acad. Sci. U.S.A. 2007;104:17849–17854. doi: 10.1073/pnas.0706128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McNealy K, Mazziotta JC, Dapretto M. Cracking the language code: Neural mechanisms underlying speech parsing. J. Neurosci. 2006;26:7629–7639. doi: 10.1523/JNEUROSCI.5501-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knowlton BJ, Mangels JA, Squire LR. A neostriatal habit learning system in humans. Science. 1996;273:1399–1402. doi: 10.1126/science.273.5280.1399. [DOI] [PubMed] [Google Scholar]
- 10.Poldrack RA, Prabhakaran V, Seger CA, Gabrieli JD. Striatal activation during acquisition of a cognitive skill. Neuropsychology. 1999;13:564–574. doi: 10.1037//0894-4105.13.4.564. [DOI] [PubMed] [Google Scholar]
- 11.Poliak S, Salomon D, Elhanany H, Sabanay H, Kiernan B, Pevny L, Stewart CL, Xu X, Chiu SY, Shrager P, Furley AJ, Peles E. Juxtaparanodal clustering of Shaker-like K+ channels in myelinated axons depends on Caspr2 and TAG-1. J. Cell Biol. 2003;162:1149–1160. doi: 10.1083/jcb.200305018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corfas G, Velardez MO, Ko CP, Ratner N, Peles E. Mechanisms and roles of axon–Schwann cell interactions. J. Neurosci. 2004;24:9250–9260. doi: 10.1523/JNEUROSCI.3649-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan GC, Doke TF, Ashburner J, Wood NW, Frackowiak RS. Normal variation in fronto-occipital circuitry and cerebellar structure with an autism-associated polymorphism of CNTNAP2. NeuroImage. 2010;53:1030–1042. doi: 10.1016/j.neuroimage.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Badre D, D'Esposito M. Is the rostro-caudal axis of the frontal lobe hierarchical? Nat. Rev. Neurosci. 2009;10:659–669. doi: 10.1038/nrn2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Badre D, D'Esposito M. Functional magnetic resonance imaging evidence for a hierarchical organization of the prefrontal cortex. J. Cogn. Neurosci. 2007;19:2082–2099. doi: 10.1162/jocn.2007.19.12.2082. [DOI] [PubMed] [Google Scholar]
- 16.Koechlin E, Ody C, Kouneiher F. The architecture of cognitive control in the human pre-frontal cortex. Science. 2003;302:1181–1185. doi: 10.1126/science.1088545. [DOI] [PubMed] [Google Scholar]
- 17.Bookheimer S. Functional MRI of language: New approaches to understanding the cortical organization of semantic processing. Annu. Rev. Neurosci. 2002;25:151–188. doi: 10.1146/annurev.neuro.25.112701.142946. [DOI] [PubMed] [Google Scholar]
- 18.Rizzolatti G, Craighero L. The mirror-neuron system. Annu. Rev. Neurosci. 2004;27:169–192. doi: 10.1146/annurev.neuro.27.070203.144230. [DOI] [PubMed] [Google Scholar]
- 19.Groen WB, Zwiers MP, van der Gaag RJ, Buitelaar JK. The phenotype and neural correlates of language in autism: An integrative review. Neurosci. Biobehav. Rev. 2008;32:1416–1425. doi: 10.1016/j.neubiorev.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 20.Dapretto M, Davies MS, Pfeifer JH, Scott AA, Sigman M, Bookheimer SY, Iacoboni M. Understanding emotions in others: Mirror neuron dysfunction in children with autism spectrum disorders. Nat. Neurosci. 2006;9:28–30. doi: 10.1038/nn1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hadjikhani N, Joseph RM, Snyder J, Tager-Flusberg H. Anatomical differences in the mirror neuron system and social cognition network in autism. Cereb. Cortex. 2006;16:1276–1282. doi: 10.1093/cercor/bhj069. [DOI] [PubMed] [Google Scholar]
- 22.Oberman LM, Hubbard EM, McCleery JP, Altschuler EL, Ramachandran VS, Pineda JA. EEG evidence for mirror neuron dysfunction in autism spectrum disorders. Brain Res. Cogn. Brain Res. 2005;24:190–198. doi: 10.1016/j.cogbrainres.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 23.Uddin LQ, Davies MS, Scott AA, Zaidel E, Bookheimer SY, Iacoboni M, Dapretto M. Neural basis of self and other representation in autism: An fMRI study of self-face recognition. PLoS One. 2008;3:e3526. doi: 10.1371/journal.pone.0003526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Courchesne E, Pierce K. Brain overgrowth in autism during a critical time in development: Implications for frontal pyramidal neuron and interneuron development and connectivity. Int. J. Dev. Neurosci. 2005;23:153–170. doi: 10.1016/j.ijdevneu.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 25.Geschwind DH, Levitt P. Autism spectrum disorders: Developmental disconnection syndromes. Curr. Opin. Neurobiol. 2007;17:103–111. doi: 10.1016/j.conb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 26.Casey BJ, Nigg JT, Durston S. New potential leads in the biology and treatment of attention deficit-hyperactivity disorder. Curr. Opin. Neurol. 2007;20:119–124. doi: 10.1097/WCO.0b013e3280a02f78. [DOI] [PubMed] [Google Scholar]
- 27.Karlsgodt KH, Sun D, Jimenez AM, Lutkenhoff ES, Willhite R, van Erp TG, Cannon TD. Developmental disruptions in neural connectivity in the pathophysiology of schizophrenia. Dev. Psychopathol. 2008;20:1297–1327. doi: 10.1017/S095457940800062X. [DOI] [PubMed] [Google Scholar]
- 28.Scott-Van Zeeland AA, Dapretto M, Ghahremani DG, Poldrack RA, Bookheimer SY. Reward processing in autism. Autism Res. 2010;3:53–67. doi: 10.1002/aur.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. A default mode of brain function. Proc. Natl. Acad. Sci. U.S.A. 2001;98:676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glahn DC, Winkler AM, Kochunov P, Almasy L, Duggirala R, Carless MA, Curran JC, Olvera RL, Laird AR, Smith SM, Beckmann CF, Fox PT, Blangero J. Genetic control over the resting brain. Proc. Natl. Acad. Sci. U.S.A. 2010;107:1223–1228. doi: 10.1073/pnas.0909969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Broyd SJ, Demanuele C, Debener S, Helps SK, James CJ, Sonuga-Barke EJS. Default-mode brain dysfunction in mental disorders: A systematic review. Neurosci. Biobehav. Rev. 2009;33:279–296. doi: 10.1016/j.neubiorev.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 32.Herbert MR, Ziegler DA, Deutsch CK, O'Brien LM, Kennedy DN, Filipek PA, Bakardjiev AI, Hodgson J, Takeoka M, Makris N, Caviness VS., Jr. Brain asymmetries in autism and developmental language disorder: A nested whole-brain analysis. Brain. 2005;128:213–226. doi: 10.1093/brain/awh330. [DOI] [PubMed] [Google Scholar]
- 33.Dosenbach NU, Nardos B, Cohen AL, Fair DA, Power JD, Church JA, Nelson SM, Wig GS, Vogel AC, Lessov-Schlaggar CN, Barnes KA, Dubis JW, Feczko E, Coalson RS, Pruett JR, Jr., Barch DM, Petersen SE, Schlaggar BL. Prediction of individual brain maturity using fMRI. Science. 2010;329:1358–1361. doi: 10.1126/science.1194144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geschwind DH. Autism: Many genes, common pathways? Cell. 2008;135:391–395. doi: 10.1016/j.cell.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Courchesne E, Redcay E, Morgan JT, Kennedy DP. Autism at the beginning: Micro-structural and growth abnormalities underlying the cognitive and behavioral phenotype of autism. Dev. Psychopathol. 2005;17:577–597. doi: 10.1017/S0954579405050285. [DOI] [PubMed] [Google Scholar]
- 36.Arking DE, Cutler DJ, Brune CW, Teslovich TM, West K, Ikeda M, Rea A, Guy M, Lin S, Cook EH, Jr., Chakravarti A. A common genetic variant in the neurexin super-family member CNTNAP2 increases familial risk of autism. Am. J. Hum. Genet. 2008;82:160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fair DA, Cohen AL, Power JD, Dosenbach NU, Church JA, Miezin FM, Schlaggar BL, Petersen SE. Functional brain networks develop from a “local to distributed” organization. PLoS Comput. Biol. 2009;5:e1000381. doi: 10.1371/journal.pcbi.1000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gottesman II, Gould TD. The endophenotype concept in psychiatry: Etymology and strategic intentions. Am. J. Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 39.Elia J, Gai X, Xie HM, Perin JC, Geiger E, Glessner JT, D'arcy M, deBerardinis R, Frackelton E, Kim C, Lantieri F, Muganga BM, Wang L, Takeda T, Rappaport EF, Grant SF, Berrettini W, Devoto M, Shaikh TH, Hakonarson H, White PS. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol. Psychiatry. 2010;15:637–646. doi: 10.1038/mp.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friedman JI, Vrijenhoek T, Markx S, Janssen IM, van der Vliet WA, Faas BH, Knoers NV, Cahn W, Kahn RS, Edelmann L, Davis KL, Silverman JM, Brunner HG, van Kessel AG, Wijmenga C, Ophoff RA, Veltman JA. CNTNAP2 gene dosage variation is associated with schizophrenia and epilepsy. Mol. Psychiatry. 2008;13:261–266. doi: 10.1038/sj.mp.4002049. [DOI] [PubMed] [Google Scholar]