Abstract
Mitsugumin 23 (MG23) is a 23 kDa transmembrane protein localized to the sarcoplasmic/endoplasmic reticulum and nuclear membranes in a wide variety of cells. Although the characteristics imply the participation in a fundamental function in intracellular membrane systems, the physiological role of MG23 is unknown. Here we report the biochemical and biophysical characterization of MG23. Hydropathicity profile and limited proteolytic analysis proposed three transmembrane segments in the MG23 primary structure. Chemical cross-linking analysis suggested a homo-oligomeric assembly of MG23. Ultrastructural observations detected a large symmetrical particle as the predominant component and a small asymmetric assembly as the second major component in highly purified MG23 preparations. Single-particle three-dimensional reconstruction revealed that MG23 forms a large bowl-shaped complex equipped with a putative central pore, which is considered an assembly of the small asymmetric subunit. After reconstitution into planar phospholipid bilayers, purified MG23 behaved as a voltage-dependent, cation-conducting channel, permeable to both K+ and Ca2+. A feature of MG23 gating was that multiple channels always appeared to be gating together in the bilayer. Our observations suggest that the bowl-shaped MG23 can transiently assemble and disassemble. These building transitions may underlie the unusual channel gating behavior of MG23 and allow rapid cationic flux across intracellular membrane systems.
The endoplasmic/sarcoplasmic reticulum (ER/SR) is a multifunctional organelle responsible for important cellular processes, including protein maturation, lipid metabolism, Ca2+ signaling, and stress response. The ER/SR serves as an intracellular Ca2+ store, and activation of Ca2+ release channels, namely, inositol trisphosphate and ryanodine receptors, controls physiological functions such as muscle contraction, secretion, metabolism, and transcription.1,2 In addition, the ER is the site for synthesis and maturation of both membrane and secretory proteins, enforcing protein glycosylation, disulfide bridging, folding, and subunit assembly. When misfolded proteins accumulate within the lumen, the ER stress response is activated according to severity, leading to the recruitment of ER chaperones, inhibition of protein synthesis, and induction of apoptotic cell death.3,4 The activity of molecular chaperones, protein-processing enzymes, and metabolic enzymes of the ER largely depends upon the high luminal Ca2+ level.
Uptake of Ca2+ into and release of Ca2+ from intracellular stores are electrogenic processes. Therefore, active Ca2+ fluxes may be synchronized with the movements of other ionic species that compensate for charge imbalance across the ER/SR membrane.5,6 We have recently identified TRIC channel subtypes that function as monovalent cation channels and probably support release of Ca2+ from the ER/SR of various cell types.7−10 It is likely that the vital function of the ER/SR requires rapid and flexible control of the ionic balance between the luminal and cytoplasmic sides. To understand the ionic homeostasis across the ER/SR membrane, it is important to further characterize the functional properties of its constituent ion channels and transporters in the intracellular membrane system.
Skeletal and cardiac muscle SR is specialized as the intracellular Ca2+ store for controlling contraction and abundantly contains Ca2+-handling proteins such as Ca2+-ATPase, calsequestrin, and ryanodine receptors.(2) Muscle SR is, therefore, an ideal model system for studying Ca2+ store functions. To understand the molecular basis of Ca2+ stores, we have searched for novel SR proteins using monoclonal antibodies (mAbs) and previously identified mitsugumin 23 (MG23) with a mature molecular size of 23 kDa.(11) Although MG23 is abundantly expressed in the SR and nuclear membranes of striated muscle cells, its expression is also detected in a wide variety of cell types. The ubiquitous distribution suggests that MG23 may contribute to a common function in intracellular membrane systems. A recent study demonstrated that mutant thymocytes lacking MG23 became resistant to DNA damage-induced apoptosis, suggesting a role in the generation of ER-derived cell death signals.(12) The physiological function of MG23, however, is still unknown. In this report, we provide biochemical and biophysical data suggesting that MG23 forms a massive homomultimeric complex, which can conduct cations, including Ca2+, across the intracellular membrane systems.
Materials and Methods
Antibody and Topology Analysis
For producing mAbs, two synthetic peptides corresponding to the N-terminal and C-terminal MG23 sequence were conjugated with a carrier protein and repeatedly injected into mice to generate hybridoma cells.(11) Immunochemical experiments established two clones, mAb7 (mAb-N) and mAb251 (mAb-C), which specifically recognize the corresponding antigen epitopes. To examine the transmembrane topology of MG23, we prepared SR vesicles from rabbit skeletal muscle(13) and isolated ER vesicles from HEK293 cells transfected with MG23 expression plasmids.(14) After the treatment of the vesicles with proteinase, the digestion profiles of native and recombinant MG23 were examined using mAbs as described previously.(15) For further details, see the Supporting Information.
Affinity Purification of Native and Recombinant MG23
For purification of native MG23, microsomal proteins were solubilized with NP-40 from rabbit muscle SR vesicles and reacted with affinity resin conjugated with mAb-N. After the resin had been extensively washed, MG23 was recovered with a buffer containing the epitope peptide. For production of recombinant MG23 using a methylotrophic yeast system (Invitrogen), a His tag sequence (Hisx6) was inserted into the rabbit MG23 cDNA at the site immediately downstream of the N-terminal signal sequence. The total microsome was prepared from yeast cells expressing His-tagged MG23, and the recombinant protein was solubilized with NP-40 or n-dodecyl β-d-maltoside (DDM) and purified using combined Ni and mAb-C affinity chromatography. For further details, see the Supporting Information. The purified MG23 preparations were used for chemical cross-linking, ultrastructural imaging, and channel recording.
Particle Image Analysis
After affinity purification, MG23 was further purified by density gradient centrifugation. For immunodecoration of MG23, mAbs were reacted with the purified preparations. MG23 particles with or without the mAb decoration were applied to thin carbon films, negatively stained, and examined using electron microscopy (EM). For three-dimensional (3D) reconstruction of MG23 structures,7,16 particle images for the large bowl-shaped and asymmetric small structures were automatically picked up from the electron micrographs.17,18 The particle images in each library were aligned rotationally and translationally19−21 and classified into clusters.22,23 The resulting class averages were used as new references, and the cycle from alignment to classification was repeated until convergence. On the basis of the symmetry of averaged images, 6-fold symmetry was imposed in the following computation of the large particles. No symmetry was assumed for the small particles. After convergence, a Euler angle was assigned to each average image,(24) and a primary 3D model was reconstructed using simultaneous iterative reconstruction techniques.(25) To optimize 3D reconstruction, reprojection images were created from the 3D model and used for realignment of the original particle images.(26) The aligned particles were classified again, and a Euler angle of each average was assigned so as to be the best correlation to the 3D reprojection image. The computational steps were repeated until a stable 3D model was obtained. For further details, see the Supporting Information.
Planar Phospholipid Bilayer Measurements
Native and recombinant MG23 proteins were solubilized and purified with either NP-40 or DDM. For detergent exchange in purified MG23 preparations, gel filtration chromatography was performed using a buffer solution containing 16 mM CHAPS and 16 mM 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC). Then, CHAPS was dialyzed using a dialysis membrane with a cutoff size of 12 kDa (Medicell International Ltd.) against a buffer containing 0.1 M NaCl, 0.1 mM EGTA, 25 mM PIPES (pH 7.4), 0.15 mM CaCl2, and 2.5 mM DTT (exchanged every 4 h for 16 h). Purified native and recombinant MG23 proteins were fused with planar phosphatidylethanolamine lipid bilayers as previously described.(27) The trans chamber was held at ground and the cis chamber at potentials relative to ground. To examine if MG23 could behave as an ion channel permeable to monovalent cations or anions, we used symmetrical (in both cis and trans chambers) bathing solutions containing either 260 mM KCl, 20 mM HEPES, and 15 μM free Ca2+ (pH 7.2) or 260 mM K-PIPES and 15 μM free Ca2+ (pH 7.2). To investigate if MG23 is permeable to Ca2+, we perfused the cis chamber with 250 mM HEPES, 80 mM Tris, and 15 μM free Ca2+ (pH 7.2) and filled the trans chamber with a solution containing 250 mM glutamic acid and 10 mM HEPES (pH 7.2) with Ca(OH)2 (free Ca2+ concentration of 65 mM). The free Ca2+ concentration and pH of solutions were measured at 22 °C using a calcium electrode (Orion 93-20) and Ross-type pH electrode (Orion 81-55) as described previously.(27) Single-channel events were recorded on digital audiotape. MG23 recordings were filtered at 600 kHz (−3 dB) and digitized at 20 kHz using Pulse (HEKA, Elektronik Lambrecht/Pfalz). The amplitude of channel openings was determined using WinEDR version 3.05 (J. Dempster, University of Strathclyde, Glasgow, U.K.) using manually controlled cursors. Noise analysis was performed with WinEDR version 3.05. The current fluctuations across the bilayer were subdivided into multiple segments in time with each segment containing N samples. The mean current for each segment was calculated with the formula
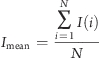 |
where I(i) is the amplitude of the ith current of the N samples in the segment. The mean current values were then plotted versus time. The relative Ca2+/K+ permeability ratio (PCa2+/PK+) of MG23 was calculated using the Fatt−Ginsborg equation:(28)
where R, T, and F have their usual meanings and Erev is the zero-current reversal potential. The value of RT/F used was 25.4 mV at 22 °C. Erev was determined under bi-ionic conditions (cis, 210 mM K+; trans, 65 mM Ca2+) and was taken as the voltage at which no current was detected. Erev was corrected for the liquid junction potentials arising between the different solutions. The junction potential was calculated to be −3 mV using Clampex version 10.2 (Molecular Devices, Sunnyvale, CA).
Results
Three Transmembrane Segments Proposed for MG23
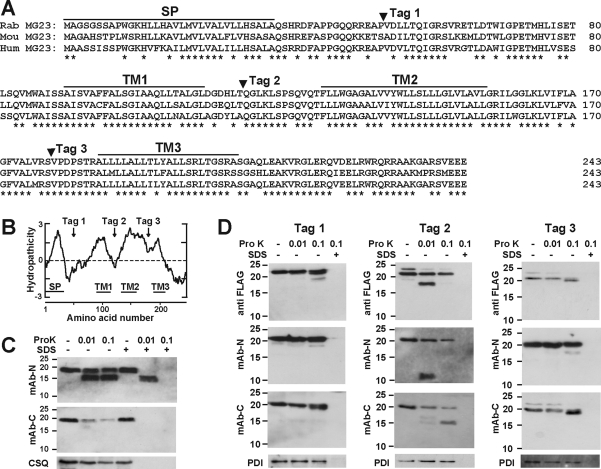
After the N-terminal signal peptide of 33 amino acid residues is removed from the primary translational product (Figure 1A), mature MG23 is localized as a 23 kDa membrane protein in the skeletal muscle SR.(11) No mobility shift after the treatment of glycosidase and thiol-reducing agents in sodium dodecyl sulfate−polyacrylamide gel electrophoresis (SDS−PAGE) has suggested that mature MG23 is not subjected to obvious post-translational modifications such as glycosylation and disulfide linkage.(11) Although the Prosite database search detected no conserved motif sequences or domain structures, the hydropathicity profile suggests multiple membrane-spanning segments in MG23 (Figure 1B). First, we examined the transmembrane topology of MG23 in the muscle SR. MG23 was detected as a protein band with a relative molecular mass of ∼19 kDa via SDS−PAGE. This band remained even after extensive digestion with proteinase K and was shown to be highly proteinase resistant in SR vesicles prepared under isotonic conditions. However, after proteinase treatments, we detected a cleaved product of ∼16 kDa, which was recognized by mAb-N but not by mAb-C (Figure 1C). Therefore, the proteinase-insensitive N-terminus is assigned to the luminal side, and conversely, the proteinase-sensitive C-terminus probably faces into the cytoplasm.
Figure 1.
Membrane topology analysis of MG23. (A) Signal sequence (SP) and proposed transmembrane segments in MG23 aligned sequences. Amino acid residues are numbered from the initiating methionine, and sets of identical residues among the animal species are marked with asterisks. The sites inserted with a FLAG tag for the topology analysis are also indicated. Our topology analysis predicted three transmembrane segments (TM1−TM3); the termini of each segment are tentatively assigned. Rab, rabbit; Mou, mouse; Hum, human. (B) Hydropathicity profile of MG23. The hydropathicity was calculated using the Kyte−Doolittle algorithm with a window size of 19 residues. The N-terminal signal peptide (SP), putative transmembrane segments (TM1−TM3), and FLAG tag sites (Tag 1−3) are indicated. (C) Topology analysis of MG23 using skeletal muscle SR vesicles. The SR vesicles were treated with proteinase K at several concentractions (Pro K, micrograms per milliliter) in the presence or absence of 0.1% SDS. Proteolytic profiles were examined via Western blotting using mAb-N, mAb-C, or the antibody to calsequestrin (CSQ, SR luminal marker protein as a control). (D) Topology analysis of FLAG-tagged MG23 in ER vesicles. HEK293 cells were transfected with expression plasmids carrying FLAG-tagged MG23 cDNAs and harvested for the preparation of ER vesicles under isotonic conditions. The ER vesicles were treated with proteinase K in the presence or absence of SDS. Proteolytic profiles were examined via immunoblotting using mAb-N, mAb-C, mAb against the FLAG tag, or the antibody to protein disulfide isomerase (PDI, ER luminal marker protein). Size markers are indicated in kilodaltons.
We further introduced the FLAG tag sequence at hydrophilic sites in the MG23 primary structure and prepared ER vesicles from cultured cells expressing the FLAG-tagged MG23 proteins for proteolytic analysis (Figure 1D). The region around residue 119 (Tag 2 site) was resistant to proteinase digestion but became proteinase-sensitive when the FLAG tag was introduced. Therefore, the Tag 2 site is reasonably assigned to the cytoplasmic side and may be associated with the membrane environment in the original form of mature MG23. On the other hand, no obvious difference in digestion pattern was observed by insertion of the tag at the Tag 1 and Tag 3 sites, suggesting their localization on the luminal side of the ER/SR. Taken together, the proteolytic analysis data predict three transmembrane segments, which are arranged in MG23 according to the positive-inside rule.(29) In this model (Figure 3C), of the 210 amino acid residues that composed the mature MG23, the cytoplasmic region contains 59 residues (28%), the transmembrane region carries 65 residues (31%), and the luminal region possesses 86 residues (41%).
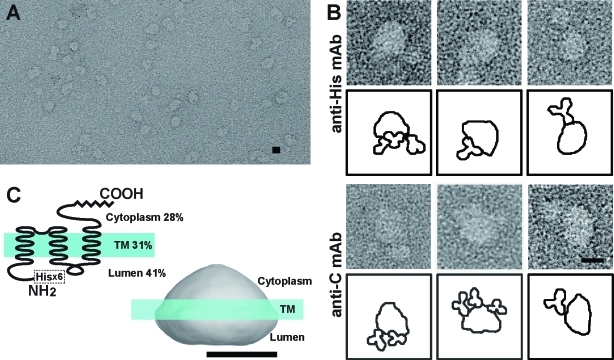
Figure 3.
EM analysis of MG23 particles. (A) Raw EM images of recombinant MG23 particles. After adsorption to the glow-discharged carbon film, negatively stained samples were imaged by EM. (B) Immunodecoration of MG23 particles. Recombinant MG23 bearing anti-His mAbs (top panels) and mAb-C (bottom panels) were imaged by EM. (C) Predicted membrane topology of the MG23 monomer from the hydropathicity profile and limited proteolytic analysis (left) and the membrane position (blue line) deduced from the volume calculation of the reconstructed MG23 (right). Scale bars are 10 nm.
Homo-Oligomeric Assembly of MG23
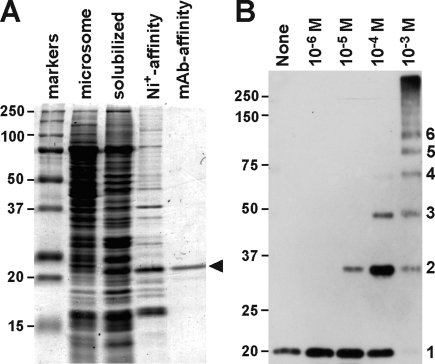
Native MG23 was solubilized from rabbit SR vesicles and purified using mAb affinity chromatography (Figure S1A of the Supporting Information). Although purified preparations contained a few impurities, including Ca2+-ATPase (∼100 kDa protein), no proteins were coenriched with MG23 during the affinity purification. Moreover, recombinant His-tagged MG23 was expressed in metanotrophic yeast (Figure 2A) and highly purified using combined affinity chromatography (>95% purity via SDS−PAGE). Both preparations were used to characterize the structural and electrophysiological properties of MG23 in this study. When native and recombinant MG23 preparations were reacted with chemical cross-linkers, immunoblotting detected the generation of homo-oligomeric products (Figure 2B). For example, cross-linking by disuccinimidyl glutarate generated oligomeric complexes, up to a hexamer, in both native and recombinant MG23 preparations (Figure S1B of the Supporting Information). The observations suggest that MG23 forms at least a homohexamer as a putative self-assembly unit.
Figure 2.
Purification and cross-linking of MG23. (A) Purification of recombinant MG23 from cDNA-transfected yeast cells. The cell lysate fraction, microsomal fraction, Ni affinity-purified fraction, and immunoaffinity-purified fraction were analyzed via SDS−PAGE. Proteins were visualized by Coomassie blue staining, and recombinant MG23 is marked with an arrowhead. (B) Cross-linking of purified recombinant MG23. An affinity-purified recombinant MG23 preparation was treated with the cross-linker disuccinimidyl glutarate at concentrations of 10−6−10−3 M, and the resulting products were examined by immunoblotting using mAb-N; monomeric to hexameric products are numbered. Size markers are indicated in kilodaltons.
MG23 Particle Imaging
After the negative staining, purified recombinant MG23 was examined by EM (Figure 3A). Most particles were round-shaped or bowl-shaped, which probably represent vertical or lateral views, respectively. We frequently detected round-shaped particles with electron-dense rim, interpreted to be the views of the MG23 particle perpendicular to the membrane. Putative side views clearly show asymmetry with respect to the membrane; one end is wider and the other narrower. To examine the topological orientation to the SR/ER membrane, recombinant MG23 particles were reacted with mAb to the N-terminal His tag (mAb-His) and mAb-C, and the resulting MG23−mAb complexes were analyzed by EM (Figure 3B). Although we frequently detected MG23 particles bearing multiple antibodies, mAb-His predominantly attached to the wider side of the particle, while mAb-C reacted with the narrower side. The EM observation, together with the proteolytic topology assay, suggests that the narrow side of the MG23 particle faces the cytoplasm while the wide side is oriented to the ER/SR lumen (Figure 3C).
3D Reconstruction of Bowl-Shaped MG23 Particles
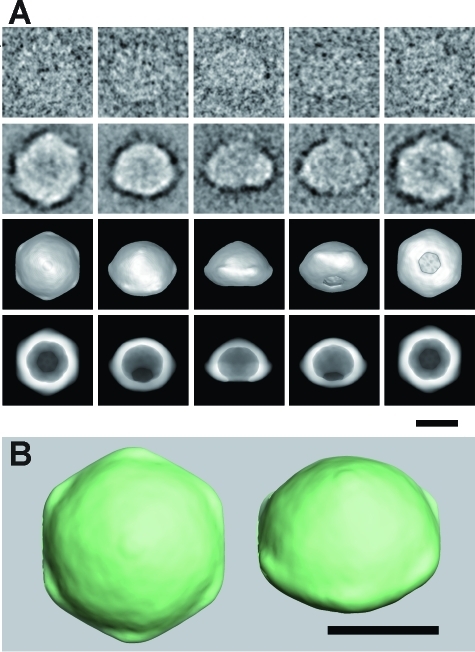
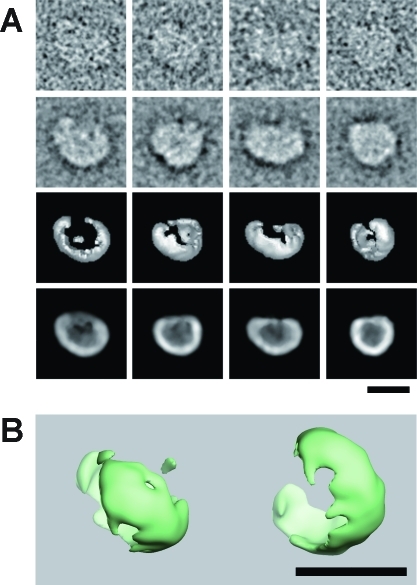
To understand the molecular shape of MG23, we reconstructed its 3D structure by the single-particle reconstruction technique. Purified recombinant MG23 protein was adsorbed to the grid, blotted with the filter paper, negatively stained, and recorded by EM. MG23 particles were selected from the digitized films using our program, and a total of 11175 images were picked up for 3D reconstruction (Figure 4). The surface representation demonstrates that MG23 is a bowl-shaped molecule, 12 nm in height, 16 nm in side length, and 17 nm diagonally at the widest transmembrane region. Because of the symmetry of the averaged top view (Figure 4A, left-most panel, second row), we applied 6-fold symmetry in this reconstruction. Projection from the final density map (Figure 4A, row 4) compares well with the corresponding raw particle images (Figure 4A, row 1), indicating that our 3D reconstruction is consistent with the data set. The presumed position of the membrane is indicated by the transparent blue band in Figure 3C, so that the volume ratio across the membrane fits to the topology model based on the hydropathicity and protein chemical data. In height, the transmembrane region of the bowl-shaped structure is ∼3 nm, the cytoplasmic domain extends ∼5 nm, and the luminal domain extends ∼4 nm. In the serial sections of this structure (Figure S2 of the Supporting Information), the thickness of the lateral wall is 28−35 Å in the transmembrane region. A plot of the Euler angles of 163 adopted class averages shows the nearly random orientation of MG23 on the grid surface (Figure S3A of the Supporting Information). According to the Fourier shell correlation (FSC) function, the resolution limit for the reconstruction is 2.0 nm by the correlation coefficient of 0.5 criterion (Figure S3B of the Supporting Information). The final reconstruction included 9172 particle images (82% of all the selected images).
Figure 4.
Reconstruction of the larger-sized MG23 particle. (A) First row, representative raw images; second row, corresponding class averages; third row, surface representations of the 3D reconstruction; fourth row, projections of the 3D reconstruction. Consistency among data sets is very high in size, shape, and inner structure, indicating successful 3D reconstruction from the original images of recombinant MG23 particles. (B) Surface representation of two MG23 structures from top (left) and oblique (right) views. Dimensions of the larger-sized bowl-shaped structure are 12 nm (height), 16 nm (wide side length), and 17 nm (diagonally at the widest transmembrane region). The scale bars are 10 nm.
3D Reconstruction of Smaller-Sized MG23 Particles
During the EM analysis described above, we realized that the recombinant MG23 preparations always contained considerable amounts of asymmetric particles that were relatively smaller than the bowl-shaped particle (Figure 3A). We also detected similar EM images showing the coexistence of the larger and smaller particles in native MG23 preparations purified from rabbit skeletal muscle (data not shown). Therefore, we also tried to reconstruct the 3D structure of the smaller-sized particles. A total of 1707 images of the smaller particles were collected from the digitized films and used for 3D reconstruction. Because no symmetry was observed in the averaged images or the raw images, we reconstructed the smaller-sized molecule assuming no intramolecular symmetry. The surface representation demonstrates that the smaller asymmetric structure has a roughly crescent-shaped appearance, 14 nm in height, 11 nm in side length, and 6 nm in narrow side length (Figure 5). The almost random orientation of the particles on the grid surface was also shown by a plot of the Euler angles of the 117 adopted class averages (Figure S3C of the Supporting Information). According to the FSC function, the resolution limit for the smaller-sized structure is 3.6 nm by the correlation coefficient of 0.5 criterion (Figure S3D of the Supporting Information). The final reconstruction included 1699 particle images (99% of all the selected images).
Figure 5.
Reconstruction of the smaller-sized MG23 particle. (A) First row, representative raw images; second row, corresponding class averages; third row, surface representations; fourth row, projections of the 3D reconstruction. (B) Dimensions of the smaller-sized crescent-shaped molecule are 14 nm (height), 11 nm (wide side length), and 6 nm (narrow side length). The massive bowl-shaped particle in Figure 3 is considered to be an assembly of multiples of this small subunit. The scale bars are 10 nm.
Considering that the MG23 preparations were subjected to the EM analysis after affinity purification and size fractionation using sucrose gradient centrifugation, which were shown to be highly purified by SDS−PAGE, the concomitance of the large and small particles likely suggests that MG23 is easily disassembled and reassembled during purification and storage. We considered that the smaller particles constitute a partial domain of the bowl-shaped assembly (see Discussion and Figure S4 of the Supporting Information).
MG23 Conducts Cations
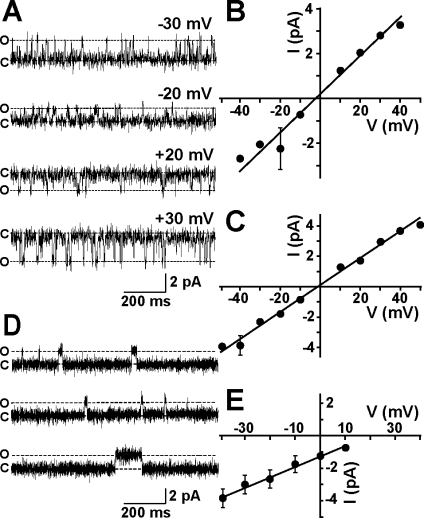
To investigate the possibility that MG23 plays a role in the transport of ions across the ER/SR, we reconstituted purified recombinant MG23 into planar lipid bilayers in symmetrical solutions of 260 mM KCl. Most incorporations into the bilayer consisted of the apparent insertion of multiple channels; however, on rare occasions, we observed what appeared to be opening events to a single consistent single-channel level. A representative experiment is shown in Figure 6A. The current−voltage relationship of these channel events (Figure 6B) yielded a conductance of 85.1 ± 5.3 pS (n = 3; mean ± standard deviation). A similar conductance of 88.2 ± 2.9 pS (n = 5) was observed in symmetrical solutions of 260 mM K-PIPES (Figure 6C), indicating that MG23 preferentially conducts cations.
Figure 6.
Single-channel behavior of MG23. (A) Representative single-channel recordings of native MG23 from rabbit skeletal muscle in solutions of symmetrical 260 mM KCl and 20 mM HEPES (pH 7.2). The holding potentials are indicated, and O and C represent the open and closed channel levels, respectively. (B) Single-channel current−voltage relationship of recombinant MG23 in symmetrical 260 mM KCl solutions (n = 3). (C) Single-channel current−voltage relationship of recombinant MG23 in symmetrical 260 mM K-PIPES solutions (n = 5). (D) Channel gating of recombinant MG23 with Ca2+ as the permeant ion. The bathing solutions contained 250 mM HEPES, 80 mM Tris (pH 7.2), and 15 μM free Ca2+ in the cis chamber and 250 mM glutamic acid and 10 mM HEPES (pH 7.2) with Ca(OH)2 (free Ca2+ concentration of 65 mM) in the trans chamber. The holding potential was 0 mV. Channel openings at holding potentials more positive than 10 mV could not be resolved. (E) Current−voltage relationship of MG23 under the conditions described in panel D (n = 5). The data represent means ± the standard deviation.
MG23 is expressed in intracellular membranes, including the SR/ER Ca2+-store membranes, and therefore, it is important to investigate if it is permeable to Ca2+. To test this, we incorporated recombinant MG23 into bilayers using solutions that are typically used to monitor Ca2+ currents through ryanodine receptor channels:(27) 250 mM Tris/HEPES and 15 μM Ca2+ on the cis side and 65 mM Ca2+ on the trans side. Under these conditions, we observed single-channel current fluctuations indicating that Ca2+ is flowing in the trans to cis direction through MG23 (Figure 6D). No channel activity was observed in solutions of symmetrical 250 mM Tris/HEPES, demonstrating that the channel is not permeable to Tris or HEPES (Figure S5 of the Supporting Information). The current−voltage relationship in these Ca2+-containing solutions exhibits a unitary conductance of 62.5 ± 10.2 pS (n = 5). Our observations therefore demonstrate that MG23 can form an ion channel that is permeable to both K+ and Ca2+. After incorporation of MG23 into bilayers, current−voltage relationships were obtained under bi-ionic conditions (cis, 210 mM K+; trans, 65 mM Ca2+). The reversal potential was calculated to be −6.9 ± −3.7 mV (n = 3) after correction for the junction potentials arising between the different solutions (Figure S6 of the Supporting Information). Using the Fatt−Ginsborg equation, the PCa2+/PK+ for MG23 was then calculated as 1.1 ± 0.2 (n = 3), which suggests that MG23 is approximately equally permeable to Ca2+ and K+.
Distinctive Characteristics and Voltage Dependence of MG23 Channel Gating
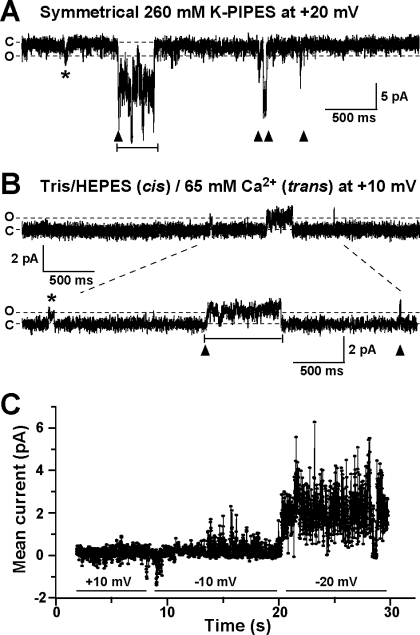
In the majority of experiments with MG23 preparations, multiple active channels were observed in the bilayer. This occurred even when we saw what appeared to be single fusion events of the liposomes into the bilayer. The presence of multiple channels and the frequent brief open events usually produced “messy-looking” or “flickery” single-channel recordings. Another distinctive characteristic of MG23 gating was the fact that multiple channels appeared to gate in unison so that many channels opened and closed together in a coupled or coordinated manner. Both the flickery gating and the coordinated channel openings can be seen in the two types of experiments: one in which K+ is the permeant ion (Figure 7A) and another in which Ca2+ is the permeant ion (Figure 7B). In both channel measurements, we observed single opening events (asterisks), coordinated gating of multiple channels (arrowheads), and long sojourns of coordinated gating (bar). We have not identified any tendency for a preferred number of MG23 channels to gate together in synchrony; this seems to be a random process. For example, two channels may open and close together (as in Figure 7B), or greater numbers of channels may gate in a synchronous fashion.
Figure 7.
Distinguishing gating characteristics of MG23. (A) Coordinated gating of recombinant MG23 channels in symmetrical solutions of 260 mM K-PIPES (pH7.2) at a holding potential of 20 mV. C indicates the zero current level (all channels closed), and O indicates the current level of a single MG23 channel opening; the dashed lines aid the visualization of these levels across the trace. An asterisk denotes a single MG23 channel opening. The arrowheads indicate where coordinated gating of multiple MG23 channels occurs, and the bar shows a long coordinated gating event. (B) Typical example of the coordinated MG23 channel gating in solutions where trans Ca2+ was the only permeant ion (250 mM Tris/HEPES, cis; 65 mM Ca2+, trans) at a holding potential of 10 mV. The dashed lines between the top and bottom traces indicate the portion of the trace that has been reproduced below on an expanded time scale to visualize the individual channel openings more clearly. (C) Noise analysis illustrates the voltage dependence of MG23 channels in symmetrical solutions of 260 mM K-PIPES (pH 7.2). As the holding potential became increasingly negative, K+ current across the bilayer became more elevated.
Because coordinated gating of multiple channels occurred randomly and gating was usually very rapid giving rise to flickering events, it was very difficult to measure the single-channel open probability (Po) of MG23 channels. To examine the voltage dependence of MG23 channel gating, we therefore used noise analysis (see Figure 7C for a typical example). At 10 mV (channel openings are downward deflections), Po was fairly low, although there were several coordinated channel opening events. At −10 mV (channel openings are upward deflections), there were more channel openings, and at −20 mV, the channels became very active and it was obvious that there were many channels present in the bilayer. The strong voltage dependence of MG23 is an important distinguishing feature of this channel and was observed in every experiment. Typically, channel activity was low around 0 mV but rapidly increased as the holding potential was made more negative. Holding potentials above −30 mV tended to break the bilayer because of the large currents.
Discussion
We successfully purified MG23 proteins from both rabbit muscle SR and recombinant yeast using affinity chromatography. The original EM images of the MG23 preparations themselves clearly suggested that the predominant large particles are morphologically identical in native and recombinant MG23 preparations. However, even after the size fractionation of highly purified MG23 by sucrose gradient centrifugation, MG23 particles were not uniform in size. In the MG23 preparations, the predominant component was the bowl-shaped particle with 6-fold symmetry (Figure 4), but we also detected the smaller crescent-shaped particles as the second most common component (Figure 5). The application of chemical cross-linkers to MG23 preparations reproducibly generated the homohexamer as a putative basic self-assembly unit (Figure 2). However, it is unlikely that these larger and smaller structures observed using EM represent hexameric and monomeric MG23, respectively. This is because the MG23 monomer at 23 kDa, composed of 210 amino acids, is too small for EM observation (for reference, see the images of 150 kDa mAbs in Figure 3B). We therefore propose that the crescent-shaped particle is a hexameric minimum structural subunit of MG23, and that multiple subunits subsequently constitute a larger assembly such as the bowl-shaped structure. With the available data, we could not determine the exact number of crescent-shaped subunits that would make up the full particle, but biochemical and structural analysis points to a number composed of two to six subunits. Fitting of more than six crescent-shaped subunits to the bowl-shaped structure seemed impossible. We present a docking of six crescent-shaped subunits as the most plausible model (Figure S4 of the Supporting Information). The 3D map of the crescent-shaped subunit (Figure 5B) is contoured at an isosurface containing a volume corresponding to 138 kDa (23 kDa × 6), and the bowl-shaped particle (Figure 4B) at 828 kDa (138 kDa × 6) using Imagic V,(19) assuming a hexagonal assembly of the crescent-shaped subunit. However, this simulation could not exclude other possibilities of subunit construction in the bowl-shaped particle.
MG23 is expressed in various cell types and localized in the SR/ER and nuclear membranes.(11) Our bilayer experiments demonstrate that recombinant MG23 prepared using DDM or NP-40 as a detergent forms cation channels displaying little difference in permeability between K+ and Ca2+. MG23 gating behavior is very unusual; the simultaneous openings (and therefore apparent coordination) of seemingly random numbers of MG23 channels were a consistent feature of the gating of this channel (Figure 7). As described above, particle image analysis implies that the crescent-shaped subunits are unstably assembled into the bowl-shaped particle that is equipped with a putative central pore. Although we can only speculate about how MG23 conducts ions, the unusual gating features could be caused by the proposed instability of the bowl-shaped particle. For example, MG23 may operate as a single conduction pathway with a central pore in the bowl-shaped assembly. During the disintegration and re-formation of the assembly, the proposed channel pore might display multiple conductance states and could pass huge currents superficially observed as simultaneous gating events. Alternatively, it may be possible that the crescent-shaped subunits form the unitary ion channel and can gate independently in the bilayer. In response to the structural transitions of the bowl-shaped assembly, the resulting conformational changes might affect ionic conduction through each crescent-shaped channel or could induce the simultaneous openings of the multiple-channel components. On the basis of our reconstructed 3D particle images, it is not possible to locate the ion conduction pathway of the MG23 channel. However, if MG23 is to follow the structural conventions of ion channels with known X-ray crystal structures, the apparent thickness at the transmembrane region of the crescent-shaped subunit (Figure S2 of the Supporting Information) suggests that it may be too narrow to form a unitary conduction pathway, hence favoring the idea of a single central pore in the bowl-shaped particle.
A novel ER/SR cation channel such as MG23, exhibiting no obvious selectivity for monovalent over divalent cations, would be expected to contribute significantly to the regulation of cellular Ca2+ homeostasis. There are many aspects of intracellular Ca2+ release that are not fully understood, and characterizing the biophysical properties of SR proteins such as MG23 will be important in resolving these issues. For example, electrogenic release of Ca2+ from and reuptake of Ca2+ into the ER/SR require synchronized counterion movements to balance the buildup of charge asymmetry across the ER/SR membrane,5,6,30,31 yet the molecular machinery responsible for the proposed counterion fluxes remains undefined. MG23 could play some part in that process. The purified MG23 channel displays a consistent voltage dependence of activation, indicating that it incorporates into the bilayer in a fixed orientation. SR vesicles also form in a fixed orientation(13) and are incorporated into artificial membranes in a fixed orientation such that the cytosolic side of SR ion channels always faces into the cis chamber.(32) Because our biochemical data (Figures 1−4) indicate that purified MG23, when reconstituted into liposomes, retains the same orientation as native MG23 in SR vesicles, it is most probable that the cytosolic side also faces into the cis chamber. Thus, in a cellular environment, given the voltage dependence of MG23 (Figure 7), it would be expected that during a Ca2+ release event, the ER/SR would become negative relative to the cytosol and this would reduce the Po of the MG23 channel, although it would not close completely. If voltage was the only controlling influence on Po, MG23 would tend to be more open during refilling of the ER/SR stores or just before a Ca2+ release event. MG23 may therefore contribute to the spontaneous Ca2+ leak that is known to occur from intracellular stores in many cell types.33−35 In cardiac cells, this is termed diastolic SR Ca2+ leak, and there is growing evidence that this is increased in disease states such as heart failure and leads to an increased likelihood of fatal cardiac arrhythmias.35−37 Indeed, in cardiac cells, it has been suggested that there must be another route for the release of Ca2+ from the SR during diastole apart from ryanodine receptors;(38) further investigation is required to determine whether MG23 is that pathway. On the other hand, the voltage dependence and sporadic gating behavior of MG23 suggest that it may play a more minor role in balancing charge asymmetry during a large and rapid Ca2+ release event (such as during a cardiac Ca2+ transient) than the recently characterized TRIC channels that are activated to high Po values as negative charge develops in the SR.(9) The full magnitude of MG23’s contribution to counterion currents, however, will not be realized until we have an improved understanding of whether the function of MG23 can be regulated by soluble ligands or by other mechanisms such as phosphorylation and nitrosylation.
There is recent evidence that MG23 is involved in the process that leads to cell death because apoptotic signaling is attenuated in mutant thymocytes prepared from MG23-knockout mice.(12) It is accepted that changes in the Ca2+ content and leak of intracellular stores can affect the sensitivity to apoptotic death,(39) but the molecular identity of the leak pathway(s) has not been identified. Our data, demonstrating that MG23 is permeable to monovalent and divalent cations, suggest that MG23 could fulfill that role. It is clear that much work is required to link the unique structural features of MG23 to its equally unique behavior as an ion channel. For example, does the transient assembly or breakdown of the bowl-shaped MG23 particle underlie the sporadic activity of multiple MG23 channels opening simultaneously, or does the production of MG23 bowls and their appearance in the ER membrane lead to the destruction of ionic gradients across the ER and immediate cell death? What is the role of MG23 in nuclear membranes because it is clearly present there in high levels? Again, to answer this question, we require a much more detailed understanding of the ion conducting properties of MG23 and of how MG23 channel gating can be regulated. Investigating the phenotype of the knockout mouse will also be vital in identifying the key physiological and pathophysiological roles of the MG23 protein.
Acknowledgments
We thank S. Aoki and M. Mio for technical assistance.
Glossary
Abbreviations
- 3D
three-dimensional
- DDM
n-dodecyl β-d-maltoside
- ER
endoplasmic reticulum
- EM
electron microscopy
- FSC
Fourier shell correlation
- mAb
monoclonal antibody
- MG23
mitsugumin 23
- SR
sarcoplasmic reticulum
Supporting Information Available
Detailed methodology (Supporting Experimental Procedures), purification and cross-linking of native MG23 (Figure S1), continuous sections through the bowl-shaped MG23 particle (Figure S2), Euler angle distribution and FSC function in the 3D reconstruction of MG23 particles (Figure S3), docking model of the crescent-shaped subunits into the bowl-shaped particle (Figure S4), MG23 is not permeable to Tris and HEPES (Figure S5), and MG23 PCa2+/PK+ data (Figure S6). This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
E.V., K.M., and M.N. contributed equally to this work.
This work was supported in part by research grants from the Japan Society for the Promotion of Science, the Ministry of Education, Culture, Sports, Science and Technology, the Japan New Energy and Industrial Technology Development Organization, the British Heart Foundation, the Astellas Foundation, the Mochida Memorial Foundation, the Kanae Foundation, the Daiichi-Sankyo Foundation of Life Science, and the Takeda Science Foundation.
Supplementary Material
References
- Berridge M. J. (2002) The endoplasmic reticulum: A multifunctional signaling organelle. Cell Calcium 32, 235–249. [DOI] [PubMed] [Google Scholar]
- MacLennan D. H.; Abu-Abed M.; Kang C.-H. (2002) Structure-function relationships in Ca2+ cycling proteins. J. Mol. Cell. Cardiol. 34, 897–918. [DOI] [PubMed] [Google Scholar]
- Ron D.; Walter P. (2007) Signal integration in the endoplasmic reticulum unfolded protein respose. Nat. Rev. Mol. Cell Biol. 8, 519–529. [DOI] [PubMed] [Google Scholar]
- Kim I.; Xu W.; Reed J. C. (2008) Cell death and endoplasmic reticulum stress: Disease relevance and therapeutic opportunity. Nat. Rev. Drug Discovery 7, 1013–1030. [DOI] [PubMed] [Google Scholar]
- Meissner G. (1983) Monovalent ion and calcium ion fluxes in sarcoplasmic reticulum. Mol. Cell. Biochem. 55, 65–82. [DOI] [PubMed] [Google Scholar]
- Floyd R.; Wray S. (2007) Calcium transporters and signalling in smooth muscle. Cell Calcium 42, 467–476. [DOI] [PubMed] [Google Scholar]
- Yazawa M.; Ferrante C.; Feng J.; Mio K.; Ogura T.; Zhang M.; Lin P.-H.; Pan Z.; Komazaki S.; Kato K.; Nishi M.; Zhao X.; Weisleder N.; Sato C.; Ma J.; Takeshima H. (2007) TRIC channels are essential for Ca2+ handling in intracellular stores. Nature 448, 78–82. [DOI] [PubMed] [Google Scholar]
- Yamazaki D.; Komazaki S.; Nakanishi H.; Mishima A.; Nishi M.; Yazawa M.; Yamazaki T.; Taguchi R.; Takeshima H. (2009) Essential role of TRIC-B channel in Ca2+-handling of alveolar epithelium and perinatal lung maturation. Development 136, 2355–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt S. J.; Park K.-H.; Nishi M.; Urashima T.; Aoki S.; Yamazaki D.; Ma J.; Takeshima H.; Sitsapesan R. (2010) Charade of the SR K+-channel: Two ion-channels, TRIC-A and TRIC-B, masquerade as a single K+-channel. Biophys. J. 99, 417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X.; Yamazaki D.; Park K.-H.; Komazaki S.; Tjondrokoesoemo A.; Nishi M.; Lin P.; Hirata Y.; Brotto M.; Takeshima H.; Ma J. (2010) Ca2+ overload and sarcoplasmic reticulum instability in tric-a null skeletal muscle. J. Biol. Chem. 285, 37370–37376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi M.; Komazaki S.; Iino M.; Kangawa K.; Takeshima H. (1998) Mitsugumin 23, a novel transmembrane protein on endoplasmic reticulum and nuclear membranes. FEBS Lett. 432, 191–196. [DOI] [PubMed] [Google Scholar]
- Yamazaki T.; Sasaki N.; Nishi M.; Takeshima H. (2010) Facilitation of DNA damage-induced apoptosis by endoplasmic reticulum protein mitsugumin 23. Biochem. Biophys. Res. Commun. 392, 196–200. [DOI] [PubMed] [Google Scholar]
- Saito A.; Seiler S.; Chu A.; Fleischer S. (1984) Preparation and morphology of sarcoplasmic reticulum terminal cisterae from rabbit skeletal muscle. J. Cell Biol. 99, 875–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.; Yamazaki T.; Yazawa M.; Treves S.; Nishi M.; Murai M.; Shibata E.; Zorzato F.; Takeshima H. (2007) Calumin, a novel Ca2+-binding transmembrane protein on the endoplasmic reticulum. Cell Calcium 42, 83–90. [DOI] [PubMed] [Google Scholar]
- Feramisco J. D.; Goldstein J. L.; Brown M. S. (2004) Membrane topology of human insig-1, a protein regulator of lipid synthesis. J. Biol. Chem. 279, 8487–8496. [DOI] [PubMed] [Google Scholar]
- Mio K.; Ogura T.; Kiyonaka S.; Hiroaki Y.; Tanimura Y.; Fujiyoshi Y.; Mori Y.; Sato C. (2007) The TRPC3 channel has a large internal chamber surrounded by signal sensing antennas. J. Membr. Biol. 367, 373–383. [DOI] [PubMed] [Google Scholar]
- Ogura T.; Sato C. (2004) Automatic particle pickup method using a neural network has high accuracy by applying an initial weight derived from eigenimages: A new reference free method for single-particle analysis. J. Struct. Biol. 145, 63–75. [DOI] [PubMed] [Google Scholar]
- Ogura T.; Sato C. (2004) Auto-accumulation method using simulated annealing enables fully automatic particle pickup completely free from a matching template or learning data. J. Struct. Biol. 146, 344–358. [DOI] [PubMed] [Google Scholar]
- van Heel M.; Gowen B.; Matadeen R.; Orlova E. V.; Finn R.; Pape T.; Cohen D.; Stark H.; Schmidt R.; Schatz M.; Patwardhan A. (2000) Single-particle cryo-electron microscopy: Towards atomic resolution. Q. Rev. Biophys. 33, 307–369. [DOI] [PubMed] [Google Scholar]
- Frank J. (2006) Three-dimensional electron microscopy of macromolecular assembles: Visualization of biological molecules in their native state, Oxford University Press, New York. [Google Scholar]
- Harauz G.; van Heel M. (1986) Exact filters for general geometry 3-dimensional reconstruction. Optik 73, 146–156. [Google Scholar]
- Ogura T.; Iwasaki K.; Sato C. (2003) Topology representing network enables highly accurate classification of protein images taken by cryo-electron-microscope without masking. J. Struct. Biol. 143, 185–200. [DOI] [PubMed] [Google Scholar]
- Bretaudiere J. P.; Frank J. (1986) Reconstitution of molecule images analysed by correspondence analysis, a tool for structural interpretation. J. Microsc. (Oxford, U.K.) 144, 1–14. [DOI] [PubMed] [Google Scholar]
- Ogura T.; Sato C. (2006) A fully automatic 3D reconstruction method using simulated annealing enables accurate posterioric angular assignment of protein projections. J. Struct. Biol. 158, 371–386. [DOI] [PubMed] [Google Scholar]
- Penczek P.; Radermacher M.; Frank J. (1992) Three-dimensional reconstruction of single particles embedded in ice. Ultramicroscopy 40, 33–53. [PubMed] [Google Scholar]
- Penczek P. A.; Grassucci R. A.; Frank J. (1994) The ribosome at improved resolution: New techniques for merging and orientation refinement in 3D cryo-electron microscopy of biological particles. Ultramicroscopy 53, 251–270. [DOI] [PubMed] [Google Scholar]
- Sitsapesan R.; Montgomery R. A.; MacLeod K. T.; Williams A. J. (1991) Sheep cardiac sarcoplasmic reticulum calcium-release channels: Modification of conductance and gating by temperature. J. Physiol. 434, 469–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatt P.; Ginsborg B. L. (1958) The ionic requirements for the production of action potentials in crustacean muscle fibres. J. Physiol. 142, 516–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. (1992) Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225, 487–494. [DOI] [PubMed] [Google Scholar]
- Somlyo A. P.; Somlyo A. V. (1986) Electron probe analysis of calcium content and movements in sarcoplasmic reticulum, endoplasmic reticulum, mitochondria, and cytoplasm. J. Cardiovasc. Pharmacol. 8, S42–S47. [DOI] [PubMed] [Google Scholar]
- Somlyo A. P.; Somlyo A. V.; Shuman H.; Endo M. (1982) Calcium and monovalent ions in smooth muscle. Fed. Proc. 41, 2883–2890. [PubMed] [Google Scholar]
- Sitsapesan R.; Williams A. J. (1994) Gating of the native and purified cardiac SR Ca2+-release channel with monovalent cations as permeant species. Biophys. J. 67, 1484–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giunti R.; Gamberucci A.; Fulceri R.; Bánhegyi G.; Benedetti A. (2007) Both translocon and a cation channel are involved in the passive Ca2+ leak from the endoplasmic reticulum: A mechanistic study on rat liver microsomes. Arch. Biochem. Biophys. 462, 115–121. [DOI] [PubMed] [Google Scholar]
- Lomax R. B.; Camello C.; van Coppenolle F.; Petersen O. H.; Tepikin A. V. (2002) Basal and physiological Ca2+ leak from the endoplasmic reticulum of pancreatic acinar cells. Second messenger-activated channels and translocons. J. Biol. Chem. 277, 26479–26485. [DOI] [PubMed] [Google Scholar]
- Eisner D. A.; Kashimura T.; O’Neill S. C.; Venetucci L. A.; Trafford A. W. (2009) What role does modulation of the ryanodine receptor play in cardiac inotropy and arrhythmogenesis?. J. Mol. Cell. Cardiol. 46, 474–481. [DOI] [PubMed] [Google Scholar]
- Wehrens X. H.; Lehnart S. E.; Marks A. R. (2005) Intracellular calcium release and cardiac disease. Annu. Rev. Physiol. 67, 69–98. [DOI] [PubMed] [Google Scholar]
- Benkusky N. A.; Farrell E. F.; Valdivia H. H. (2004) Ryanodine receptor channelopathies. Biochem. Biophys. Res. Commun. 322, 1280–1285. [DOI] [PubMed] [Google Scholar]
- Santiago D. J.; Curran J. W.; Bers D. M.; Lederer W. J.; Stern M. D.; Ríos E.; Shannon T. R. (2010) Ca sparks do not explain all ryanodine receptor-mediated SR Ca leak in mouse ventricular myocytes. Biophys. J. 98, 2111–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinton P.; Giorgi C.; Siviero R.; Zecchini E.; Rizzuto R. (2008) Calcium and apoptosis: ER-mitochondria Ca2+ transfer in the control of apoptosis. Oncogene 27, 6407–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.