Abstract
Signal recognition particle (SRP) targets proteins for co-translational insertion through or into the endoplasmic reticulum membrane. Mammalian SRP slows nascent chain elongation by the ribosome during targeting in vitro. This ‘elongation arrest’ activity requires the SRP9/14 subunit of the particle and interactions of the C-terminus of SRP14. We have purified SRP from Saccharomyces cerevisiae and demonstrated that it too has elongation arrest activity. A yeast SRP containing Srp14p truncated at its C-terminus (ΔC29) did not maintain elongation arrest, was substantially deficient in promoting translocation and interfered with targeting by wild-type SRP. In vivo, this mutation conferred a constitutive defect in the coupling of protein translation and translocation and temperature-sensitive growth, but only a slight defect in protein translocation. In combination, these data indicate that the primary defect in SRP ΔC29 is in elongation arrest, and that this is a physiologically important and conserved function of eukaryotic SRP.
Keywords: elongation arrest/SRP/targeting/translation/yeast
Introduction
Signal recognition particle (SRP) is the most widespread dedicated protein targeting factor, with homologues in all three kingdoms (Walter and Johnson, 1994; Lütcke, 1995; Bui and Strüb, 1999). It plays a crucial role in ensuring targeting of presecretory and membrane proteins to the eukaryotic endoplasmic reticulum (ER), bacterial cytoplasmic and chloroplast thylakoid membranes. Canine SRP, the first identified, was purified on the basis of its translocation-promoting activity (Walter and Blobel, 1980). It binds tightly to ribosome–nascent chain complexes in which signal sequences of ER-targeted proteins are just exposed (Siegel and Walter, 1988a). This binding results in a slowing of translation, a phenomenon termed ‘elongation arrest’ (Walter and Blobel, 1981). This is proposed to increase the window of opportunity for the SRP–ribosome–nascent chain complex to interact with the SRP receptor in the ER membrane, the next step towards docking the ribosome on to the translocation apparatus and establishment of co-translational translocation.
SRP is a ribonucleoprotein consisting, in higher eukaryotes, of the 7SL RNA and six proteins. All subunits are required for full activity of SRP, but particular activities have been ascribed to individual components (Siegel and Walter, 1985, 1988b; Krieg et al., 1986; Kurzchalia et al., 1986). Elongation arrest requires the SRP9 and SRP14 proteins that bind as a heterodimer to the 5′ and 3′ ends of the 7SL RNA in the ‘Alu-domain’ of the particle (Strüb et al., 1991). Recent work (Birse et al., 1997) has elaborated the three-dimensional structure of SRP9/14; the complex is symmetrical, the two proteins adopting the same fold despite dissimilar primary sequences. SRP lacking the Alu-domain or just SRP9/14 can promote translocation but lacks elongation arrest activity (Siegel and Walter, 1985, 1986). These particles also have reduced affinity for ribosomes lacking signal sequences (Hauser et al., 1995; Powers and Walter, 1996). In contrast, SRP reconstituted with an SRP14 lacking 20 C-terminal amino acids (ΔC20) lacks elongation arrest activity but retains its affinity for ribosomes (Thomas et al., 1997). This mutant thus defines an elongation arrest-specific SRP–ribosome interaction.
Elongation arrest was first observed as a halt in translation of signal sequence-containing proteins by wheat germ ribosomes mediated by canine SRP (Walter and Blobel, 1981). In a mammalian translation system, however, canine SRP only delays production of full-length translation products (Wolin and Walter, 1989). All elongation arrest-deficient SRP derivatives have decreased translocation-promoting activity (Siegel and Walter, 1985, 1986; Hauser et al., 1995; Thomas et al., 1997), suggesting that elongation arrest is important for full SRP activity. However, mammalian SRP is the only eukaryotic SRP studied and it is therefore unknown whether elongation arrest is peculiar to it, or is a conserved function. As all studies have been undertaken in vitro, the physiological importance of elongation arrest has not been addressed.
Here we examine functions and interactions of SRP in the yeast Saccharomyces cerevisiae, an organism amenable to biochemical and genetic analysis. SRP is not essential for growth of yeast cells, although it is required for efficient ER targeting of proteins that contain strongly hydrophobic signal sequences (Ng et al., 1996), and in its absence yeast cells grow poorly (Hann and Walter, 1991). Yeast SRP contains at least six proteins (Srp72p, Srp68p, Srp54p, Sec65p, Srp21p and Srp14p), and its RNA is scR1 (Hann and Walter, 1991; Hann et al., 1992; Stirling and Hewitt, 1992; Brown et al., 1994). The proteins are homologous to those of mammalian SRP, with one exception, Srp21p. No Srp9p has been identified in yeast SRP, although a 7 kDa protein was found in fractions containing purified yeast SRP proteins (Brown et al., 1994). Recently, recombinant yeast Srp14p was shown to homodimerize and bind to RNAs containing the 5′ portion of scR1 (Strüb et al., 1999), suggesting a novel Alu-domain structure.
Here we present purification of active yeast SRP, the first characterization of a eukaryotic SRP other than the canine particle. It contains only the six proteins previously determined to be in it and no SRP9 homologue. However, it has two Srp14p subunits. Yeast SRP has elongation arrest activity, and interactions of the C-terminus of Srp14p, crucial for elongation arrest in mammalian SRP, are vital for this activity in this particle both in vitro and in vivo. Elongation arrest is therefore a conserved, functionally important feature of eukaryotic SRP.
Results
Purification of yeast SRP
We constructed a yeast strain expressing Srp72p extended at its C-terminus by a cleavage site for the tobacco etch virus (TEV) protease and two copies of the protein A immunoglobulin (Ig) binding domain (see Materials and methods). The modified protein fully complemented a lack of endogenous Srp72p and we isolated SRP from a post-ribosomal extract of this strain on IgG–Sepharose, eluting it with TEV protease. One further purification step yielded fractions (Figure 1A, lanes 8 and 9) that contained almost exclusively proteins of the sizes of known yeast SRP components, and immunoblotting confirmed that they were Srp72p, Srp68p, Srp54p, Sec65p, Srp21p and Srp14p (data not shown). Purified SRP did not contain a 7 kDa band as found previously in pools of SRP proteins (Brown et al., 1994). This, and the lack of a yeast open reading frame encoding an SRP9 homologous protein, indicates that yeast SRP does not contain SRP9. We also purified SRP containing Srp14p lacking the last 29 amino acids (ΔC29; Figure 1B). This truncation mimics the ΔC20 mutation, which dissociates ribosome binding and elongation arrest activities of mammalian SRP14 (Thomas et al., 1997), and we hoped that it would provide insight into these functions of the yeast particle.
Fig. 1. Purification of yeast SRP. (A) SRP was purified as described in Materials and methods, fractions resolved on a 10% Proseive (FMC Bioproducts) gel and stained with Coomassie Blue. Lanes 1 and 2: post-ribosomal extract and IgG–Sepharose flowthrough (1/16 000); lanes 3–5: TEV eluate, ω-aminobutyl agarose flowthrough and wash (1/80); lanes 6–12: elutions (1/20). Marker and SRP proteins are indicated. (B) Purification of SRP ΔC29 as in (A). ΔC29 contains an N-terminal triple-HA tag and thus runs above Srp21p. (C) RNA analysis. RNA isolated from purified SRP was run on a 6% acrylamide–50% urea gel and stained with ethidium bromide. Lane 1: pBR322 MspI digest markers; lanes 2 and 3: RNA from fractions shown in lanes 8 of (A) and (B), respectively (1/20).
Analysis of purified SRP for RNA revealed a prominent species (Figure 1C, lanes 2 and 3) of a size consistent with the 519 nucleotide length of scR1 (Felici et al., 1989; Hann and Walter, 1991). Northern blotting confirmed that it was scR1 (data not shown).
Purified yeast SRP is active
We examined the ability of purified SRP to promote translocation of SRP-dependent substrates into ER-derived microsomes. The synthetic mRNA species used to programme these translations encoded the SRP- dependent chimeric protein DHCαF (Ng et al., 1996) and prepro-α-factor (ppαF), which is targeted to the yeast ER independently of SRP. We also used preprolactin (ppL), used extensively in characterization of canine SRP. The ppL used contains a single amino acid substitution, glycine–alanine, at amino acid 10. This increases the efficiency of secretion of a protein containing the ppL signal sequence from yeast (Ngsee and Smith, 1990) and the translocation of ppL across the yeast ER (J.D.Brown, unpublished data). The identity of translocated species produced from these substrates was verified by virtue of their being protected from digestion by protease (Figure 2A).
Fig. 2. Activity of purified yeast SRP. (A) Confirmation of translocated species by protease protection. Translation reactions were carried out with or without addition of microsomal membranes (yRM) and subsequently incubated with 0.5 mg/ml proteinase K where indicated. (B and C) Translation reactions were carried out using extract containing (+) or immunodepleted of (–) SRP. yRM and purified SRP were added as indicated (concentrations of SRP in nanomoles). SRP was wild type except in (B) lane 13 and (C) lane 9, where SRP ΔC29 was used. (D) Translation reactions contained non-depleted extract supplemented with yRM and SRP (100 nM) as indicated. Reactions were run on 15% SDS–polyacrylamide gels and visualized by autoradiography. Untranslocated (ppL, ppαF and DHCαF) and processed, translocated (pL, gpαF and gDHCαF) species are indicated. Graphs were plotted using averaged data from three independent experiments. The translocation with non-depleted extract was set as 100%, bars, 1 SD.
SRP-depleted cytosol (Ng et al., 1996) was substantially defective in promoting translocation of DHCαF and ppL (Figure 2B, lane 7 and C, lane 3, respectively) but translocation of ppαF was unaffected (Figure 2B, lane 3). Addition of purified SRP restored translocation of DHCαF and ppL in a dose-dependent manner (Figure 2B, lanes 8–12 and C, lanes 4–8, and graphs) and it was thus active. Immunoblotting and comparison with known amounts of purified SRP allowed us to estimate the concentration of SRP in translation reactions containing non-depleted cytosol to be 10 nM (data not shown). Addition of 10 nM SRP to depleted cytosol restored translocation to ∼75% of that seen with non-depleted extract (Figure 2B and C graphs), and purification had therefore not substantially impaired the activity of SRP.
SRP containing ΔC29 had less translocation-promoting activity than the wild-type particle. Addition of 100 nM mutant particle to SRP-depleted cytosol resulted in translocation of DHCαF or ppL similar to that achieved with 2–5 nM of the wild-type particle (Figure 2B and C, lanes 11–13 and 7–9, respectively). One interpretation of this result is that the mutant particle was largely defective in binding ribosome–nascent chain complexes. We therefore performed experiments (Figure 2D) in which we added either wild-type or ΔC29 SRP to reactions that contained non-depleted cytosol. Wild-type SRP increased translocation of DHCαF and ppL (Figure 2D, lanes 4 and 8 and graph). However, the mutant SRP decreased translocation of these proteins (lanes 3 and 7) but not ppαF (data not shown). The simplest explanation of this result is that the mutant SRP competed with the endogenous SRP for binding available ribosome–nascent chain complexes, but in most cases failed to target them efficiently to the membrane. Thus, the SRP ΔC29 is not grossly deficient in ribosome–nascent chain binding.
Yeast SRP has elongation arrest activity
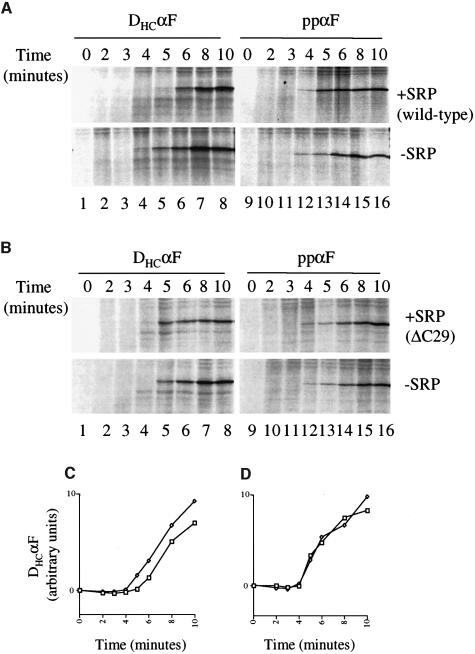
An explanation for the reduced translocation-promoting activity of the ΔC29 mutant is that it lacks elongation arrest activity and therefore cannot retain nascent chains in a short translocation competent state. We tested yeast SRP for elongation arrest activity in time-course experiments comparing the kinetics of production of full-length translation product in the presence or absence of added SRP. We used DHCαF as an SRP-dependent substrate and ppαF as a negative control. The comparison between these two substrates was particularly useful as they are of similar length and composition, differing chiefly in the signal sequence (Ng et al., 1996). Wild-type SRP reproducibly delayed production of full-length DHCαF by 1–2 min; it was just detectable at 4 min without addition of SRP, and at 6 min when SRP was added (Figure 3A, compare lanes 4, 5 and 6). In contrast, full-length ppαF was produced with similar kinetics whether SRP was added or not, appearing after 4 min (Figure 3A, lane 12). In equivalent experiments with SRP containing ΔC29 (Figure 3B), we detected no delay in the production of either DHCαF or ppαF, suggesting that this mutant was indeed defective in elongation arrest. Quantification and averaging of data from three experiments (Figure 3C and D) confirmed that addition of wild-type SRP, but not SRP containing ΔC29, delayed the initial appearance of full-length DHCαF, but not its rate of accumulation.
Fig. 3. Yeast SRP mediates elongation arrest. (A and B) In vitro translation reactions were carried out as in Figure 2 using mRNAs encoding DHCαF or ppαF (left and right panels, respectively) with or without 100 nM added SRP. SRP was wild type (A) or ΔC29 (B). Samples were stopped at the times indicated. All panels are from the same experiment, and are equivalent exposures processed identically. (C and D) Quantified data from three experiments performed with DHCαF were averaged and plotted. SRP was wild type (C) or ΔC29 (D). Squares, plus SRP; diamonds, without added SRP.
To demonstrate further the significance of the effect of SRP on translation, we carried out 40 translation reactions programmed with DHCαF mRNA, of which half contained additional SRP. For each condition 10 reactions were stopped at 7 min, when the reaction is in its linear phase of product accumulation and 10 were stopped at 30 min when the maximum amount of product has been produced (data not shown). Quantification of reaction products and analysis by t-test confirmed that at 7 min there was a significant (98% confidence level) difference in the levels of product between reactions, which were lower with added SRP than without it (see Materials and methods). At 30 min there was no significant difference, and SRP had no effect on the total product made. Analogous experiments using mRNA encoding ppαF revealed no statistically significant difference in the amount of translation product in the presence of additional SRP. Thus, yeast SRP does indeed promote elongation arrest during translation of DHCαF mRNA. In equivalent experiments using SRP containing ΔC29 we could, as in the single time-course experiment, detect no difference in the translation of either DHCαF or ppαF.
Yeast SRP contains two Srp14ps
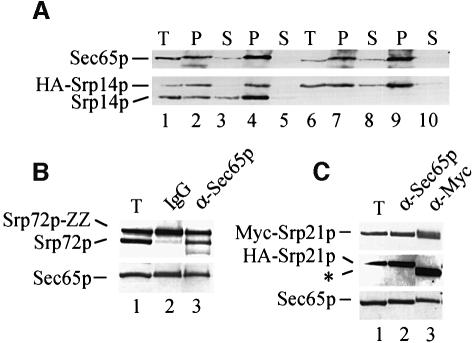
Conservation of elongation arrest but lack of SRP9 in yeast SRP prompted us to investigate its composition. In particular we analysed the copy number of Srp14p and other proteins in the particle, using strains carrying genes encoding two different copies of the protein of interest. For Srp14p we used a strain containing a fully functional (see Figures 5–7) triple haemagglutinin (HA) epitope-tagged Srp14p and the untagged protein. Immunoprecipitation of the two Srp14p variants with antibodies against Sec65p from an extract of this strain indicated that they were both efficiently assembled into SRP (Figure 4A, lanes 4 and 5). Immunoprecipitation with anti-HA antibodies, under conditions where almost all the tagged protein was bound by antibody, resulted in co-isolation of approximately half the wild-type protein (Figure 4A, lanes 2 and 3). This is consistent with the particle containing two Srp14p proteins, the two variants associating randomly, resulting in a 1:2:1 ratio of tagged only:tagged plus untagged: untagged only. SRP was not immunoprecipitated with anti-HA antibodies when there was no tagged Srp14p in the extract (data not shown). Similar experiments with extracts from strains carrying two Srp72p or Srp21p variants did not result in co-isolation of both copies of either protein with reagents that bound only one (Figure 4B and C, lane 2). Thus, the particle is monomeric in cell extracts and contains one copy of each of these proteins. Taken together with the studies of Strüb et al. (1999), which demonstrated in vitro binding of a dimer of recombinant Srp14p to scR1 RNA, the conclusion is that the protein composition of the Alu-domain of yeast SRP differs from the equivalent domain of mammalian SRP, most likely containing a homodimer of Srp14p.
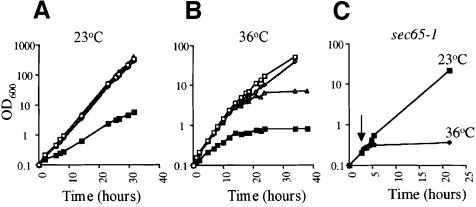
Fig. 5. Truncation of Srp14p results in delayed onset temperature sensitivity. (A and B) Strains expressing HA-Srp14p (open squares), DC11 (diamonds), ΔC29 (triangles) or lacking Srp14p (squares with crosses) were grown in liquid culture at either 23 (A) or 36°C (B). (C) sec65-1 cells were grown at 23°C and half transferred to 36°C (arrow).
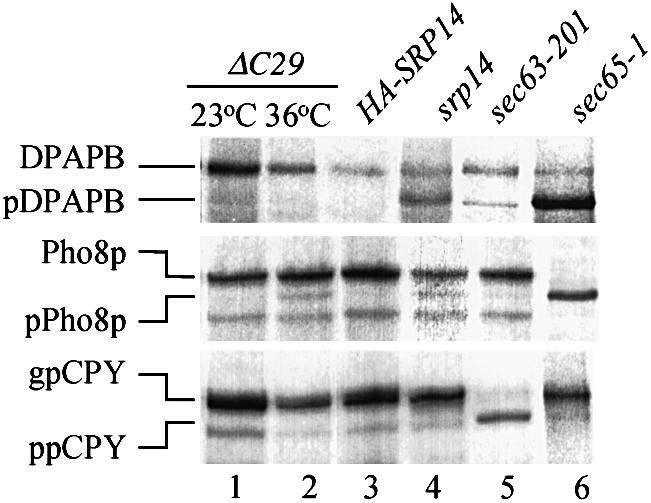
Fig. 7. Cells expressing ΔC29 have slight translocation defects for Pho8p, but not DPAPB or CPY, at 36°C. Immunoprecipitations were carried out with antibodies against the indicated proteins from extracts of cells pulse-labelled for 6 min (see Materials and methods). Immunoprecipitates were electrophoresed through 10% Proseive gels and visualized by autoradiography. Labelling was at 23°C (srp14-ΔC29, srp14), 30°C (sec63-201) or following transfer from 23 to 36°C for 30 min (sec65-1) or 16 h (srp14-ΔC29, HA-SRP14). Dipeptidyl aminopeptidase B: DPAPB, glycosylated luminal; pDPAPB, cytoplasmic. Alkaline phosphatase: Pho8p, glycosylated luminal; pPho8p, cytoplasmic. Carboxypeptidase Y: gpCPY, glycosylated lumenal; ppCPY, cytoplasmic. In this experiment a band appeared in the sec63-201 sample at the position of pDPAPB. This was not seen in other experiments.
Fig. 4. Yeast SRP contains two Srp14ps. (A) Extracts of strains expressing either wild-type and HA-Srp14p (lanes 1–5) or just HA-Srp14p (lanes 6–10) were immunoprecipitated with anti-HA (lanes 2 and 7) or anti-Sec65p antibodies (lanes 4 and 9). Material corresponding to 50% used in each immunoprecipitation (lanes 1 and 6) and of each supernate (lanes 3, 5, 8 and 10) and all immunoprecipitated material were analysed. (B and C) As (A) except that strains used expressed protein A-tagged Srp72p and wild-type Srp72p (B) or Myc-Srp21p and HA-Srp21p (C). Proteins were isolated using IgG–Sepharose or the antibody indicated. *, Ig chains recognized by secondary antibodies.
Cells expressing Srp14p-ΔC29 are temperature sensitive
Our in vitro data are consistent with the primary defect of the ΔC29 mutant being in elongation arrest. The competition experiments (Figure 2D) indicated that it bound ribosome–nascent chain complexes, but we could not exclude the possibility that it was partially defective in this activity. However, since strains expressing the mutant grew well in culture at 30°C (data not shown), readily allowing us to produce sufficient cells for purification of the particle, we considered that the severe targeting defects of the purified ΔC29 particle in vitro may be an exaggeration of the in vivo phenotype. We therefore took advantage of the yeast system and studied the mutant in vivo to see if we could obtain a more accurate measure of its defects.
Srp14-ΔC29 cells grew poorly on plates at 36°C (data not shown) and we tested their growth in liquid culture. At 23°C, srp14-ΔC29 cells grew at the same rate as wild type and those expressing full-length HA-tagged Srp14p, but slowed and stopped growing after ∼16 h at 36°C (Figure 5A and B and data not shown). This contrasted dramatically with the almost immediate growth defect of the only previously published thermo-sensitive SRP mutant, sec65-1 (Figure 5C). However, a yeast strain carrying an srp14 deletion showed a similar growth profile to srp14-ΔC29 cells (Figure 5B). This conditional growth defect of yeast lacking an SRP component had not been noted previously, and to ensure that this was not specific to srp14 mutants we examined growth of strains lacking other SRP subunits. These strains were similarly temperature sensitive, ceasing growth at 36°C after 16–20 h (data not shown).
ΔC29 is stable and SRP associated
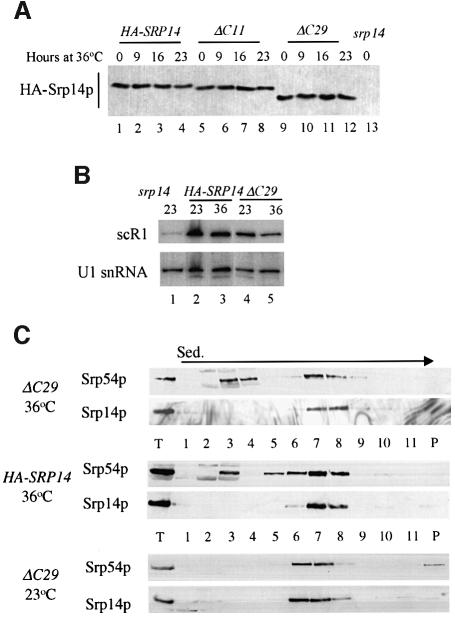
Next we studied the amount and integrity of SRP in ΔC29 cells. This would determine whether the slowing of growth of srp14-ΔC29 cells at 36°C was due to newly synthesized SRP or ΔC29 protein being unstable, resulting in decreased SRP activity over time. We compared the level of Srp14p-ΔC29 with full-length protein and that of a truncation, ΔC11, which lacks a yeast-specific extension to Srp14p but does not affect growth (Figure 5A and B). The amounts of the various Srp14ps did not change dramatically after transfer to 36°C, even beyond times at which growth of srp14-ΔC29 cells was seriously affected (Figure 6A, lanes 1–12). Srp14p-ΔC29 was therefore stable. Next, we examined scR1 since lack of Srp14p results in a dramatic decrease (10- to 20-fold) in its steady-state level (Brown et al., 1994). The amount of scR1 RNA remained relatively constant at 36°C (Figure 6B, lanes 2–5), suggesting that ΔC29 was not only present but also associated with SRP at 36°C, and we tested this directly by performing sucrose density gradient sedimentation on cell extracts. In extracts of srp14-ΔC29 cells grown at 23 or 36°C for 16 h (when growth begins to slow), virtually all the Srp14p co-sedimented with another SRP protein, Srp54p (Figure 6C). Thus, the growth defect of srp14-ΔC29 cells cannot be accounted for by loss of SRP, but is rather a consequence of a functional defect in the mutant particle.
Fig. 6. SRP containing ΔC29 is stable at 36°C. (A and B) Protein or RNA samples were prepared from cells expressing Srp14p variants grown at 23°C or following transfer to 36°C for the time indicated. (A) Equal amounts of protein samples (assessed by Bradford assay) were analysed by imunoblotting. (B) Equal amounts of RNA (assessed by measuring OD260) were denatured, electrophoresed through a 6% acrylamide–50% urea gel, blotted on to a nylon membrane and hybridized sequentially with 32P-labelled DNA fragments of SCR1 and SNR19. 36°C samples were taken after 16 h growth at this temperature. (C) Whole-cell extracts from strains grown at the temperatures indicated for 16 h were prepared and fractionated on 10–30% w/v sucrose gradients. One quarter of each gradient fraction and 1/10th load were analysed for SRP proteins. In this experiment, SRP in srp14-ΔC29 strain extracts sedimented at slightly different rates. This was not seen in other experiments.
Translocation defects associated with ΔC29
We examined srp14-ΔC29 cells for defects in targeting to the ER membrane. If the decreased translocation in in vitro assays containing the ΔC29 particle were due to overall decreased activity then we expected this to be revealed in vivo through the persistence of cytosolic forms of SRP-targeted proteins lacking ER-specific signal sequence cleavage and/or glycosylation. We pulse-labelled srp14-ΔC29 cells with [35S]methionine/cysteine following growth at either 23 or 36°C for 16 h, and immunoprecipitated proteins from cell lysates (see Materials and methods). srp14-ΔC29 cells had no defects for any of the substrates tested at 23°C (Figure 7, lane 1), only ER-modified forms being visible. At 36°C a slight defect was revealed in the translocation of the SRP-dependent substrate Pho8p compared with cells expressing full-length HA-tagged Srp14p, a small amount of non-glycosylated precursor being visible (Figure 7, compare lanes 2 and 3). We did not, however, see a defect for DPAPB, typically severely affected in cells compromised for SRP activity. Control strains gave the expected results. Thus, the sec65-1 mutant revealed a virtually complete failure to translocate both Pho8p and DPAPB (Figure 7, lane 6), and an srp14-null strain revealed partial translocation defects for these proteins (Figure 7, lane 4). Such attenuated defects are typical of cells permanently lacking SRP function, and are the result of adaptive changes that occur in such strains (Ogg et al., 1992). A strain compromised for the SRP-independent pathway (sec63-201; Ng et al., 1996) had the expected severe translocation defect for CPY (Figure 7, lane 5).
The results obtained with the srp14-ΔC29 mutant were striking. It does not have severe defects in SRP-dependent targeting and we concluded that SRP containing Srp14p-ΔC29 must bind ribosome–nascent chain complexes efficiently and target them to the ER membrane in vivo. The limiting factor in co-translational targeting is translation beyond the point at which SRP can no longer remain ribosome bound (Siegel and Walter, 1988a). Therefore, if the primary defect in ΔC29 is in elongation arrest rather than signal sequence recognition, and for the substrates tested (Pho8p and DPAPB) this ‘point of no return’ was passed occasionally, then slight translocation defects, such as those seen, would result. We therefore employed an alternative measure of targeting by SRP in vivo, in particular one that could probe elongation arrest activity. This was the ubiquitin-assisted translocation assay (UTA) (Johnsson and Varshavsky, 1994), specifically designed to measure the coupling of translation and translocation. UTA reporters (Figure 8A) encode a signal sequence followed by a variable length of ‘spacer’, then ubiquitin and finally a marker, the cytosolic enzyme Ura3p in the reporters used here. Close coupling of translation and translocation results in the whole protein being translocated into the ER, and the cells remain ura– in an otherwise ura3 genetic background (Ura3p is required in the cytosol to function). If targeting is not efficient or closely coupled to translocation, the ubiquitin moiety is exposed in the cytosol, folds rapidly, and ubiquitin-dependent proteases cleave C-terminally to it leaving Ura3p in the cytosol—cells are then ura+.
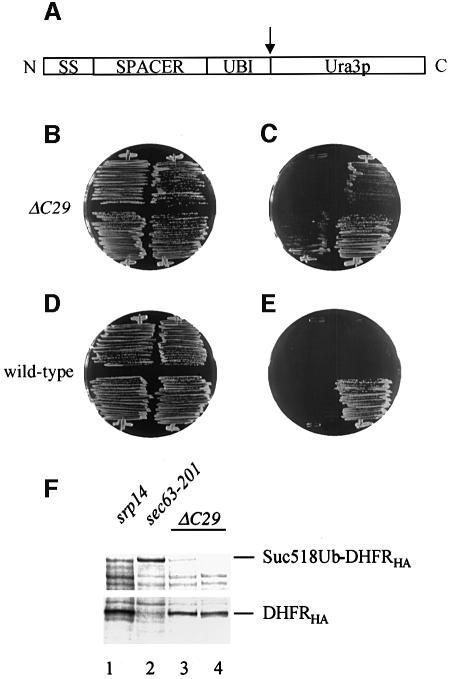
Fig. 8. Translocation defects of srp14-ΔC29 cells in the UTA assay. (A) Organization of UTA constructs. The protein is depicted N- to C-terminus; signal sequence (SS), spacer (SPACER), ubiquitin (UBI) and Ura3p. The cleavage site for cytosolic ubiquitin-dependent proteases is indicated (arrow). (B and C) srp14-ΔC29 cells streaked on to a –trp (B; selecting for the plasmids) and a –ura (C; selecting for cytosolic Ura3p) plate were incubated for 2 days at 30°C. They contained empty vector (top left), Suc2277 (top right), Dap2300 (bottom left) or the control substrate Suc223, which has a 23 amino acid spacer (bottom right). (D and E) As for (B) and (C) but with wild-type cells. As seen previously (Johnsson and Varshavsky, 1994), Suc223 gave a Ura+ phenotype in wild-type cells. (F) Anti-HA immunoprecipitations from extracts of pulse-labelled cells expressing Suc2518UbDHFRHA (lanes 1–3) or Suc223UbDHFRHA control, which only yields the DHFRHA fragment (lane 4) (Johnsson and Varshavski, 1994). Labelling of cells and analysis are as in Figure 7.
We compared growth of srp14-ΔC29, wild-type and control strains carrying a UTA reporter with the invertase (Suc2) signal sequence and a 277 amino acid spacer (Suc2277) on plates lacking uracil (Figure 8B–E; Table I). Translocation of invertase is partially blocked when the SRP-dependent pathway is disrupted (Hann and Walter, 1991; Ogg et al., 1992; Rothe and Lehle, 1998), indicating that it is targeted at least partly by SRP. Wild-type cells carrying Suc2277 were ura–, indicating efficient translocation of the fusion protein. srp14-ΔC29 cells were, however, ura+. This was seen at temperatures from 14 to 34°C and thus the UTA reporter revealed a constitutive defect in the srp14-ΔC29 mutant, not revealed by immunoprecipitation of the endogenous SRP-dependent substrates (Figure 7). A sec65-1 strain was also ura– at permissive growth temperatures, although it grew considerably less well than srp14-ΔC29 cells. This was expected, as the primary selection used to isolate the sec65-1 mutation was inefficient translocation of a reporter protein across the ER membrane at 23°C (Stirling et al., 1992). The SRP-independent pathway mutant sec63-201 was ura–, indicating that the reporter was not being targeted through this pathway, whereas cells lacking either SRP or SRP receptor activity were ura+.
Table I. Detection of translocation defects (phenotypes of cells carrying UTA constructs) using the UTA assay.
| Construct | TR3 wild type | JDY375 ΔC29 | JDY365 sec63-201 | JDY66 srp14 | JDY219 srp102 | RSY547 sec65-1 |
|---|---|---|---|---|---|---|
| Suc2277 | – | + | – | ++ | ++ | +/– |
| Dap2300 | – | + | – | ++ | ++ | +/– |
| Mfα139Suc2275 | ++ | ++ | ++ | ++ | ++ | ++ |
| Suc223 | ++ | ++ | ++ | ++ | ++ | ++ |
| pRS314 | – | – | – | – | – | – |
Strains carrying constructs indicated were streaked on to –ura plates and growth at 30°C was compared with that on –trp plates. –, none; +/–, slight; +, good; ++, as on –trp plate.
To extend this analysis we used two further reporters. One contained, in place of invertase-derived sequences, the first 300 amino acids of DPAPB (Dap2300). Since DPAPB is SRP dependent, this construct should also be a reporter for SRP-dependent targeting. Indeed, Dap2300 gave an identical spectrum of growth on plates lacking uracil such as Suc2277, confirming our findings (Figure 8B–E; Table I). The second construct encoded the first 39 amino acids (including the signal sequence) of ppαF followed by 275 amino acids of invertase prior to ubiquitin (Mfα139Suc2275). In a previous analysis this yielded ura– wild-type cells (Johnsson and Varshavsky, 1994). However, in our wild-type strain, Mfα139Suc2275 yielded a ura+ phenotype. Thus, although ppαF is efficiently targeted in this strain (Hann and Walter, 1991), the ppαF signal sequence is recognized too late to establish the close coupling of translation and translocation required for translocation of intact fusion protein.
Finally, since growth is an indirect measure of the translocation efficiency of the UTA reporters, we investi gated this directly using Suc2 signal sequence-containing constructs in which the URA3 sequences were replaced with those encoding dihydrofolate reductase and an HA tag (Johnsson and Varshavski, 1994). We pulse-labelled cells expressing these constructs at 23°C and performed immunoprecipitations with anti-HA antibodies on extracts from them. Figure 8F shows that such a reporter was, as expected, translocated intact in the SRP-independent pathway mutant (sec63-201; Figure 8F, lane 2), but was mostly cleaved in both srp14-null and srp14-ΔC29 cells, a large proportion of the immunoprecipitated material corresponding to the DHFRHA portion of the fusion (Figure 8F, lanes 1 and 3). This result is in stark contrast to that obtained with the endogenous proteins in Figure 7. Cumulatively, these data suggest that the slight translocation defect seen in srp14-ΔC29 cells is due to a failure to couple translation and translocation tightly, the defect predicted for an elongation arrest mutant.
Discussion
We isolated intact, active yeast SRP, and this is the only purification and characterization of a eukaryotic SRP other than the canine particle. Using this material we determined that yeast SRP has elongation arrest activity. Our data also indicate that this function of SRP resides in the Alu-domain of the yeast particle, as it does in mammalian SRP, since an Srp14p truncation mutant was deficient in elongation arrest both in vivo and in vitro. This mutant had low translocation-promoting activity in vitro, but in vivo the translocation of SRP-dependent proteins was, in general, efficient, indicating that this particle was not inherently deficient in signal sequence binding or targeting per se.
Purified yeast SRP contains only the components previously identified. Its composition differs in three ways from the mammalian particle. First, it contains Srp21p, a protein with no known homologues; secondly, it has no SRP9 component; and thirdly it has two copies of Srp14p, most likely as an Srp14p homodimer in the particle, replacing SRP9/14 (Strüb et al., 1999). The overall organization of the particle remains to be determined. Of particular interest will be identification of the portion of scR1 RNA corresponding to the conserved Srp54p binding site, or domain IV, found in all SRP RNAs, and the binding site of the yeast-specific Srp21p.
Purified yeast SRP restored translocation of DHCαF and ppL into microsomes when added to yeast cytosol immunodepleted of SRP. The purified material retained most of its activity, and the slight loss may have been due to the partial degradation of scR1 RNA that we have not been able to avoid (Figure 1C). The increase in translocation with increasing SRP concentration was non-linear. Addition of SRP to 5–10 times that in cytosolic translation extracts did not increase translocation efficiency by >1.5-fold that obtained with non-SRP depleted extract. SRP-dependent translocation (maximum ∼25%) was not as efficient as that of ppαF by the SRP-independent pathway (usually ∼60%) or as catalysed by mammalian SRP in higher eukaryotic systems. Thus, something other than SRP is limiting in our assays. Since only ∼20% of Sec61p, the main component of the translocation apparatus, is ribosome bound in yeast (e.g. Wilkinson et al., 2000), the majority of translocation sites in yeast ER membranes may not be available to SRP-dependent targeting.
Yeast SRP is able, like its mammalian counterpart, to slow translation during targeting. This major finding is the first demonstration of conservation of an activity of SRP other than those mediated by the signal sequence binding protein SRP54. We demonstrated this in time-course experiments and using statistical analysis of multiple samples taken at two time points. Our results are similar to those obtained by Wolin and Walter (1989) when they examined the elongation arrest activity of canine SRP in a mammalian translation system. Combined, these studies using homologous systems indicate that the halt in translation mediated by canine SRP in a wheat germ translation system (Walter and Blobel, 1981) is an artifact of the heterologous system.
Co-translational signal sequence binding by SRP is a feature conserved from eukaryotes to prokaryotes (Luirink et al., 1992; Valent et al., 1995). Although not tested previously for yeast SRP, available evidence suggested an obligatory co-translational interaction with the signal sequence (Hansen and Walter, 1988; Ogg and Walter, 1995). Our observation of elongation arrest indicates that yeast SRP does indeed interact co-translationally with signal sequences and lends credence to the supposition (Ng et al., 1996) that SRP is the first selector of signal sequences in yeast.
Srp14p-ΔC29 proved an invaluable tool. First and foremost SRP ΔC29 was deficient in elongation arrest assays—it did not delay the production of full-length DHCαF in vitro and had a severe defect in promoting translocation of UTA reporters intact in vivo. The elongation arrest function of SRP is maintained in the same component of SRP, despite the assembly of yeast Srp14p into a homodimer rather than an SRP9/14 heterodimer.
As the only Srp14p in cells, ΔC29 resulted in neither the slow growth nor the readily detected translocation defects seen with SRP-deficient strains. srp14-ΔC29 cells grew well at most temperatures and we only detected translocation defects for one endogenous protein after prolonged growth at high temperatures. However, SRP containing ΔC29 was compromised for targeting activity in vitro, having at most 10% of the activity of the wild-type particle. This is a greater defect than that observed with elongation arrest deficient mammalian SRP, but follows the same trend, indicating the importance of elongation arrest for targeting. The difference in targeting efficiency of the ΔC29 mutant between in vivo and in vitro suggests that the time required for SRP–ribosome–nascent chain complexes to become docked to the translocation site in vivo might be much less than in vitro, leading to a high proportion of targeting events being successful in the absence of elongation arrest. Alternatively, the in vivo situation may be more forgiving in its requirement for a complete coupling between translation and translocation than the in vitro assays. The low translocation-promoting activity of ΔC29 did not appear to be a result of purification, since cytosol from srp14-ΔC29 cells was also defective in promoting translocation of SRP- dependent substrates (data not shown).
Our data do not formally exclude the possibility that yeast SRP containing ΔC29 is less efficient at binding signal sequences than wild type, but we consider this highly unlikely. The competition that we observed between ΔC29 and wild-type SRP in in vitro assays indicates that the mutant retains ribosome–nascent chain binding. The contrast between efficient translocation of endogenous SRP-dependent substrates and cleavage of UTA reporters in srp14-ΔC29 strains also sheds light on this. Efficient translocation, as seen for DPAPB, implies, before all else, signal sequence recognition. However, the UTA construct Dap2300, containing the same signal sequence, gave a ura+ phenotype and UTA constructs were largely cleaved in srp14-ΔC29 cells. The simplest interpretation of these data is that in srp14-ΔC29 cells signal sequences are recognized by SRP but translation is not paused, and the cleavable reporters containing ubiquitin reveal this defect.
The temperature-sensitive phenotype of srp14-ΔC29 cells reveals the importance of elongation arrest in vivo. It may indicate that at high temperature some protein(s) necessary for growth requires elongation arrest to be targeted to the ER membrane. However, the similar phenotype of SRP null and srp14-ΔC29 cells argues against this, and other possibilities are that these proteins may be dealt with by the SRP-independent targeting pathway at lower temperatures, but cannot be targeted via this route at higher temperatures, or that they are only essential for growth at high temperatures. Targeting of short proteins, or those that fold rapidly, might be predicted to be the most seriously affected by lack of elongation arrest. In addition, lack of elongation arrest may have a more pronounced effect on the first translational initiation on each mRNA. Once membrane localization has been achieved with one ribosome, the targeting problem would essentially be reduced in complexity from four dimensions to three, improving the efficiency of the reaction and of translocation.
Reconstitution of the SRP14 ΔC20 mutant into mammalian SRP results in a different tertiary structure of the Alu-domain RNA from the wild-type particle. In particular, nucleotides in a putative pseudoknot in the mammalian SRP RNA Alu-domain (Zwieb et al., 1996) are altered with respect to their sensitivity to chemical modification, consistent with a more open conformation (Thomas et al., 1997). This pseudoknot is suggested to mimic tRNA and form a major part of the ribosome interface of the Alu-domain, proposed to be at the elongation factor binding site (discussed in Bui and Strüb, 1999). No direct evidence for this has yet been found, but the structure of SRP, revealed by electron microscopy, is a rod long enough (6 × 24 nm; Andrews et al., 1985) to span the ribosomal large subunit and interact at both nascent chain exit site (SRP54) and elongation factor binding site (the Alu-domain).
We have yet to examine how ΔC29 affects regions of scR1 RNA to which it binds, but there is currently no suggestion of a pseudoknot in this region of scR1. Our attention is focused on the conserved single-stranded loop within the Srp14p binding site (Strüb et al., 1991, 1999), and the C-terminal portion of Srp14p itself. Mutations in the single-stranded loop have been shown to affect targeting by both mammalian and Schizosaccharomyces pombe SRP (Liao et al., 1992; Chang et al., 1997). The overall symmetry in the SRP9/14 fold (Birse et al., 1997) conceptually allows replacement of SRP9 with Srp14p in the yeast complex without dramatic structural alteration. However, how truncation of two copies of yeast Srp14p has the same effect as truncating one protein in mammalian SRP9/14 is unclear. Amino acids within the C-terminus of SRP14 are predicted to be part of the SRP9–SRP14 interface (Birse et al., 1997) and presumably would have a similar position in the yeast Srp14p dimer. However, these amino acids may have other roles in the complete SRP, or in their absence the overall structure of the Alu-domain might be altered to affect its interaction with the ribosome. The challenge is to define interactions important for elongation arrest and to understand how these conspire to slow protein synthesis.
Materials and methods
Yeast strains
The following were derived from TR1 (trp1, lys2, his3, ura3, ade2, MATa/α; Parker et al., 1988): TR3 (MATα), JDY66 (MATα, srp14::HIS3) (Brown et al., 1994); JDY219 (MATa, srp102::HIS3), JDY349, JDY417 and JDY432 [MATα, srp14::HIS3 plus pJDY69 (HA-SRP14), pJEY42 (HA-srp14-ΔC29) and pJEY59 (HA-srp14-ΔC11), respectively], JDY115 [srp21::HIS3 plus pJDY67 (HA-SRP21) and pJEY106 (Myc-SRP21)], JDY365 [MATa, trp1-Δ99, his3-Δ200, ura3-Δ99, leu2-Δ1, ade2-101, cir°, sec63-ΔC27-HIS3, a truncation equivalent to the original sec63-201 allele (Ng and Walter, 1996)], JDY375 (MATα, srp14-ΔC29-his5+), JDY440 [MATa/MATα srp72Δ::HIS3/SRP72, pJEY85 (SRP72-ZZ)], JDY480 (MATα, srp72Δ::HIS3, srp14-ΔC29-his5+, pJEY85), this study; and RSY547 (MATa, sec65-1, trp1-Δ1, his3, ura3-52, leu2-3, –112, ade2-1) was obtained from Dr R.Schekman, University of California, Berkeley.
Antibodies
Anti-Srp14p and Sec65p (Brown et al., 1994), anti-Srp54p (Hann and Walter, 1991), anti-Srp68p (amino acids 48–264) and anti-Srp72p (amino acids 7–107) antibodies were raised in rabbits against glutathione S-transferase (GST) fusion proteins and were affinity purified on equivalent MBP fusion proteins. Anti-Pho8p, DPAPB and CPY antibodies (Brown et al., 1994). Anti-HA monoclonal antibody HA.11 (BAbCO, CA).
Plasmid construction
Epitope and protein A tags. SRP72. pJEY85 that encoded the modified Srp72p was produced in a three-way ligation using fragments consisting of the SRP72 promoter and gene, the TEV protease cleavage site and two copies of the protein A–IgG binding domain (from plasmid pZZ-HIS5, a kind gift of Sean Munro, MRC Laboratory of Molecular Biology, Cambridge) and pRS314 (Sikorski and Heiter, 1989).
SRP14. A triple-HA epitope tag was introduced immediately following the SRP14 initiator methionine codon in pRS314 producing plasmid pJDY69. The srp14-ΔC29 allele in plasmid pJEY42 was generated by replacing a SphI–EcoRI fragment of pJDY69 with a fragment consisting of a SphI site then a stop codon and a NheI site at one end followed by flanking DNA of SRP14 and an EcoRI site, thus replacing the last 29 codons of SRP14 with histidine. ΔC11 (in pJEY59) was made by replacing the SRP14 promoter HA-tag and gene in pJEY42 with a PCR fragment that introduced a stop at codon 136 followed by a NheI site. An integrative version of srp14-ΔC29 was constructed by replacing the TEV–protein A fragment of pZZ-HIS5 with the BamHI–EcoRI srp14-ΔC29 fragment.
SRP21. Three HA or nine Myc epitope tags were introduced immediately after the initiator methionine codon in pRS314 and pRS313 (Sikorski and Heiter, 1989), producing plasmids pJDY67 and pJEY106, respectively.
Plasmids for synthesis of mRNA substrates for translations were pDJ100 (ppαF; Hansen et al., 1986), pJD96 (DHCαF), derived from pJD82 (Ng et al., 1996) with a codon bias optimized DPAPB signal sequence, and the ppαF 5′ untranslated region inserted prior to the initiator methionine, and pJD71 (ppL), derived from pSPBP6 (Siegel and Walter, 1988b) and encoding a glycine–alanine substitution at amino acid 10. UTA constructs apart from Dap2300 were kindly provided by Nils Johnsson (Max-Delbruck Laboratory, Cologne). In Dap2300, a SalI–ClaI fragment of Suc2277 (Johnsson and Varshavsky, 1994) was replaced with the first 300 amino acids of DPAPB.
Preparation and analysis of yeast extracts
Native yeast extract preparation and sucrose gradient sedimentation were as described (Brown et al., 1994). All samples were in buffer A [20 mM HEPES–KOH pH 7.5, 500 mM KOAc pH 7.5, 2 mM MgOAc, 14% v/v glycerol, 2 mM dithiothreitol, 0.02% v/v Nikkol (Nikko Chemical Co., Tokyo, Japan), 0.5 mM PMSF]. Native immunoprecipitations were carried out from an amount of extract containing 100 µg of protein in buffer A. Protein A-tagged proteins were recovered on IgG–Sepharose. Cell labelling with [35S]promix (Amersham Pharmacia Biotech), and preparation of non-native extracts and immunoprecipitations were as described (Ng et al., 1996).
Purification and analysis of SRP
Postribosomal extract (in buffer A) from 150–200 g of cells grown to OD600 ∼3.0 was incubated in batch with 2 ml of IgG–Sepharose beads that were then packed into a column and washed with buffer A. SRP was released by incubation for 1 h in 200 U/ml TEV protease (Gibco-BRL). The eluate at 100 mM KOAc was passed over a 0.5 ml column of ω-aminobutyl agarose (Sigma), which was washed with buffer A at 350 mM KOAc and eluted at 800 mM KOAc. Peak fractions were pooled and salt concentration reduced to 200 mM KOAc (SRP buffer) on Bio-Gel (Bio-Rad Laboratories). In vitro translations and immunodepletion of SRP were as described (Ng et al., 1996). Reactions were stopped by addition of 15% w/v TCA. Quantification of translation products was carried out using PhosphorImaging and ImageQuant software (Molecular Dynamics). Statistical analysis was performed using Student’s t-test (two-tailed).
Acknowledgments
Acknowledgements
The authors thank Nils Johnsson and members of the Brown laboratory for discussion and suggestions, and Jean Beggs, David Tollervey, Kevin Hardwick and Susan Farrington for critical reading of the manuscript. J.D.B. also thanks Peter Walter (University of California San Francisco) in whose laboratory the seeds of this work were sown. This work was supported by a Wellcome Trust Research Career Development Award to J.D.B.
References
- Andrews D.W., Walter,P. and Ottensmeyer,F.P. (1985) Structure of the signal recognition particle by electron microscopy. Proc. Natl Acad. Sci. USA, 82, 785–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse D.E.A., Kapp,U., Strüb,K., Cusack,S. and Aberg,A. (1997) The crystal structure of the signal recognition particle Alu RNA binding heterodimer, SRP9/14. EMBO J., 16, 3757–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.D., Hann,B.C., Medzihradszky,K.F., Niwa,M., Burlingame,A.L. and Walter,P. (1994) Subunits of the Saccharomyces cerevisiae signal recognition particle required for its functional expression. EMBO J., 13, 4390–4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui N. and Strüb,K. (1999) New insights into signal recognition and elongation arrest activities of the signal recognition particle. Biol. Chem., 380, 135–145. [DOI] [PubMed] [Google Scholar]
- Chang D.Y., Newitt,J.A., Hsu,K., Bernstein,H.D. and Maraia,R.J. (1997) A highly conserved nucleotide in the Alu domain of SRP RNA mediates translation arrest through high affinity binding to SRP9/14. Nucleic Acids Res., 25, 1117–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felici F., Cesareni,G. and Hughes,J.M.X. (1989) The most abundant small cytoplasmic RNA of Saccharomyces cerevisiae has an important function required for normal cell growth. Mol. Cell. Biol., 9, 3260–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann B.C. and Walter,P. (1991) The signal recognition particle in S.cerevisiae. Cell, 67, 131–144. [DOI] [PubMed] [Google Scholar]
- Hann B., Stirling,C.J. and Walter,P. (1992) SEC65 gene product is a subunit of the yeast signal recognition particle required for its integrity. Nature, 356, 532–533. [DOI] [PubMed] [Google Scholar]
- Hansen W. and Walter,P. (1988) Prepro-carboxypeptidase Y and a truncated form of pre-invertase, but not full-length pre-invertase, can be posttranslationally translocated across microsomal vesicle membranes from Saccharomyces cerevisiae. J. Cell Biol., 106, 1075–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen W., Garcia,P.D. and Walter,P. (1986) In vitro protein translocation across the yeast endoplasmic reticulum: ATP-dependent post-translational translocation of the prepro-α factor. Cell, 45, 397–406. [DOI] [PubMed] [Google Scholar]
- Hauser S., Bacher,G., Dobberstein,B. and Lütcke,H. (1995) A complex of the signal sequence binding protein and the SRP RNA promotes translocation of nascent proteins. EMBO J., 14, 5485–5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson N. and Varshavsky,A. (1994) Ubiquitin-assisted dissection of protein transport across membranes. EMBO J., 13, 2686–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg U.C., Walter,P. and Johnson,A.E. (1986) Photocrosslinking of the signal sequence of nascent preprolactin to the 54-kilodalton polypeptide of the signal recognition particle. Proc. Natl Acad. Sci. USA, 83, 8604–8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzchalia T.V., Wiedmann,M., Girshovich,A.S., Bochkareva,E.S., Bielka,H. and Rapoport,T.A. (1986) The signal sequence of nascent preprolactin interacts with the 54K polypeptide of the signal recognition particle. Nature, 320, 634–636. [DOI] [PubMed] [Google Scholar]
- Liao X., Selinger,D., Althoff,S., Chiang,A., Hamilton,D., Ma,M. and Wise,J.A. (1992) Random mutagenesis of Schizosaccharomyces pombe SRP RNA: lethal and conditional lesions cluster in presumptive protein binding sites. Nucleic Acids Res., 20, 1607–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luirink J., High,S., Wood,H., Giner,A., Tollervey,D. and Dobberstein,B. (1992) Signal sequence recognition by an Escherichia coli ribonucleoprotein complex. Nature, 359, 741–743. [DOI] [PubMed] [Google Scholar]
- Lütcke H. (1995) Signal recognition particle (SRP), a ubiquitous initiator of protein translocation. Eur. J. Biochem., 228, 531–550. [DOI] [PubMed] [Google Scholar]
- Ng D.T. and Walter,P. (1996) ER membrane protein complex required for nuclear fusion. J. Cell Biol., 132, 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng D.T.W., Brown,J.D. and Walter,P. (1996) Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J. Cell Biol., 134, 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngsee J.K. and Smith,M. (1990) Changes in a mammalian signal sequence required for efficient protein secretion by yeasts. Gene, 86, 251–255. [DOI] [PubMed] [Google Scholar]
- Ogg S.C. and Walter,P. (1995) SRP samples nascent chains for the presence of signal sequences by interacting with ribosomes at a discrete step in translation elongation. Cell, 81, 1075–1084. [DOI] [PubMed] [Google Scholar]
- Ogg S., Poritz,M. and Walter,P. (1992) The signal recognition particle receptor is important for growth and protein secretion in Saccharomyces cerevisiae. Mol. Biol. Cell, 3, 895–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T. and Walter,P. (1996) The nascent polypeptide-associated complex modulates interactions between the signal recognition particle and the ribosome. Curr. Biol., 6, 331–338. [DOI] [PubMed] [Google Scholar]
- Rothe C. and Lehle,L. (1998) Sorting of invertase signal peptide mutants in yeast dependent and independent on the signal-recognition particle. Eur. J. Biochem., 252, 16–24. [DOI] [PubMed] [Google Scholar]
- Siegel V. and Walter,P. (1985) Elongation arrest is not a prerequisite for secretory protein translocation across the microsomal membrane. J. Cell Biol., 100, 1913–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel V. and Walter,P. (1986) Removal of the Alu structural domain from signal recognition particle leaves its protein translocation activity intact. Nature, 320, 81–84. [DOI] [PubMed] [Google Scholar]
- Siegel V. and Walter,P. (1988a) The affinity of signal recognition particle for presecretory proteins is dependent on nascent chain length. EMBO J., 7, 1769–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel V. and Walter,P. (1988b) Each of the activities of signal recognition particle (SRP) is contained within a distinct domain: analysis of biochemical mutants of SRP. Cell, 52, 39–49. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Heiter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling C.J. and Hewitt,E.W. (1992) The Saccharomyces cerevisiae SEC65 gene encodes a component of the yeast signal recognition particle with homology to human SRP19. Nature, 356, 534–537. [DOI] [PubMed] [Google Scholar]
- Stirling C.J., Rothblatt,J., Hosobuchi,M., Deshaies,R. and Schekman,R. (1992) Protein translocation mutants defective in the insertion of integral membrane proteins into the endoplasmic reticulum. Mol. Biol. Cell, 3, 129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strüb K., Moss,J. and Walter,P. (1991) Binding sites of the 9- and 14-kilodalton heterodimeric protein subunit of the signal recognition particle (SRP) are contained exclusively in the Alu domain of SRP RNA and contain a sequence motif that is conserved in evolution. Mol. Cell. Biol., 11, 3949–3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strüb K., Fornallaz,M. and Bui,N. (1999) The Alu domain homolog of the yeast signal recognition particle consists of an Srp14p homodimer and a yeast-specific RNA structure. RNA, 5, 1333–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas Y., Bui,N. and Strüb,K. (1997) A truncation in the 14 kDa protein of the signal recognition particle leads to tertiary structure changes in the RNA and abolishes the elongation arrest activity of the particle. Nucleic Acids Res., 25, 1920–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valent Q.A., Kendall,D.A., High,S., Kusters,R., Oudega,B. and Luirink,J. (1995) Early events in preprotein recognition in E.coli: interaction of SRP and trigger factor with nascent polypeptides. EMBO J., 14, 5494–5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P. and Blobel,G. (1980) Purification of membrane-associated protein complex required for protein translocation across the endoplasmic reticulum. Proc. Natl Acad. Sci. USA, 77, 7112–7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P. and Blobel,G. (1981) Translocation of proteins across the endoplasmic reticulum. III. Signal recognition protein (SRP) causes signal sequence and site specific arrest of chain elongation that is released by microsomal membranes. J. Cell Biol., 91, 557–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P. and Johnson,A.E. (1994) Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell Biol., 10, 87–119. [DOI] [PubMed] [Google Scholar]
- Wilkinson B.M., Tyson,J.R., Reid,P.J. and Stirling,C.J. (2000) Distinct domains within yeast Sec61p involved in post-translational translocation and protein dislocation. J. Biol. Chem., 275, 521–529. [DOI] [PubMed] [Google Scholar]
- Wolin S.L. and Walter,P. (1989) Signal recognition particle mediates a transient elongation arrest of preprolactin in reticulocyte lysate. J. Cell Biol., 109, 2617–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb C., Muller,F. and Larsen,N. (1996) Comparative analysis of tertiary structure elements in signal recognition particle RNA. Fold. Des., 1, 315–324. [DOI] [PubMed] [Google Scholar]