Abstract
The general transcription factor TFIID and its individual subunits (TAFIIs) have been the focus of many studies, yet their functions in vivo are not well established. Here we characterize the requirement of yeast TAFIIs for the derepression of the ribonucleotide reductase (RNR) genes. Promoter mapping studies revealed that the upstream repressing sequences, the damage-responsive elements (DREs), rendered these genes dependent upon TAFIIs. DREs are the binding sites for the sequence-specific DNA binding-protein Crt1 that represses transcription by recruiting the Ssn6–Tup1 co-repressor complex to the promoter. We demonstrate that deletion of SSN6, TUP1 or CRT1 alleviated the TAFII dependence of the RNR genes, indicating that TAFII dependence requires the co-repressor complex. Furthermore, we provide evidence that Crt1 specifies the TAFII dependence of these genes. Our studies show that TFIID interacts with the repression domain of Crt1, suggesting that the derepression mechanism involves an antagonism between TFIID and the co-repressor complex. Our results indicate that yeast TAFIIs have other functions in addition to core promoter selectivity, and describe a novel activity: the derepression of promoters.
Keywords: Crt1/repression/Ssn6–Tup1/TFIID/transcription
Introduction
The general transcription factor TFIID has been the focus of years of biochemical study because of its central role in transcription. Some of the proposed activities of this complex include co-activator, core promoter selectivity, kinase and histone modification (for review see Burley and Roeder, 1996; Verrijzer and Tjian, 1996; Lee and Young, 1998). The role of many of these activities in gene regulation has yet to be elucidated.
Initial analyses of yeast TAFII mutants revealed that mutation or depletion of seven different TAFIIs resulted in a cessation of growth, but yet the transcription of many cellular genes was unaffected (Apone et al., 1996; Moqtaderi et al., 1996; Walker et al., 1996). These studies, and a recent genome-wide gene expression analysis using TAFII mutants (Holstege et al., 1998), indicate that TAFIIs are not universally required for transcription in vivo. Likewise, conditional expression or mutation of metazoan TAFIIs resulted in similar phenotypes, indicating that a restricted transcriptional requirement is not unique to yeast TAFIIs (Wang and Tjian, 1994; Suzuki-Yagawa et al., 1997; Zhou et al., 1998; Martin et al., 1999; Metzger et al., 1999; Soldatov et al., 1999). The debate on the necessity for TAFIIs in transcription was revived by studies reporting broad transcriptional defects in a number of TAFII mutants, some grouped by their sequence and structural similarities to the core histones (Apone et al., 1998; Holstege et al., 1998; Krishnamurthy et al., 1998; Michel et al., 1998; Moqtaderi et al., 1998; Komarnitsky et al., 1999; Sanders et al., 1999; Reese et al., 2000). However, the exact fraction of the genome affected by the inactivation of these mutants, and what accounts for their distinct phenotypes, is unclear and controversial (for reviews see Hahn, 1998; Green, 2000). Moreover, the interpretation of data obtained using certain TAFII mutants is complicated by their presence in histone acetyltransferase (HAT) complexes (Grant et al., 1998a; Ogryzko et al., 1998; Wieczorek et al., 1998) and potential allele-specific defects (Michel et al., 1998).
The characterization of genes whose expression is sensitive to TAF145 mutations revealed that the core promoter sequences, but not the upstream activating sequences, rendered them TAF145 dependent (Shen et al., 1997; Walker et al., 1997; Tsukihashi et al., 2000). These findings indicate that TAFIIs are acting as promoter selectivity factors and not as co-activators for these genes, which is consistent with the core promoter selectivity functions of some metazoan TAFIIs (for reviews see Hoffmann et al., 1997; Verrijzer and Tjian, 1996).
Most, if not all, promoters are under constant repression by nucleosomes (Roth, 1995) and transcriptional repressor proteins (Herschbach and Johnson, 1993; Lee and Young, 1998). The transcription machinery, including TFIID, must contend with these activities and factors. Unfortunately, the mechanism by which TFIID counteracts these two classes of transcriptional repressors is poorly understood. It has been proposed that the HAT activity of yTAFII145/dTAFII250 may play an important role in modifying histones near promoters of repressed target genes (Mizzen et al., 1996), although the exact function of this activity in gene regulation has not been determined. An important step in addressing these questions is to identify genes that are regulated predominantly by transcriptional repression and whose expression is dependent upon TAF145.
Here we report the characterization of TAFII-dependent genes that are regulated by transcriptional repression and chromatin structure, the DNA damage-regulated ribonucleotide reductase (RNR) genes. The RNR genes are repressed in the absence of DNA damage by the gene-specific repressor Crt1 and the general co-repressor complex Ssn6–Tup1 (Zhou and Elledge, 1992; Huang et al., 1998), and we show that the TAFII dependence of the RNR genes is mediated by these three factors. Our analysis defines a function for TAFIIs: the derepression of regulated promoters.
Results
Yeast TAFIIs are required for the transcription of the DNA damage-induced ribonucleotide reductase genes
We investigated the requirement of yeast TAFIIs for the regulation of DNA damage-induced genes using strains that conditionally express their gene products from the GAL1 promoter (Walker et al., 1996). Cells containing the sole copy of TAF145, TAF90, TAF68/61 or TAF47 under the control of the GAL1 promoter were transferred to dextrose-containing medium and then treated with the DNA-damaging agent methylmethane sulfonate (MMS) or the replication inhibitor hydroxyurea (HU). The effects of TAFII depletion on the expression of the ribonucleotide reductase genes were examined by northern blotting. We focused on RNR2, RNR3 and RNR4 because they are the prototypical models to study DNA damage-induced transcription and they have been characterized extensively (Elledge and Davis, 1989, 1990; Yagle and McEntee, 1990; Elledge et al., 1992; Huang and Elledge, 1997). Depletion of yeast TAFII145p, TAFII90p or TAFII68p strongly reduced, or eliminated, the DNA damage-induced expression of all three RNR genes (Figure 1A). In contrast, depletion of yeast TAFII47p did not result in any significant changes in expression, indicating that RNR gene transcription is dependent upon specific TAFIIs and that depleting an essential gene product does not cause the defects observed in the other three conditional strains. Immunoblotting confirmed that within 8 h upon transfer from galactose- to dextrose-containing medium, each TAFII was depleted (Figure 1B). These data also show that the levels of TATA box-binding protein (TBP) did not change significantly over the course of the experiment; thus, the loss of RNR transcription does not result from reduced TBP levels. Our results clearly demonstrate that the DNA damage-induced transcription of the RNR genes is dependent upon certain TAFII genes.
Fig. 1. Depletion of TAFIIs impairs DNA damage-dependent expression of the RNR genes. (A) Strains containing yeast TAF145 (YSW94), TAF90 (LYC-1), TAF68 (YJR18) and TAF47 (YJR11) under the control of the dextrose-repressible GAL1 promoter were grown in galactose to an OD of 0.5–1.0 (GAL), collected, washed and resuspended in dextrose-containing medium. After 10 h in dextrose medium, the cultures were divided into three flasks, and MMS (0.03%) or HU (0.1 M) was added to separate cultures. After an additional 1.5 and 3 h induction period, cells were collected for RNA isolation. Note that the total amount of time in dextrose for these experiments was 11.5 and 13 h, respectively. (B) Western blot. The levels of TAFII (left) and TBP (right) protein were analyzed after 0, 8 and 12 h in dextrose-containing medium. The asterisk on the left of the TAFII145 blot indicates a cross-reacting protein migrating near TAFII145.
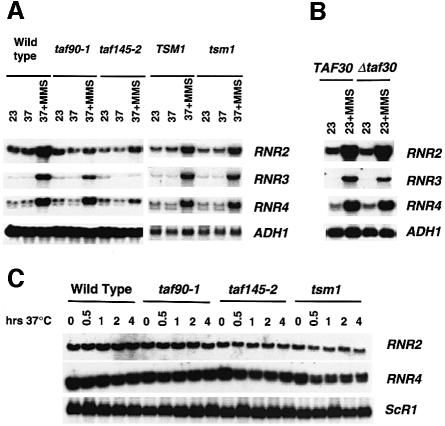
We next analyzed the expression of the RNR genes in temperature-sensitive mutants of TAF90, TAF145 and TAF170/TSM1, and in a strain containing a deletion of TAF30/ANC1. For the experiment shown in Figure 2A, cultures of TAFII mutants were shifted to the restrictive temperature for 60 min and were then treated with MMS (0.015%) for 2 h. Analysis of the mRNA levels of RNR2, RNR3 and RNR4 revealed that all three were compromised, but to various degrees. Inactivation of taf145-2 had the strongest effect on the expression of all three genes, but the inactivation of taf90-1 showed weaker effects on RNR2 and RNR3 compared with the TAF145 mutant (Figure 2A). Moreover, the expression of RNR4 in the TAF90 mutant seemed relatively unaltered. The weaker phenotype observed in the TAF90 mutant may result from its incomplete inactivation prior to the addition of MMS because longer exposure of the temperature-sensitive mutant to the restrictive temperature prior to MMS addition (not shown), or depletion of its protein (Figure 1), had stronger effects on the expression of all three RNR genes. Inactivation of a TSM1 mutant had little effect on the induced expression of all three RNR genes, once again indicating that their expression requires specific TAFIIs. The induction of these genes was unaffected whenever the mutant strains were grown at the permissive temperature, and results similar to those described above were also obtained using other DNA damage-inducing agents (not shown).
Fig. 2. Analysis of the expression of the ribonucleotide reductase genes in TAFII mutants. (A) Wild type (SW87 and WCS131) and strains containing temperature-sensitive mutations in yeast TAF90 (LY20), taf145-2 (YSW93) and tsm1 (WCS132) were grown at the permissive temperature of 23°C (23) and then shifted to 37°C for 1 h (37). Afterwards, MMS was added to 0.015% and the cells were collected after a 2 h induction period (37 + MMS). (B) Cells containing a deletion of TAF30/ANC1 (DDY555) were grown at 23°C and treated with MMS for 2 h. (C) Analysis of the uninduced levels of RNR2 and RNR4 gene transcription. Strains were grown at 23°C and then shifted to 37°C. Aliquots were withdrawn prior to temperature shift (0) and at 30 min, 1, 2 and 4 h.
Yeast TAF30/ANC1, a non-essential gene, was isolated recently as a gene dosage suppressor of Δrad53 and Δmec1 lethality (Desany et al., 1998); therefore, the expression of the RNR genes was examined in a TAF30/ANC1 null mutant. The results show that TAF30/ANC1 is not required for the expression of the RNR genes (Figure 2B). This result suggests that TAF30/ANC1 may affect the cell cycle arrest functions of RAD53 and MEC1, rather than the DNA damage-regulated transcription pathway, which is consistent with the cell cycle regulatory functions of TAFIIs (for a review see Green, 2000).
We often found that inactivation of taf145-2 resulted in RNR gene mRNA levels that are equivalent to those observed in untreated cells (Figure 2A and data not shown), suggesting that TAFIIs are required specifically for their induced expression. We therefore examined the effects of TAFII inactivation on the uninduced levels of RNR transcription. As expected, the results of Figure 1C show that TAFIIs are not required for the uninduced level of expression of the RNR genes.
TAFII mutations directly affect the RNR promoters
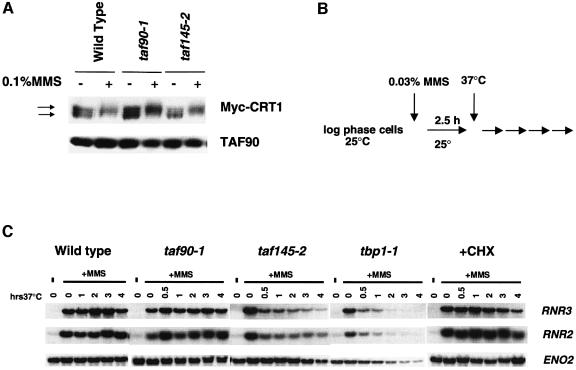
Expression of the RNR genes requires a functional DNA damage signaling pathway that is activated by DNA damage (Elledge et al., 1992; Kiser and Weinert, 1996). Activation of this pathway causes the phosphorylation of Crt1, a transcriptional repressor, and the derepression of the RNR genes (Huang et al., 1998). The defects described above could result from a defective signaling pathway, rather than from a direct effect on the RNR promoters. To rule out this possibility, we examined the MMS-induced phosphorylation of Crt1 in the TAF90 and TAF145 mutants at the restrictive temperature. Inactivation of the TAF90 or TAF145 mutant did not affect the DNA damage-induced phosphorylation of Crt1 (Figure 3A), indicating that the kinase signaling pathway is intact throughout the course of our experiments.
Fig. 3. TAFIIs directly regulate the RNR genes. (A) Analysis of the MMS-induced phosphorylation of Crt1. Wild-type (SW87), taf90-1 (LY20) and taf145-2 (YSW93) cells transformed with pMH190 (GAL-3MYC-CRT1, ARS/CEN, URA3) were grown in SC-URA plus raffinose, and Crt1 was induced for 2 h by the addition of galactose (Huang et al., 1998). Cultures were then incubated for 1 h at 37°C, followed by a 2 h treatment with MMS (0.1%). Crt1 was detected by western blotting using anti-myc monoclonal antibodies. TAFII90p served as a loading control. (B) Effects of the inactivation of TAFII mutants on active RNR genes. Cells were treated with 0.03% MMS for 2.5 h at 23°C and then shifted to 37°C. Aliquots of cells were withdrawn at the times indicated in the figure. A separate culture of wild-type cells was treated with 100 µg/ml cycloheximide at the time of temperature shift (+CHX). (C) Northern blot of RNR2 and RNR3 mRNA.
To address this issue using another strategy, we compared the loss of RNR3 and RNR2 mRNA caused by the inactivation of TAFII mutants with that of a TBP mutant (tbp1-1). The TBP is required for the transcription of all genes; thus, the loss of transcription in this mutant is indicative of a direct effect on the promoter (Cormack and Struhl, 1992). Since TAFIIs are required for the induced expression of these genes, we treated the cells with MMS at the permissive temperature to activate the genes prior to shifting the cultures to 37°C (Figure 3B). Transferring wild-type cells to 37°C after MMS treatment resulted in a small, transient increase in RNR transcription. However, in both the taf145-2 and tbp1-1 strains, the level of RNR2 and RNR3 mRNA declined sharply within 30 min (Figure 3C). This rapid loss of message is consistent with a direct effect on the promoter, since the time required to produce most secondary effects involves a substantial reduction in both a transcript and its translation product. Nonetheless, since the half-lives of the regulatory proteins in the DNA damage pathway are not known, we simulated a secondary effect using the protein synthesis inhibitor cycloheximide. Blocking protein synthesis provides an estimation of the time required to deplete essential regulatory factors, and this strategy has been used previously to distinguish a primary from a secondary effect (Cormack and Struhl, 1992). In a separate experiment, we verified that cycloheximide treatment did not affect the half-life of the RNR2 or RNR3 mRNA (not shown). Inhibition of protein synthesis did not affect the level of RNR2 or RNR3 mRNA until after 3 h (Figure 3C), a time significantly longer than the initial effects observed upon the inactivation of the taf145-2 or tbp1-1 mutant.
Surprisingly, we found that activated RNR3 and RNR2 genes are insensitive to TAF90 mutations. Furthermore, while the level of RNR transcription decreased dramatically in the TAF145 mutant, it remained significantly above the uninduced level (compare lane 14 with lanes 19 and 20 of Figure 3C). This is in contrast to the results obtained when taf145-2 is inactivated prior to MMS addition: under these conditions, we observe promoter activity similar to the uninduced level (see Figure 2A). A possible explanation for these results is that TAFIIs are required for overcoming a rate-limiting step in the derepression process. Given the results of Figure 3, we conclude that TAFII mutations directly affect the RNR promoters.
TAFII dependence maps to the damage-responsive elements (DREs)
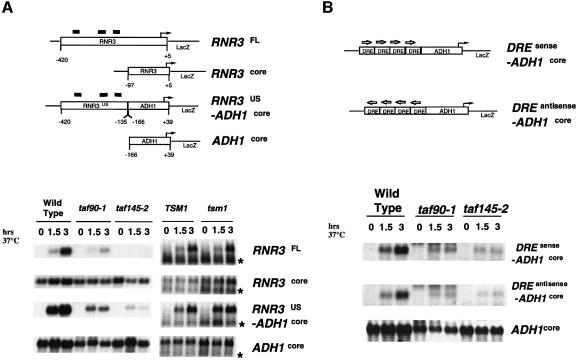
We next sought to identify the region(s) of the RNR3 promoter that renders it sensitive to TAFII mutations. We focused on the RNR3 gene because it is the best characterized of the RNR gene family, and its promoter elements were identified by previous mapping studies (Endo-Ichikawa et al., 1996; Huang et al., 1998). As an initial step, we verified that the TAFII-dependent transcription that was observed on the chromosomal copy of the RNR3 gene can be recapitulated on a plasmid-borne copy. The promoter region of RNR3 (sequences from –420 to +5, RNR3FL-lacZ) was fused to the lacZ gene and introduced into yeast on a low-copy number plasmid. The results show that, like the chromosomal copy of RNR3, transcription from the RNR3FL-LacZ transgene required functional TAF90 and TAF145, but not TSM1 (Figure 4A). Confident in this strategy, we constructed chimeric promoters using sequences from RNR3 and ADH1. The expression of ADH1 is not affected by TAFII mutations (Apone et al., 1996; Walker et al., 1996; Shen and Green, 1997). The sequences used in the construction of these promoters were chosen based on previous promoter mapping studies of ADH1 (Shen and Green, 1997) and RNR3 (Endo-Ichikawa et al., 1996; Huang et al., 1998). We first asked whether the core promoter alone conferred TAFII-dependent transcription on RNR3 by analyzing the expression of a lacZ reporter gene driven by the RNR3 core promoter (RNR3core-lacZ; Figure 4A). Consistent with our results of Figure 1C, demonstrating that the uninduced levels of RNR gene transcription are not unaffected by TAFII mutations, inactivation of taf90-1, taf145-2 or tsm1 alleles had no effect on the expression of RNR3core-lacZ (Figure 4A). Next, we analyzed a promoter composed of the upstream regulatory sequences from RNR3 and the core promoter from the TAFII-independent ADH1 gene (RNR3US-ADH1core; Figure 4A). The regulatory region of RNR3 used in this construct has three DREs (X-boxes), which were shown to confer DNA damage-dependent transcription on the CYC1 promoter in previous studies (Endo-Ichikawa et al., 1996; Huang et al., 1998). The expression from the RNR3US-ADH1core chimeric promoter was dependent upon MMS treatment in the wild-type cells, and required functional TAF90 and TAF145 (Figure 4A).
Fig. 4. The TAFII-dependent region of RNR3 maps to the damage-responsive elements (DREs). (A) Schematic representations of the chimeric promoters (above). The positions of the DREs are indicated by the black bars above the RNR3 promoter and are located at –213, –261 and –323 relative to the major transcription start site. The TATA boxes are located at RNR3, –75, ADH1, –91. Yeast strains transformed with the plasmids indicated on the right hand side of the panel were grown at 23°C to midlog (0), transferred to 37°C for 45 min, and MMS was added to a final concentration of 0.015% to cells containing RNR3FL-LacZ, RNR3core-LacZ and RNR3US-ADH1coreLacZ. Cells were withdrawn for RNA isolation after 1.5 and 3 h. LacZ RNA was detected by northern blotting. Note that the lacZ transcript marked with an asterisk originates from the TSM1 expression plasmid, pRS313-TSM1. (B) The same as (A), but using a construct containing a synthetic tandem array of four DREs (Xs) from the RNR2 promoter positioned upstream of the ADH1 core promoter.
Detailed analyses of the RNR3 gene revealed that it is regulated primarily by a transcriptional repression mechanism mediated by three DREs (Zhou and Elledge, 1992; Endo-Ichikawa et al., 1996; Huang et al., 1998); therefore, it is likely that the requirement for TAFIIs maps to these regulatory sequences. Nonetheless, the region of RNR3 used in our initial mapping studies is quite large, and it is possible that TAFIIs are functioning as co-activators for an unidentified transcription factor that binds to this region. Therefore, to map definitely the TAFII dependence to the DREs, we constructed a promoter containing a tandem array of consensus DREs (X-boxes) positioned upstream of the ADH1 core promoter (DRE-ADH1core; Figure 4B). The results of Figure 4B show that the expression of the DRE-ADH1core promoter in wild-type cells was dependent upon the addition of MMS to the culture medium and, more importantly, they also show that its expression was TAF145 and TAF90 dependent. On the basis of these results, we conclude that the TAFII dependence of the RNR genes is mediated by the DREs.
The Ssn6–Tup1 complex is required for the TAFII dependence of the RNR genes
Mapping the TAFII-dependent region to the DREs indicates that the transcription factors that function at these sequences confer TAFII dependence to the RNR genes. In the absence of DNA damage-induced signals, the RNR genes are repressed by the Ssn6–Tup1 co-repressor complex, which is recruited to the promoter by the sequence-specific DNA-binding protein Crt1 (Huang et al., 1998). It is feasible that TAFIIs control RNR gene transcription by antagonizing Ssn6–Tup1-mediated repression and, if this is so, deleting SSN6 or TUP1 should alleviate the TAFII dependence. Double TAFIIts–/Δssn6 and TAFIIts–/Δtup1 mutants were shifted to 37°C to inactivate the TAFII mutants, and the levels of RNR2 and RNR4 mRNA were measured by northern blotting. Deletion of SSN6 (Figure 5A) or TUP1 (not shown) resulted in the derepression of the RNR genes in the wild-type and the mutant strains at the permissive temperature. More importantly, shifting the TAFIIts–/Δssn6 mutants to the restrictive temperature did not significantly affect the levels of RNR2 and RNR4 mRNA, when compared with the Δssn6 strain (Figure 5A). A slight (∼2-fold) heat shock-induced reduction was noted in all cells. This experiment shows that deletion of SSN6 or TUP1 alleviates the requirement for TAFIIs.
Fig. 5. TAFII-dependent transcription of the RNR genes requires the Ssn6–Tup1 co-repressor complex. (A) Analysis of RNR transcription in Δssn6 and double TAFts–/Δssn6 mutants. Strains were grown at 23°C and then transferred to 37°C. Aliquots of cells were withdrawn for RNA isolation at 1, 2 and 3 h following temperature shift. (B) Analysis of Ssn6–Tup1-regulated stress-responsive genes in conditional expression strains. Strains were grown in galactose (GAL), collected, washed, transferred into dextrose-containing medium for 10 h and then separated into three flasks. At that time, MMS (0.03%) or NaCl (1 M) was added to individual flasks. Aliquots of cells were removed after 1.5 and 3 h and used to isolate RNA.
The results presented above indicate that the Ssn6–Tup1 complex is required for the TAFII dependence of the RNR promoters; therefore, we explored the possibility that the transcription of all SSN6/TUP1-regulated genes relies upon TAF90 and TAF145 function. DDR48 and CTT1 are two SSN6/TUP1-regulated genes that are induced by heat shock and high osmolarity (Schmitt and McEntee, 1996; Marquez et al., 1998). DDR48 is also inducible by MMS, but is regulated by a mechanism different from RNR3 (Maga et al., 1986; Zhou and Elledge, 1993; Kiser and Weinert, 1996; Schmitt and McEntee, 1996). Since both genes are induced by heat shock, we used the TAFII depletion strategy described in Figure 1. Depletion of TAFII90p or TAFII145p did not affect the induction of either CTT1 or DDR48 (Figure 5B); thus, SSN6/TUP1 intrinsically cannot confer TAFII-dependent transcription to all promoters.
TAFIIs target the Crt1 repressor protein
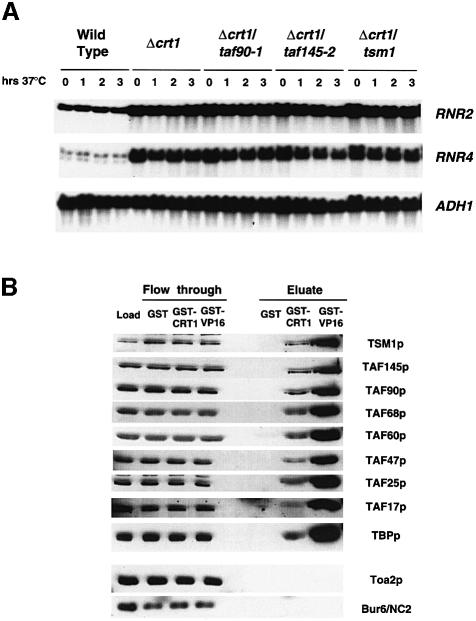
Both DDR48 and RNR3 are repressed by the Ssn6–Tup1 complex and are regulated by the DNA damage signaling pathway, but their distinct expression patterns are mediated by different sequence-specific DNA-binding proteins. The expression of DDR48 is dependent upon Msn2/4, and that of RNR3 upon Crt1 (Schmitt and McEntee, 1996; Huang et al., 1998), suggesting that the TAFII dependence of the RNR genes is specified by Crt1. We tested this hypothesis by analyzing the expression of RNR2 and RNR4 in double TAFIIts–/Δcrt1 strains using the strategy described in Figure 5A. Yeast strains containing a deletion of CRT1 have elevated levels of RNR mRNA compared with wild-type cells (Figure 6A, compare lane 1 with lane 5), which is in agreement with the findings of a published report (Huang et al., 1998). Similarly to what was observed in double TAFIIts–/Δssn6 and TAFIIts–/Δtup1 strains, shifting the TAFIIts–/Δcrt1 mutants to 37°C failed to result in significant reductions in the expression of RNR2 and RNR4 (Figure 6A). Identical results were obtained in the double mutants treated with MMS (data not shown).
Fig. 6. Crt1 mediates the TAFII dependence of the RNR genes. (A) Deletion of CRT1 relieves the TAFII requirement. Strains were treated as described in Figure 5A. (B) TFIID and Crt1 interact in vitro. Yeast whole-cell extracts were chromatographed on GST, GST–Crt1 and GST–VP16 affinity columns. After extensive washing, the bound proteins were eluted with high salt and were detected by western blotting. The relative amounts of material loaded onto the gel are as follows: load, 1/300 of the total; eluate, 1/35 of the total. Note that the amounts of the smaller TAFIIs (TAFII47, TAFII25 and TAFII17) in the eluates appear lower than they actually are because of the spreading of the signal caused by the high salt in the eluate fractions.
TFIID is believed to function by interacting with sequence-specific DNA-binding proteins and general transcription factors (Burley and Roeder, 1996; Lee and Young, 1998), and it is likely that it regulates the RNR genes by interacting with Crt1. We explored the likelihood of this mechanism by examining the ability of a glutathione S-transferase (GST)–Crt1 affinity column to retain subunits of TFIID from whole-cell extracts. GST and GST–VP16 columns were used as negative and positive controls, respectively. As expected, both the GST–Crt1 and GST–VP16 columns retained subunits of TFIID, namely TAFII170/Tsm1p, TAFII145p, TAFII90p, TAFII68p, TAFII60p, TAFII47p, TAFII25p, TBPp and TAFII17p (Figure 6B). However, neither column retained a subunit of TFIIA (Toa2) or yeast NC2 (Bur6p), two TBP-binding transcription factors that are distinct from TFIID. The retention of TAFIIs on the VP16 column was significantly higher, but since the quantity of fusion protein contained on each column was equalized based on protein content, rather than molar amounts, it is difficult to judge the relative affinity of TFIID for Crt1 versus VP16. The stoichiometry of the TAFIIs eluting from the GST–Crt1 column seemed to be equivalent because the fraction of each TAFII bound to the column (eluate) was similar to that contained in the starting material (load). Since both Crt1 and TFIID bind to DNA, we verified that this interaction is not dependent on nucleic acids by repeating the chromatography in the presence of the DNA-intercalating agent ethidium bromide (data not shown). These results support the idea that Crt1 mediates the TAFII dependence of the RNR gene promoters.
TFIID binds to a region of Crt1 required for Ssn6–Tup1 binding and repression
We next sought to map the region of Crt1 that is required for TFIID binding. Crt1 is a member of a family of highly conserved DNA-binding proteins characterized by the similarities within their DNA-binding and C-terminal dimerization/oligomerization domains (Emery et al., 1996). A schematic diagram of Crt1 is presented in Figure 7A. GST ‘pull-down’ experiments were performed in whole-cell extracts using full-length Crt1 and its derivatives, and the binding of TFIID was monitored by immunoblotting for TAFII145p and TBP. The results of Figure 7B show that all of the derivatives containing the first 240 amino acids of Crt1 retained TAFII145p and TBP as well as full-length protein; however, the DNA-binding domain and the entire C-terminus were dispensable for binding. The N-terminus of Crt1 contains a proline-rich domain, and since proline-rich domains in some gene-regulatory proteins interact with human TFIID (Chiang and Roeder, 1995), we tested whether or not this region binds to yTFIID. However, the proline-rich domain of Crt1 is not sufficient for interacting with TFIID because a derivative containing this region and amino acids up to the DNA-binding domain [GST–Crt1(77–240)] failed to retain TFIID subunits in this assay (Figure 7B). From this analysis, we conclude that TFIID interacts with the N-terminus of Crt1.
Fig. 7. Mapping of Crt1. (A) Domains of Crt1. Amino acids 1–40 are dispensable for Crt1 function (Huang et al., 1998). The locations of the proline-rich domain, 77–155; DNA-binding domain, 246–318; a conserved ‘B’ box and a region displaying homology to numerous yeast and metazoan genes, 411–579 are indicated in the diagram. (B) Mapping of the TFIID interaction region. A 50 µg aliquot of GST derivatives bound to glutathione–agarose beads was incubated with yeast whole-cell extracts, washed and eluted as described in Materials and methods. TFIID binding was detected by western blotting for TAFII145p and TBP. (C) Mapping of the Ssn6–Tup1 interaction region. A 20 µg aliquot of GST derivatives bound to glutathione–agarose beads was incubated with in vitro translated Ssn6 and Tup1, washed and eluted as described in Materials and methods. The binding of Ssn6 and Tup1 was detected by fluorography of SDS–polyacrylamide gels. (D) Identification of the repression domain of Crt1. Plasmids expressing Crt1–LexA DNA-binding domain fusion proteins were transformed into BY4705 cells containing the reporter plasmid JK101 (Brent and Ptashne, 1985) and grown in liquid SC-raffinose medium. β-galactosidase activities were measured in protein extracts prepared from at least three independent isolates. The results shown are from the same experiment; however, the average fold repression and SEMs from three independent experiments were: LexA–Crt1(1–811), 16.5 ± 1.8; LexA–Crt1(1–240) 37.5 ± 3.2; LexA–Crt1(1–350) 21.4 ± 1.6.
We next determined whether the region of Crt1 that binds to TFIID is important for its function as a transcriptional repressor. Crt1 binds to Ssn6 and Tup1; however, the region required for this function is not known (Huang et al., 1998). We therefore used the GST–Crt1 derivatives described above to identify the region that binds to Ssn6 and Tup1. The results shown in Figure 7C demonstrate that both Ssn6 and Tup1 bound to the N-terminus of Crt1(1–240). Moreover, there is a correlation between the region required for Ssn6 and Tup1 and TFIID binding because neither GST–Crt1(77–240) nor GST–Crt1(155–240) was capable of interacting with Ssn6 or Tup1. In these experiments, Ssn6 and Tup1 were co-translated; however, identical results were obtained when Ssn6 or Tup1 binding was analyzed individually (not shown).
The repression domain of Crt1 was identified by analyzing the ability of LexA DNA-binding domain–Crt1 fusion proteins to repress a promoter containing LexA operator sites in vivo (Brent and Ptashne, 1985). We found that the entire coding region of Crt1 repressed the reporter gene >15-fold when fused to the LexA DNA-binding domain [LexA–Crt1(1–811); Figure 7D]. Derivatives containing only the N-terminus, LexA–Crt1(1–240), or the N-terminus and DNA-binding domain, LexA–Crt1(1–350), were more effective than LexA–Crt1(1–811), repressing the reporter gene up to 40- and 25-fold, respectively. We speculate that the weaker repression by LexA–Crt1(1–811) compared with the shorter derivatives may result from the presence of Crt1’s DNA-binding and/or oligomerization domain, which may interfere with its ability to interact with the promoter. An intact N-terminus of Crt1 is required for its transcriptional repression function because neither LexA–Crt1(77–240) nor LexA–Crt1(155–240) repressed the reporter gene significantly (Figure 7D). None of the derivatives tested enhanced transcription from the reporter gene. Western blotting of cell extracts using antibodies to the LexA DNA-binding domain revealed that all derivatives accumulated to levels within 2-fold of each other (not shown). Our mapping studies of Crt1 indicate that the N-terminus of Crt1 is required for transcriptional repression, Ssn6–Tup1 binding and TFIID binding.
Discussion
We report that TAF68, TAF90 and TAF145 are required for the induction of the RNR genes by DNA damage. Unlike the TAF145-dependent ribosomal protein and cyclin genes, expression of the RNR genes requires multiple TAFIIs. However, despite showing an overall sensitivity to TAFII mutations and depletion, the individual RNR genes showed different sensitivities to these conditions. RNR4 seemed to be the least sensitive and, interestingly, it has the weakest consensus and the fewest DREs in its promoter region compared with the other two genes (Huang et al., 1998); thus, it is less tightly regulated by Crt1 and DNA damage. This is consistent with our mapping of the TAFII dependence to the DREs and Crt1 function. The regulation of the RNR genes is also different from that of the previously characterized TAF145- dependent genes (Shen and Green, 1997; Walker and Green, 1997; Tsukihashi et al., 2000) in that the TAFII-dependent region maps to the upstream repression sequences, the DREs. Our results indicate that TFIID has other functions in addition to promoter recognition and selectivity in yeast, which have not been firmly established by previous work. Three recent studies identified TAF17 and TAF68 as putative co-activators of Gcn4-mediated transcription (Apone et al., 1998; Krishnamurthy et al., 1998; Moqtaderi et al., 1998); however, it is likely that this activity is attributed to their function in the SAGA HAT complex. Mutation of SAGA-specific components affects the Gcn4-mediated expression of HIS3 (Tr +13) and HIS4 (for review see Grant et al., 1998b), but depletion or mutation of TAFIIs not contained in SAGA does not (Moqtaderi et al., 1996, 1998; our unpublished data). Moreover, the SAGA complex, but not TFIID, can interact with Gcn4 (Drysdale et al., 1998; Grant et al., 1998a). In light of these observations, it is unlikely that TFIID functions as a co-activator of Gcn4-mediated transcription. We argue that the effects we observe on the RNR promoters are mediated by TFIID based upon the following criteria: (i) expression of these genes is affected by mutation or depletion of the TFIID subunit TAF145; (ii) their expression is not affected by the deletion of SAGA-specific components (our unpublished data); and (iii) TFIID binds to Crt1 in vitro. Our conclusion is dependent upon the widely accepted view that TAFII145p is specific for TFIID. It is a formal possibility that TAFII145p functions outside of the context of TFIID but, without evidence to support this, our interpretations are valid.
A seemingly obvious explanation for the TAFII dependence of the RNR promoter is that TFIID has limited access to the repressed promoter, and it requires multiple TAFII–DNA contacts to stabilize its interaction with the core promoter (for reviews see Burley and Roeder, 1996; Verrijzer and Tjian, 1996; Lee and Young, 1998). Mutant TFIID may be unable to access the promoter under repressive conditions but, when repression is relieved, it can do so. Our data suggest that the mechanism is not this simple. If true, we expect that all strongly repressed promoters, especially Ssn6–Tup1-regulated promoters, would have a similar requirement for TAFIIs, and that the core promoter would mediate the TAFII dependence. Our results indicate that neither of these is true.
We present evidence that the TAFII dependence of the RNR genes is specified by Crt1. Studies on Crt1 clearly indicate that it is a transcriptional repressor and has no detectable gene activation activities (Huang et al., 1998; Figure 7D) and, thus, TAFIIs are acting as antirepressors, rather than as co-activators. To our knowledge, this is the first description of such activities of TAFIIs in vivo. TFIID interacts within the same region of Crt1 that is required for repression and Ssn6–Tup1 binding, and deletion of any of these three genes alleviates the TAFII dependence of the RNR promoters. Therefore, the mechanism is likely to involve competition (functional or physical) between TFIID and the Ssn6–Tup1–Crt1 complex at the promoter. How the interaction of TFIID with Crt1 inhibits its activity is not clear at this time; however, it may do so by introducing a conformational change in the repression domain of Crt1 and affecting its function. Studies have shown that the binding of hTAF31 to the activation domains of VP16 and p53 causes them to fold into a conformation conducive to gene activation (Uesugi et al., 1997; Uesugi and Verdine, 1999). Given the architectural and, in some cases, strategical similarities between transcriptional activators and repressors, a TAFII-induced conformational change in the repression domain of Crt1 is feasible.
How TAFIIs mediate the derepression of the RNR genes is an open question; however, the mechanism is certainly highly interdependent upon the functions of the co-repressor complex. Ssn6–Tup1 is reported to repress transcription factors directly (Herschbach and Johnson, 1993; Kuchin and Carlson, 1998; Papamichos-Chronakis et al., 2000), and it is feasible that TFIID is antagonizing interactions between Ssn6–Tup1–Crt1 and components of the pre-initiation complex, such as subunits of the RNA polymerase holoenzyme complex (Kuchin and Carlson, 1998; Papamichos-Chronakis et al., 2000). Ssn6–Tup1 can repress genes by a chromatin-mediated mechanism (Roth, 1995), and genetic analysis implicates histone modification in the derepression of the RNR2 gene (Edmondson et al., 1996). A role for TFIID in the activation of the ADH2 gene has been proposed (Komarnitsky et al., 1998) and, interestingly, the expression of ADH2 requires extensive chromatin remodeling at its promoter (Verdone et al., 1996). Given this genetic evidence, TAFIIs may be involved in regulating chromatin structure. There are two models that support a chromatin-mediated mechanism. First, TFIID itself modifies histones around the promoter through TAF145p’s HAT activity (Mizzen et al., 1996). Alternatively, TAFIIs may play a role in the recruitment of HAT complexes, or other chromatin remodeling activities, to the promoter. TBP binds to some SAGA acetyltransferase complex components, and recruitment of SAGA to promoters by TBP/TFIID has been presented as a targeting mechanism (for review see Grant et al., 1998b). Future studies aimed at analyzing the recruitment of chromatin-modifying complexes by TFIID, isolation and characterization of HAT-defective mutants of TAF145 and the delineation of the chromatin structure of the RNR3 promoter will further our understanding of the antirepression activities of TFIID.
Materials and methods
Yeast strains and genetic manipulations
The strains used in this study are as follows: AW41 (Ray et al., 1991); tbp1-1 (Cormack and Struhl, 1992); DDY547 and DDY555 (Welch et al., 1993); LYC-1 and LY20 (Apone et al., 1996); YSW87, YSW94, YJR11 and YJR18 (Walker et al., 1996); YJR195, as YSW93, ssn6Δ::URA3; YJR196, as YSW93, tup1Δ::URA3; YJR199, as AW41, ssn6Δ::URA3; YJR200, as AW41, tup1Δ::URA3; YJR201, as LY20, ssn6Δ::URA3; YJR202, as LY20, tup1Δ::URA3; YJR220, as YSW87, tup1Δ::URA3; YJR221, as YSW87, ssn6Δ::URA3; YJR352, as YSW87, crt1Δ::URA3; YJR353, as LY20, crt1Δ::URA3; YJR354, as YSW93, crt1Δ::URA3; YJR 355, as AW41, crt1Δ::URA3; WCS131, Matα, ade2-101, his3-200, leu2-Δ1, lys2-801, trp1Δ63, ura3-52, tsm1::TRP1, [pRS313-TSM1]; WCS132, as WCS131 except [pRS313-tsm1-1]; BY4705, Mat α, ade2Δ::hisg, his3Δ200, leu2Δ0, lys2Δ0, met15Δ0, trp1Δ63, ura3Δ0. The SSN6 and TUP1 deletion strains were constructed by transforming with AvrII-digested pRS406-Δssn6 or pRS406-Δtup1 (Cooper et al., 1994). Deletion of SSN6 or TUP1 in the TAFII mutants did not result in any obvious enhanced synthetic phenotypes compared with the single mutants alone (not shown). CRT1 knockouts were constructed by transforming cells with EagI–XhoI-digested pBScrt1Δ::URA3, and verified by Southern blotting and examination of RNR gene expression.
Temperature shift experiments were typically conducted as follows: cells are grown to midlog, and the culture transferred to a shaking water bath at 37°C. Cultures were typically pre-incubated at 37°C for 45–120 min and then treated with DNA-damaging or replicative stress agents. Aliquots were taken for RNA preparation at various times thereafter. TAFII depletion experiments were conducted as described previously (Walker et al., 1996). Aliquots were removed for RNA isolation and protein extract preparation.
Plasmid constructions
All promoter fragments were amplified by PCR from genomic DNA and inserted into the polycloning site of pRS316. For the RNR3 and ADH1 genes, +1 indicates the start site of transcription determined by primer extension analysis (Tornow and Santangelo, 1990; Yagle and McEntee, 1990). DRE-ADH1-lacZ was constructed by inserting a synthetic double-stranded oligonucleotide (5′-tcgagTCGCCATGGCAACactTCGCCATGGCAACctgTCGCCATGGCAACtgaTCGCCATGGCAACt-3′). PCR primer sequences and a detailed description of the construction are available upon request. pGST–Crt1 and its derivatives were generated by inserting the coding sequence of CRT1 into pRET3aGSTN2, which contains the GST gene driven by the T7 promoter (a gift from Dr Song Tan). Regions of CRT1 were amplified by PCR and cloned into pEG202 (Brent and Ptashne, 1985) to produce LexA DNA-binding domain fusions. The coding regions of SSN6 and TUP1 were amplified by PCR and cloned into BS-KS+ for use in in vitro transcription/translation.
RNA analysis, western blotting and affinity chromatography
Methods for protein extraction used for western blotting, and RNA isolation and analysis were described in Walker et al. (1996) and Apone et al. (1996). The methods for affinity chromatography and yeast whole-cell extract preparation were described previously (Reese et al., 1994). Columns of 0.5 ml (0.8 mg/ml GST protein) were equilibrated with 0.15 M potassium acetate buffer T (Reese et al., 1994) supplemented with 0.003% NP-40 and protease inhibitors, and 1 ml of yeast whole-cell extract (∼5 mg/ml protein) was passed 5–10 times over the column. After extensive washing with 0.15 M buffer T, proteins were eluted with 1 M NaCl in buffer T supplemented with 0.003% NP-40 and protease inhibitors. In some experiments, chromatography was performed in the presence of 300 µg/ml ethidium bromide. GST pull-downs were performed similarly except that all steps were carried out in batch. Briefly, ∼50 µg of GST, GST–Ssn6 and GST–Crt1 protein were incubated with whole-cell extracts for 1.5 h at 4°C, washed four times for 10 min and eluted. The Ssn6–Tup1 interaction assays were performed as follows: 20 µg of GST–Crt1 (or mutant derivatives) were incubated with 10 µl of co-translated Ssn6 and Tup1 in 90 µl of binding buffer [20 mM HEPES–KOH pH 7.5, 150 mM potassium acetate, 1 mM EDTA, 1 mM dithiothreitol, 5% (v/v) glycerol and 0.01% NP-40]. After a 60 min incubation at 4°C, the beads were collected by low speed centrifugation and washed four times for 10 min each with 300 µl of binding buffer. The bound proteins were eluted with SDS–PAGE loading buffer, separated by SDS–PAGE, stained, treated with En3Hance (Dupont-NEN), dried and exposed to film.
Acknowledgments
Acknowledgements
We thank Drs Michael Green, Winston Shen, Steve Elledge, Bob Simpson, Rick Young, Steve Hahn, Song Tan and Kevin Struhl for strains, antibodies and plasmids. The authors acknowledge the technical assistance of Aihong Pan and Michael Stitzel in generating the Δssn6 and Δtup1 strains. We thank Drs Simpson, Jerry Workman, Tan, Dave Gilmour, Davis Ng, Craig Cameron, Patrick Grant, Danny Reinberg, Mike Hampsey and members of the Reese laboratory and the Pennsylvania State gene regulation group for advice and comments on this work. This research was supported by funds provided by the National Institutes of Health (GM58672) to J.C.R.
References
- Apone L.M., Virbasius,C.M., Reese,J.C. and Green,M.R. (1996) Yeast TAFII90 is required for cell-cycle progression through G2/M but not for general transcription activation. Genes Dev., 10, 2368–2380. [DOI] [PubMed] [Google Scholar]
- Apone L.M., Virbasius,C.A., Holstege,F.C., Wang,J., Young,R.A. and Green,M.R. (1998) Broad, but not universal, transcriptional requirement for yTAFII17, a histone H3-like TAFII present in TFIID and SAGA. Mol. Cell, 2, 653–661. [DOI] [PubMed] [Google Scholar]
- Brent R. and Ptashne,M. (1985) A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell, 43, 729–736. [DOI] [PubMed] [Google Scholar]
- Burley S.K. and Roeder,R.G. (1996) Biochemistry and structural biology of transcription factor IID (TFIID). Annu. Rev. Biochem., 65, 769–799. [DOI] [PubMed] [Google Scholar]
- Chiang C.M. and Roeder,R.G. (1995) Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science, 267, 531–536. [DOI] [PubMed] [Google Scholar]
- Cooper J.P., Roth,S.Y. and Simpson,R.T. (1994) The global transcriptional regulators, SSN6 and TUP1, play distinct roles in the establishment of a repressive chromatin structure. Genes Dev., 8, 1400–1410. [DOI] [PubMed] [Google Scholar]
- Cormack B.P. and Struhl,K. (1992) The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell, 69, 685–696. [DOI] [PubMed] [Google Scholar]
- Desany B.A., Alcasabas,A.A., Bachant,J.B. and Elledge,S.J. (1998) Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev., 12, 2956–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale C.M. et al. (1998) The Gcn4p activation domain interacts specifically in vitro with RNA polymerase II holoenzyme, TFIID, and the Adap–Gcn5p coactivator complex. Mol. Cell. Biol., 18, 1711–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson D.G., Smith,M.M. and Roth,S.Y. (1996) Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev., 10, 1247–1259. [DOI] [PubMed] [Google Scholar]
- Elledge S.J. and Davis,R.W. (1989) DNA damage induction of ribonucleotide reductase. Mol. Cell. Biol., 7, 2783–2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S.J. and Davis,R.W. (1990) Two genes differentially regulated in the cell cycle and by DNA-damaging agents encode alternative regulatory subunits of ribonucleotide reductase. Genes Dev., 4, 740–751. [DOI] [PubMed] [Google Scholar]
- Elledge S.J., Zhou,Z. and Allen,J.B. (1992) Ribonucleotide reductase: regulation, regulation, regulation. Trends Biochem. Sci., 17, 119–123. [DOI] [PubMed] [Google Scholar]
- Emery P., Durand,B., Mach,B. and Reith,W. (1996) RFX proteins, a novel family of DNA binding proteins conserved in the eukaryotic kingdom. Nucleic Acids Res., 24, 803–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo-Ichikawa Y., Kohno,H., Furukawa,T., Ueda,T., Ogawa,Y., Tokunaga,R. and Taketani,S. (1996) Requirement of multiple DNA–protein interactions for inducible expression of RNR3 gene in Saccharomyces cerevisiae in response to DNA damage. Biochem. Biophys. Res. Commun., 222, 280–286. [DOI] [PubMed] [Google Scholar]
- Grant P.A., Schieltz,D., Pray-Grant,M.G., Steger,D.J., Reese,J.C., Yates,J.R.R. and Workman,J.L. (1998a) A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell, 94, 45–53. [DOI] [PubMed] [Google Scholar]
- Grant P.A., Sterner,D.E., Duggan,L.J., Workman,J.L. and Berger,S.L. (1998b) The SAGA unfolds: convergence of transcription regulators in chromatin-modifying complexes. Trends Cell Biol., 8, 193–197. [DOI] [PubMed] [Google Scholar]
- Green M.R. (2000) TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem. Sci., 25, 59–63. [DOI] [PubMed] [Google Scholar]
- Hahn S. (1998) The role of TAFs in RNA polymerase II transcription. Cell, 95, 579–582. [DOI] [PubMed] [Google Scholar]
- Herschbach B.M. and Johnson,A.D. (1993) Transcriptional repression in eukaryotes. Annu. Rev. Cell Biol., 9, 479–509. [DOI] [PubMed] [Google Scholar]
- Hoffmann A., Oelgeschlager,T. and Roeder,R.G. (1997) Considerations of transcriptional control mechanisms: do TFIID–core promoter complexes recapitulate nucleosome-like functions? Proc. Natl Acad. Sci. USA, 94, 8928–8935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege F.C., Jennings,E.G., Wyrick,J.J., Lee,T.I., Hengartner,C.J., Green,M.R., Golub,T.R., Lander,E.S. and Young,R.A. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell, 95, 717–728. [DOI] [PubMed] [Google Scholar]
- Huang M. and Elledge,S.J. (1997) Identification of RNR4, encoding a second essential small subunit of ribonucleotide reductase in Saccharomyces cerevisiae. Mol. Cell. Biol., 17, 6105–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M., Zhou,Z. and Elledge,S.J. (1998) The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell, 94, 595–605. [DOI] [PubMed] [Google Scholar]
- Kiser G.L. and Weinert,T.A. (1996) Distinct roles of yeast MEC and RAD checkpoint genes in transcriptional induction after DNA damage and implications for function. Mol. Biol. Cell, 7, 703–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky P.B., Klebanow,E.R., Weil,P.A. and Denis,C.L. (1998) ADR1-mediated transcriptional activation requires the presence of an intact TFIID complex. Mol. Cell. Biol., 18, 5861–5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky P.B., Michel,B. and Buratowski,S. (1999) TFIID-specific yeast TAF40 is essential for the majority of RNA polymerase II-mediated transcription in vivo. Genes Dev., 13, 2484–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy N., Jackson,B.M., Rhee,E. and Hinnebusch,A.G. (1998) yTAF61 has a general role in RNA polymerase II transcription and is required by GCN4 to recruit the SAGA coactivator complex. Mol. Cell, 2, 683–692. [DOI] [PubMed] [Google Scholar]
- Kuchin S. and Carlson,M. (1998) Functional relationships of Srb10–Srb11 kinase, carboxy-terminal domain kinase CTDK-I, and transcriptional corepressor Ssn6–Tup1. Mol. Cell. Biol., 18, 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.I. and Young,R.A. (1998) Regulation of gene expression by TBP-associated proteins. Genes Dev., 12, 1398–1408. [DOI] [PubMed] [Google Scholar]
- Maga J.A., McClanahan,T.A. and McEntee,K. (1986) Transcriptional regulation of DNA damage responsive (DDR) genes in different rad mutant strains of Saccharomyces cerevisiae. Mol. Gen. Genet., 205, 276–284. [DOI] [PubMed] [Google Scholar]
- Marquez J.A., Pascual-Ahuir,A., Proft,M. and Serrano,R. (1998) The Ssn6–Tup1 repressor complex of Saccharomyces cerevisiae is involved in the osmotic induction of HOG-dependent and -independent genes. EMBO J., 17, 2543–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J., Halenbeck,R. and Kaufmann,J. (1999) Human transcription factor hTAFII150 (CIF150) is involved in transcriptional regulation of cell cycle progression. Mol. Cell. Biol., 19, 5548–5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger D., Scheer,E., Soldatov,A. and Tora,L. (1999) Mammalian TAFII30 is required for cell cycle progression and specific cellular differentiation programmes. EMBO J., 18, 4823–4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B., Komarnitsky,P. and Buratowski,S. (1998) Histone-like TAFs are essential for transcription in vivo. Mol. Cell, 2, 663–673. [DOI] [PubMed] [Google Scholar]
- Mizzen C.A. et al. (1996) The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell, 87, 1261–1270. [DOI] [PubMed] [Google Scholar]
- Moqtaderi Z., Bai,Y., Poon,D., Weil,P.A. and Struhl,K. (1996) TBP-associated factors are not generally required for transcriptional activation in yeast. Nature, 383, 188–191. [DOI] [PubMed] [Google Scholar]
- Moqtaderi Z., Keaveney,M. and Struhl,K. (1998) The histone H3-like TAF is broadly required for transcription in yeast. Mol. Cell, 2, 675–682. [DOI] [PubMed] [Google Scholar]
- Ogryzko V.V., Kotani,T., Zhang,X., Schlitz,R.L., Howard,T., Yang,X.J., Howard,B.H., Qin,J. and Nakatani,Y. (1998) Histone-like TAFs within the PCAF histone acetylase complex. Cell, 94, 35–44. [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis M., Conlan,R.S., Gounalaki,N., Copf,T. and Tzamarias,D. (2000) Hrs1/Med3 is a Cyc8–Tup1 corepressor target in the RNA polymerase II holoenzyme. J. Biol. Chem., 275, 8397–8403. [DOI] [PubMed] [Google Scholar]
- Ray B.L., White,C.I. and Haber,J.E. (1991) The TSM1 gene of Saccharomyces cerevisiae overlaps the MAT locus. Curr. Genet., 20, 25–31. [DOI] [PubMed] [Google Scholar]
- Reese J.C., Apone,L., Walker,S.S., Griffin,L.A. and Green,M.R. (1994) Yeast TAFIIS in a multisubunit complex required for activated transcription. Nature, 371, 523–527. [DOI] [PubMed] [Google Scholar]
- Reese J.C., Zhang,Z. and Kurpad,H. (2000) Identification of a novel TFIID subunit, TSG2/TAF48. J. Biol. Chem., 275, 17391–17398. [DOI] [PubMed] [Google Scholar]
- Roth S.Y. (1995) Chromatin-mediated transcriptional repression in yeast. Curr. Opin. Genet. Dev., 5, 168–173. [DOI] [PubMed] [Google Scholar]
- Sanders S.L., Klebnaow,E.R. and Weil,P.A. (1999) TAF25p, a non-histone-like subunit of TFIID and SAGA complexes, is essential for total mRNA gene transcription in vivo. J. Biol. Chem., 274, 18847–18850. [DOI] [PubMed] [Google Scholar]
- Schmitt A.P. and McEntee,K. (1996) Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 93, 5777–5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W.C. and Green,M.R. (1997) Yeast TAFII145 functions as a core promoter selectivity factor, not a general coactivator. Cell, 90, 615–624. [DOI] [PubMed] [Google Scholar]
- Soldatov A., Nabirochkina,E., Georgieva,S., Belenkaja,T. and Georgiev,P. (1999) TAFII40 protein is encoded by the e(y)1 gene: biological consequences of mutations. Mol. Cell. Biol., 19, 3769–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki-Yagawa Y., Guermah,M. and Roeder,R.G. (1997) The ts13 mutation in the TAFII250 subunit (CCG1) of TFIID directly affects transcription of D-type cyclin genes in cells arrested in G1 at the nonpermissive temperature. Mol. Cell. Biol., 17, 3284–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornow J. and Santangelo,G.M. (1990) Efficient expression of the Saccharomyces cerevisiae glycolytic gene ADH1 is dependent upon a cis-acting regulatory element (UASRPG) found initially in genes encoding ribosomal proteins. Gene, 90, 79–85. [DOI] [PubMed] [Google Scholar]
- Tsukihashi Y., Miyake,T., Kawaichi,M. and Kokubo,T. (2000) Impaired core promoter recognition caused by novel yeast TAF145 mutations can be restored by creating a canonical TATA element within the promoter region of the TUB2 gene. Mol. Cell. Biol., 20, 2385–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uesugi M. and Verdine,G.L. (1999) The α-helical FXXPhiPhi motif in p53: TAF interaction and discrimination by MDM2. Proc. Natl Acad. Sci. USA, 96, 14801–14806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uesugi M., Nyanguile,O., Lu,H., Levine,A.J. and Verdine,G.L. (1997) Induced α helix in the VP16 activation domain upon binding to a human TAF. Science, 277, 1310–1313. [DOI] [PubMed] [Google Scholar]
- Verdone L., Camilloni,G., Di Mauro,E. and Caserta,M. (1996) Chromatin remodeling during Saccharomyces cerevisiae ADH2 gene activation. Mol. Cell. Biol., 16, 1978–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrijzer C.P. and Tjian,R. (1996) TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem. Sci., 21, 338–342. [PubMed] [Google Scholar]
- Walker S.S., Reese,J.C., Apone,L.M. and Green,M.R. (1996) Transcription activation in cells lacking TAFIIS. Nature, 383, 185–188. [DOI] [PubMed] [Google Scholar]
- Walker S.S., Shen,W.C., Reese,J.C., Apone,L.M. and Green,M.R. (1997) Yeast TAFII145 required for transcription of G1/S cyclin genes and regulated by the cellular growth state. Cell, 90, 607–614. [DOI] [PubMed] [Google Scholar]
- Wang E.H. and Tjian,R. (1994) Promoter-selective transcriptional defect in cell cycle mutant ts13 rescued by hTAFII250. Science, 263, 811–814. [DOI] [PubMed] [Google Scholar]
- Welch M.D., Vinh,D.B., Okamura,H.H. and Drubin,D.G. (1993) Screens for extragenic mutations that fail to complement act1 alleles identify genes that are important for actin function in Saccharomyces cerevisiae. Genetics, 135, 265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek E., Brand,M., Jacq,X. and Tora,L. (1998) Function of TAF(II)-containing complex without TBP in transcription by RNA polymerase II. Nature, 393, 187–191. [DOI] [PubMed] [Google Scholar]
- Yagle K. and McEntee,K. (1990) The DNA damage-inducible gene DIN1 of Saccharomyces cerevisiae encodes a regulatory subunit of ribonucleotide reductase and is identical to RNR3. Mol. Cell. Biol., 10, 5553–5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z. and Elledge,S.J. (1992) Isolation of crt mutants constitutive for transcription of the DNA damage inducible gene RNR3 in Saccharomyces cerevisiae. Genetics, 131, 851–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z. and Elledge,S.J. (1993) DUN1 encodes a protein kinase that controls the DNA damage response in yeast. Cell, 75, 1119–1127. [DOI] [PubMed] [Google Scholar]
- Zhou J., Zwicker,J., Szymanski,P., Levine,M. and Tjian,R. (1998) TAFII mutations disrupt Dorsal activation in the Drosophila embryo. Proc. Natl Acad. Sci. USA, 95, 13483–13488. [DOI] [PMC free article] [PubMed] [Google Scholar]