Abstract
With recent interest in the molecular mechanisms responsible for Fabry disease, the number of known mutations in the GLA gene which encodes α-galactosidase A has expanded considerably. Combining a large database of Fabry disease-causing mutations with the recently determined crystal structure of human α-galactosidase A allows for a new understanding of the atomic defects in the protein responsible for Fabry disease. We have conducted a systematic survey of the known Fabry disease-causing mutations and analyzed the mutations in the context of the α-galactosidase A structure. We have applied quantitative methods for identifying the plausible effect of each mutation on the α-galactosidase A protein. We present the analysis of 331 different defects in the GLA gene leading to non-native proteins in patients with Fabry disease. These mutations include 278 missense mutations, 49 nonsense mutations, and 4 single amino acid deletions.
Conclusion
Over half of the residues in the protein have been found to have changes in patients with Fabry disease. Most of these genetic mutations lead to the disruption of the hydrophobic core of the protein, thus Fabry disease is primarily a disease of protein folding. Further understanding of α-galactosidase A, one of the best studied members of the lysosomal storage disease family, will lead to increased understanding of other lysosomal storage diseases and other protein folding diseases.
Keywords: α-galactosidase, Fabry disease, glycosidase, lysosomal storage disease, E.C.-3.2.1.22
The degradation of macromolecules, including glycopeptides and glycolipids, occurs in the lysosome via catabolic enzymes. For example, glycosidases cleave the oligosaccharides from glycoproteins and glycolipids into smaller components used by the cell. In humans, defects in these lysosomal enzymes cause lysosomal storage diseases (LSDs). In this family of diseases, a defect in a gene leads to the loss of functional enzyme, which then leads to the accumulation of substrates in the tissues. The symptoms of the particular disease depend upon the amount, toxicity and location of the substrate that accrues. Over forty different LSDs have been identified to date, including Tay–Sachs, Gaucher, Sandhoff and Fabry diseases.
Fabry disease, an X-linked inherited disorder affecting 1 in every 40000 males, is characterized by chronic pain, vascular degeneration, cardiac and renal abnormalities, and other symptoms [1]. The disease was first reported in 1898 [2], and the defective protein responsible for the disease was first identified to be α-galactosidase A (α-gal A, also known as GLA and α-GAL) (E.C. 3.2.1.22) in 1967 [3]. α-Galactosidase A is encoded by the GLA gene and catalyzes the removal of terminal α-galactose groups from substrates such as glycoproteins and glycolipids. In patients with Fabry disease the loss of functional enzyme leads to the buildup of substrates, primarily globotriaosylceramide, in the tissues [1]. Most patients with Fabry disease have a single point mutation in their GLA gene, and several hundred different missense and nonsense mutations have been identified in the patient population.
The desired result of clinical research on LSDs is to develop treatments that would prevent disease symptoms from ever appearing. In Gaucher disease (caused by a defect in the lysosomal enzyme β-glucosidase), enzyme replacement therapy (ERT) in paediatric patients successfully prevents disease and is currently in use in over 3500 patients from 55 countries [4,5]. Gaucher disease was the first LSD approved for treatment with recombinant ERT in 1994 [4,5], and in 2001 Fabry disease became the second. We have recently determined the three-dimensional crystal structure (at 3.25 Å resolution) of the lysosomal enzyme human α-galactosidase A, the molecule which when deficient is responsible for the development of Fabry disease (Figure 1) [6]. This structure represents one of the first structures from family 27 and clan D (including over 270 proteins from different species) in the classification of glycoside hydrolases based upon similarities in sequences, active sites and mechanisms [7]. The structure was determined both alone and in complex with its catalytic product, the α-galactose monosaccharide, indicating the atomic basis for substrate recognition. The structural analysis revealed a portion of the glycoprotein’s N-linked carbohydrate, which accounts for 5–15% of the mass of α-galalactosidase A and represents over 70 different glycoforms attached to 23 different core carbohydrate structures [8]. The carbohydrate component of the glycoprotein is required for the molecule to traffic to the lysosome via the mannose-6-phosphate receptor pathway [9].
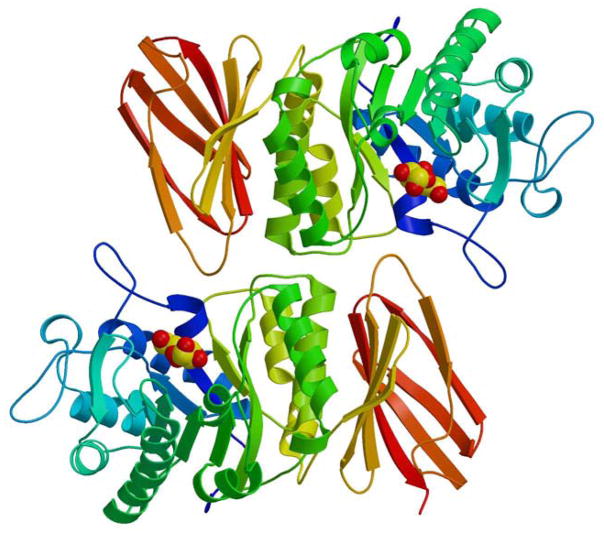
Figure 1.
The structure of α-galactosidase A. The structure of the human α-galactosidase A dimer is shown in ribbon representation. The ribbon is coloured from blue to red as the polypeptide goes from N- to C-terminus. The active site is identified by the catalytic product galactose, shown in sphere Corey-Pauling-Koltun (CPK) format. Each monomer in the homodimer contains two domains, a (β/ α)8 barrel containing the active site (blue to yellow) plus a C-terminal antiparallel β domain (yellow to red).
Following the recent determination of the structure of human α-galactosidase A, we are now able to ask fundamental questions about the structure and function of α-galactosidase A and its relationship to the human disease process. The α-galactosidase A structure revealed for the first time the locations and nature of the hundreds of different atomic defects responsible for Fabry disease. We are now in position to learn the details of the atomic basis for the loss of enzyme activity in patients with Fabry disease, and to tailor therapy based upon the specific defect in α-galactosidase A in different patients. In this report, we address the defects in the α-galactosidase A enzyme that lead to Fabry disease. As we are interested in the defects in the enzyme, in this work we do not include the mutations in the α-galactosidase A gene that lead to grossly altered gene products, such as insertions and deletions leading to frameshifts and splice defects. As Fabry disease represents one of the best studied LSDs, the lessons learned from the study of α-galactosidase A will lead to greater understanding of glycosidases and LSDs generally.
Materials and methods
Crystallographic structure determination
The methods associated with the X-ray crystallographic determination of the structure of human α-galactosidase A have been described previously [6]. Since that publication, we have extended our database of mutations associated with Fabry disease (see Tables I and II).
Table I.
Synopsis of Fabry disease-causing mutations.
| Number of mutations | Number of residues affected | |
|---|---|---|
| In full-length α-gal A | ||
| Single amino acid substitutions | 278 | 173 |
| Nonsense mutations | 49 | 49 |
| Single amino acid deletions | 4 | 4 |
| Total | 331 | 204 |
| In mature α-gal A | ||
| Single amino acid substitutions | 270 | 167 |
| Nonsense mutations | 49 | 49 |
| Single amino acid deletions | 4 | 4 |
| Total | 323 | 198 |
α-gal A, α-galactosidase A
Table II.
Details of mutations reported in patients with Fabry disease.
| Residue | Mutations found in Fabry disease patients | |||||
|---|---|---|---|---|---|---|
| Number | Type | Type | ASA | Effect on protein | Category | Reference |
| 1 | M | I | Initiation codon required | Other | [17] | |
| T | [18] | |||||
| R | [19] | |||||
| 14 | L | P | In signal sequence | Other | [20] | |
| 16 | L | P | In signal sequence | Other | [21] | |
| 19 | L | P | In signal sequence | Other | [22] | |
| 20 | A | P | In signal sequence | Other | [23] | |
| 31 | A | V | In signal sequence | Other | [18] | |
| 32 | L | P | 7.1 | Signal sequence cleavage site | Other | [24] |
| 34 | N | S | 1.7 | Bifurcated H-bond to N224 | Buried | [25] |
| K | [15] | |||||
| 35 | G | R | 6.3 | Gly phi/psi, Arg collides with N192 | Other | [26] |
| 40 | P | S | 0.0 | Helix terminus, buried and packed on W262 | Buried | [27] |
| L | [28] | |||||
| 41 | T | I | 0.8 | Buried | Buried | [15] |
| 42 | M | V | 0.7 | Buried, no room for beta branch | Buried | [29] |
| T | [19] | |||||
| L | [30] | |||||
| 43 | G | D | 0.0 | No room for side chain | Buried | [31] |
| V | [19] | |||||
| R | [32] | |||||
| 44 | W | X | 0.0 | Stop | Stop | [33] |
| C | Totally buried | Buried | [34] | |||
| 45 | L | R | 2.4 | Little room for a buried charge | Buried | [18] |
| 46 | H | R | 0.0 | Buried, next to W47 in active site | Buried | [18] |
| S | (double mutant with L45R) | [18] | ||||
| Y | [35] | |||||
| 47 | W | L | 3.8 | Active site residue | Active | [36] |
| G | [35] | |||||
| 48 | E | K | 0.0 | Buried | Buried | [37] |
| 49 | R | L | 2.9 | Mostly buried, on dimer interface | Buried | [26] |
| S | [29] | |||||
| P | [35] | |||||
| G | [32] | |||||
| 50 | F | C | 0.4 | Buried in hydrophobic pocket | Buried | [19] |
| 51 | M | K | 14.7 | Contacts to active site W47 and dimer interface | Active | [38] |
| 52 | C | S | 13.0 | Disulphide required near active site | Other | [39] |
| R | [17] | |||||
| X | Stop | Stop | [40] | |||
| 56 | C | G | 10.6 | Disulphide bond | Other | [25] |
| F | [39] | |||||
| Y | [29] | |||||
| X | Stop | Stop | [41] | |||
| 59 | E | K | 7.9 | Ion pairs across dimer interface to H406 | Other | [39] |
| 63 | C | Y | 1.7 | Buried disulphide | Other | [42] |
| 65 | S | T | 3.9 | Partially buried | Other | [43] |
| (splice variant) | [44] | |||||
| 66 | E | Q | 3.1 | Mostly buried in ion pair (double mutant with R112C) | Other | [45] |
| K | [42] | |||||
| 68 | L | F | 0.6 | Buried in hydrophobic pocket | Buried | [19] |
| 72 | M | V | 0.0 | Totally buried | Buried | [46] |
| R | [47] | |||||
| I | [32] | |||||
| 78 | S | X | 12.0 | Stop | Stop | [48] |
| 79 | E | X | 17.3 | Stop | Stop | [49] |
| 81 | W | X | 0.2 | Stop | Stop | [39] |
| S | Buried | Buried | [37] | |||
| 85 | G | D* | 3.3 | Restricted phi/psi in turn | Other | [24] |
| 86 | Y | C | 0.0 | Totally buried | Buried | [18] |
| X | Stop | Stop | [50] | |||
| 88 | Y | D | 2.4 | Partly buried in hydrophobic pocket | Buried | [36] |
| 89 | L | R | 0.1 | Buried in hydrophobic pocket | Buried | [39] |
| P | [18] | |||||
| 91 | I | T | 0.0 | Buried in hydrophobic pocket | Buried | [18] |
| 92 | D | H | 0.0 | Active site residue | Active | [29] |
| N | [18,38] | |||||
| 93 | D | G | 0.4 | Active site residue | Active | [29] |
| N | [41,51] | |||||
| V | [15] | |||||
| 94 | C | Y | 0.0 | Buried disulphide | Buried | [18] |
| S | [35] | |||||
| 95 | W | S | 0.0 | Buried in hydrophobic pocket | Buried | [28] |
| X | Stop | Stop | [37] | |||
| 97 | A | V | 2.0 | Mostly buried in turn, little room for larger side chains | Buried | [18] |
| P | [52] | |||||
| 99 | Q | X | 21.7 | Stop | Stop | [39] |
| 100 | R | K | 3.4 | Ion pairs to D155, stacked on Y151 | Other | [39] |
| T | [18] | |||||
| 103 | E | Q | 20.6 | Ion pair to R105 | Other | [42] |
| 107 | Q | X | 11.0 | Stop | Stop | [29] |
| 112 | R | C | 1.9 | Mostly buried guanidium group in ion pair | Buried | [45] |
| H | [39] | |||||
| S | [41] | |||||
| 113 | F | L | 0.0 | Completely buried in packed hydrophobic core | Buried | [18] |
| S | [35] | |||||
| 119 | Q | X | 18.0 | Stop | Stop | [29] |
| 120 | L | P | 1.5 | Side chain buried, main chain in middle of helix | Buried | [39] |
| 121 | A | T | 0.6 | No room for larger group (double mutant with L120P) | Buried | [39] |
| P | [53] | |||||
| 127 | K | X | 8.8 | Stop | Stop | [15] |
| 128 | G | E | 7.8 | Restricted phi/psi in turn | Other | [17] |
| 129 | L | P | 0.0 | Buried | Buried | [54] |
| 131 | L | P | 0.0 | Buried in hydrophobic pocket | Buried | [39] |
| 132 | G | R | 0.0 | No room for side chain | Buried | [19] |
| 134 | Y | S | 0.0 | Active site residue | Active | [18] |
| X | Stop | Stop | [28] | |||
| 135 | A | V | 0.0 | Buried, no room for larger side chain | Buried | [41,51] |
| 136 | D | H | 0.1 | Buried | Buried | [38] |
| 138 | G | R | 0.0 | Buried, no room for side chain | Buried | [18] |
| E | [32] | |||||
| 141 | T | I | 0.1 | Buried, mutation removes N139 carbohydrate | Buried | [19] |
| 142 | C | Y | 8.8 | Disulphide required near active site | Active | [55] |
| R | [40] | |||||
| X | Stop | Stop | [40] | |||
| W | [42] | |||||
| 143 | A | P | 3.0 | Larger side chain disrupts C52–C94 disulphide | Other | [39] |
| T | [18] | |||||
| 144 | G | V | 5.6 | In turn; gly restricted phi/psi | Other | [39] |
| 146 | P | S | 3.2 | Partially hydrophobic | Other | [56] |
| 147 | G | R | 1.6 | In turn; gly restricted phi/psi | Other | [42] |
| 148 | S | R | 0.7 | Buried, little room for larger side chain | Buried | [18] |
| N | [28] | |||||
| 151 | Y | X | 8.8 | Stop | Stop | [41] |
| 152 | Y | X | 1.4 | Stop | Stop | [19] |
| 153 | D | – | 13.8 | Deletion within helix changes register | Other | [41] |
| 155 | D | H | 0.0 | Buried, ion pairs to R100 | Buried | [51] |
| 156 | A | T | 1.0 | Buried, no room for larger side chain | Buried | [57] |
| V | [55] | |||||
| 157 | Q | X | 18.8 | Stop | Stop | [39] |
| 162 | W | R | 2.7 | Mostly buried in hydrophobic pocket | Buried | [25] |
| C | [58] | |||||
| X | Stop | Stop | [59] | |||
| 163 | G | V | 2.7 | In turn, restricted phi/psi | Other | [18] |
| 165 | D | V | 2.8 | Buried ion pair w/H125 | Buried | [26] |
| 166 | L | V | 0.4 | Buried, little room for branch at beta carbon | Buried | [55] |
| G | Creates hole in interior (2 nucleotide changes) | [15] | ||||
| 167 | L | P | 0.2 | Creates bend in strand β4 near active site | Active | [60] |
| 168 | K | R | 3.3 | Active site residue | Active | [19] |
| 169 | F | S | 0.0 | Buried in hydrophobic pocket | Buried | [38] |
| 170 | D | V | 0.0 | Catalytic residue in active site | Active | [18] |
| H | [37] | |||||
| 171 | G | R | 0.6 | Between two active site residues | Active | [51] |
| D | [41] | |||||
| 172 | C | Y | 11.4 | Required disulphide in active site | Active | [39] |
| R | [28] | |||||
| F | [38] | |||||
| G | [61] | |||||
| W | [62] | |||||
| 173 | Y | X | 12.6 | Stop | Stop | [15] |
| 177 | L | X | 10.6 | Stop | Stop | [41] |
| 183 | G | D | 0.3 | Buried, no room for side chain | Buried | [40] |
| S | [19] | |||||
| 187 | M | V | 0.0 | Totally buried, little room for branched beta carbon | Buried | [28] |
| T | [15] | |||||
| 191 | L | Q | 1.6 | Buried in hydrophobic pocket | Buried | [38] |
| P | [36] | |||||
| 194 | T | I | 0.2 | Buried, little room for longer side chain | Buried | [42] |
| 199 | V | M | 0.4 | buried, little room for longer side chain | Buried | [19] |
| 201 | S | F | 3.7 | Larger side chain extends into active site | Active | [41] |
| Y | [15] | |||||
| 202 | C | W | 0.0 | Buried disulphide | Buried | [56] |
| Y | [18] | |||||
| 204 | W | X | 0.5 | Stop | Stop | [28] |
| 205 | P | T | 0.0 | Buried hydrophobic | Buried | [17] |
| R | [19] | |||||
| L | [62] | |||||
| 207 | Y | S | 6.5 | Active site residue | Active | [19] |
| 215 | N | S | 7.5 | Mutation disrupts N-linked carbohydrate | Other | [63] |
| 216 | Y | D | 0.9 | Buried hydrophobic | Buried | [18] |
| 219 | I | N | 0.3 | Buried hydrophobic (2 nucleotide changes) | Buried | [39] |
| 220 | R | X | 7.6 | Stop | Stop | [64] |
| 221 | Q | X | 13.5 | Stop | Stop | [19] |
| 222 | Y | X | 5.8 | Stop | Stop | [65] |
| 223 | C | G | 0.0 | Buried disulphide | Buried | [66] |
| R | [19] | |||||
| Y | [62] | |||||
| 24 | N | D | 0.0 | Buried hydrogen bonding network | Buried | [48] |
| S | [28] | |||||
| 226 | W | X | 0.0 | Stop | Stop | [67] |
| R | Buried in hydrophobic pocket | Buried | [28] | |||
| C | [37] | |||||
| 227 | R | X | 0.4 | Stop | Stop | [63] |
| Q | Active site residue | Active | [25] | |||
| 230 | A | T | 43.1 | Affects critical D231 in active site | Active | [28] |
| 231 | D | N | 10.2 | Active site residue | Active | [68] |
| 234 | D | Y | 0.1 | Buried, no room for larger side chain | Buried | [19] |
| E | [41] | |||||
| 235 | S | C | 3.5 | Initiates a6 helix | Other | [40] |
| 236 | W | C | 1.9 | Buried | Buried | [29] |
| L | [40] | |||||
| X | Stop | Stop | [32] | |||
| R | [15] | |||||
| 239 | I | T | 0.0 | Buried | Buried | [53] |
| 242 | I | N | 0.2 | Buried in hydrophobic pocket | Buried | [49] |
| 243 | L | F | 0.2 | Buried in hydrophobic pocket, no room for Phe | Buried | [32] |
| 244 | D | N | 11.6 | Ion pairs to R356 | Other | [39] |
| H | [40] | |||||
| 245 | W | X | 3.5 | Stop | Stop | [32] |
| 247 | S | P | 2.0 | Buried on a6 helix, no room for Pro | Buried | [38] |
| C | [32] | |||||
| 250 | Q | X | 4.9 | Stop | Stop | [38] |
| 251 | E | X | 22.6 | Stop | Stop | [69] |
| 257 | A | P | 0.1 | Buried | Buried | [36] |
| 258 | G | R | 0.6 | Buried, restricted phi/psi in turn | Buried | [35] |
| 259 | P | L | 11.0 | P259 and G260 form beta turn | Other | [40] |
| R | [38] | |||||
| 260 | G | A | 0.4 | Buried in turn, restricted phi/psi | Buried | [76] |
| 261 | G | D | 0.0 | Buried in turn, no room for side chain | Buried | [49] |
| 262 | W | X | 0.0 | Stop | Stop | [70] |
| C | Completely buried | Buried | [42] | |||
| 263 | N | S | 0.9 | Buried in hydrogen bonding network | Buried | [18] |
| 264 | D | V | 4.3 | Near active site residues | Active | [25] |
| Y | [41] | |||||
| 265 | P | R | 0.8 | Buried, little room for Arg | Buried | [58] |
| 266 | D | V | 1.0 | Active site residue | Active | [25] |
| H | [28] | |||||
| N | [50] | |||||
| E | [32] | |||||
| 267 | M | I | 0.3 | Buried, little room for branch at beta carbon | Buried | [40] |
| R | [15] | |||||
| 268 | L | S | 0.0 | Completely buried | Buried | [42] |
| 269 | V | A | 0.0 | Completely buried | Buried | [63] |
| M | [15] | |||||
| 270 | I | T | 0.0 | Completely buried | Buried | [62] |
| 271 | G | C | 4.1 | In turn, restricted phi/psi, no room for large side chain | Other | [19] |
| S | [15] | |||||
| V | [15] | |||||
| 272 | N | K | 0.0 | Completely buried | Buried | [39] |
| S | [71] | |||||
| 276 | S | N | 0.8 | Buried | Buried | [42] |
| G | [41] | |||||
| 277 | W | X | 9.2 | Stop | Stop | [40] |
| 279 | Q | E | 0.3 | Buried hydrogen bonding network on dimer axis | Buried | [45] |
| H | [35] | |||||
| R | [37] | |||||
| K | [72] | |||||
| 280 | Q | H | 3.2 | Little room for larger side chain | Other | [35] |
| K | [51] | |||||
| 282 | T | N | 0.0 | Buried hydrophobic pocket | Buried | [38] |
| 283 | Q | P | 0.0 | Completely buried | Buried | [15] |
| 284 | M | T | 0.1 | Buried hydrophobic pocket | Buried | [17] |
| 285 | A | P | 0.2 | Buried | Buried | [41] |
| D | [15] | |||||
| 287 | W | X | 0.0 | Stop | Stop | [63] |
| G | Buried hydrophobic | Buried | [29] | |||
| C | [18] | |||||
| 288 | A | D | 0.0 | Buried in hydrophobic pocket on helix a7 | Buried | [39] |
| P | [19] | |||||
| 289 | I | F | 0.0 | No room for Phe | Buried | [40] |
| 290 | M | I | 0.8 | Buried | Buried | [15] |
| 292 | A | P | 0.0 | Completely buried | Buried | [34] |
| 293 | P | S | 0.3 | Buried | Buried | [36] |
| A | [19] | |||||
| T | [15] | |||||
| 294 | L | X | 0.0 | Stop | Stop | [35] |
| 296 | M | V | 0.0 | Completely buried in hydrophobic pocket | Buried | [73] |
| I | [23] | |||||
| 297 | S | F | 0.0 | Completely buried, no room for larger side chain | Buried | [25] |
| C | [32] | |||||
| 298 | N | K | 0.2 | Buried, in hydrogen bonding network | Buried | [17] |
| H | [29] | |||||
| S | [18] | |||||
| 300 | L | H | 0.0 | Completely buried | Buried | [42] |
| F | [41] | |||||
| 301 | R | Q | 3.6 | Partly buried, ion pairs with D299 | Other | [33] |
| X | Stop | Stop | [39] | |||
| P | [38] | |||||
| G | [19] | |||||
| 303 | I | N | 2.7 | Partially buried in hydrophobic pocket | Buried | [19] |
| 306 | Q | X | 14.5 | Stop | Stop | [15] |
| 312 | Q | H | 4.4 | Partially buried | Other | [15] |
| 313 | D | Y | 3.5 | Partially buried, little room for Tyr | Other | [25] |
| Polymorphism with ~60% wild-type activity | [74] | |||||
| 316 | V | E | 0.1 | Buried in hydrophobic pocket | Buried | [26] |
| 317 | I | T | 1.5 | Buried in hydrophobic pocket | Buried | [75] |
| 320 | N | K | 0.0 | Buried hydrogen bonding network | Buried | [76] |
| Y | [28] | |||||
| I | [42] | |||||
| 321 | Q | E | 5.0 | Ne2 of gln needed in H-bonding network | Other | [40] |
| X | Stop | Stop | [42] | |||
| R | [15] | |||||
| 325 | G | D | 0.6 | Buried, no room for side chain | Buried | [62] |
| 327 | Q | K | 0.6 | Buried hydrogen bonding network | Buried | [63] |
| E | [42] | |||||
| 328 | G | R | 0.2 | Buried, no room for side chain | Buried | [45] |
| A | [25] | |||||
| V | [41] | |||||
| 330 | Q | X | 1.8 | Stop | Stop | [51] |
| 333 | Q | X | 16.4 | Stop | Stop | [42] |
| 338 | E | K | 0.3 | Buried, H-bonds to W340 | Buried | [41] |
| X | Stop | Stop | [41] | |||
| 340 | W | X | 0.1 | Stop | Stop | [25] |
| R | Buried in hydrophobic pocket | Buried | [29] | |||
| 341 | E | K | 0.3 | Buried hydrogen bonding network | Buried | [77] |
| D | [19] | |||||
| 342 | R | X | 0.0 | Stop | Stop | [63] |
| Q | Totally buried | Buried | [56] | |||
| 344 | L | P | 2.3 | Mostly buried, mutation creates consecutive Pro’s | Buried | [42] |
| 345 | S | P | 14.9 | Little room for Pro | Other | [42] |
| 348 | A | P | 5.4 | Pro introduces kink in strand β11 | Other | [15] |
| 349 | W | X | 2.2 | Stop | Stop | [38] |
| 352 | A | D | 0.0 | Completely buried | Buried | [60] |
| 354 | I | K | 0.0 | Completely buried | Buried | [78] |
| 355 | N | K | 0.0 | Buried hydrogen bonding network | Buried | [32] |
| 356 | R | W | 4.6 | Ion pairs to D244 | Other | [79] |
| 357 | Q | X | 9.2 | Stop | Stop | [42] |
| 358 | E | – | 6.6 | Ion pair to K240 and hydrogen bonds to W236 | Other | [17] |
| K | [80] | |||||
| G | [32] | |||||
| A | [15] | |||||
| 360 | G | S | 0.9 | Gly specific phi/psi | Other | [51] |
| 361 | G | R | 3.8 | No room for large side chain | Other | [63] |
| 362 | P | L | 10.7 | initiates β12 strand | Other | [19] |
| 363 | R | H | 6.3 | Guanidinium packs on F337 | Other | [35] |
| C | [19] | |||||
| 365 | Y | X | 4.6 | Stop | Stop | [81] |
| 373 | G | S | 0.0 | Buried, no room for side chain | Buried | [76] |
| D | [82] | |||||
| 377 | A | D | 0.0 | Buried in hydrophobic pocket | Buried | [35] |
| 378 | C | Y | 0.0 | In disulphide | Buried | [40] |
| R | [83] | |||||
| 382 | C | W | 0.0 | In disulphide | Buried | [84] |
| Y | [37] | |||||
| 383 | F | – | 6.8 | Deletion shifts register of strand β13 | Other | [85] |
| 384 | I | N | 0.0 | Buried in hydrophobic pocket | Buried | [19] |
| 385 | T | P | 7.2 | Pro kinks strand β13 | Other | [19] |
| 386 | Q | X | 2.2 | Stop | Stop | [18] |
| P | Pro kinks strand β13 | Other | [15] | |||
| 396 | F | Y | 12.4 | Unknown; mutation not disease associated | Other | [86] |
| 398 | E | X | 16.2 | Stop | Stop | [25] |
| K | Unknown | Other | [19] | |||
| 399 | W | X | 12.6 | Stop | Stop | [18] |
| 401 | S | X | 8.3 | Stop | Stop | [19] |
| 404 | R | – | 9.5 | Deletion shifts register in strand β15 | Other | [25] |
| 407 | I | K | 0.1 | Buried | Buried | [37] |
| 409 | P | A | 0.0 | Buried; initiates strand β16 | Buried | [35] |
| T | [35] | |||||
| S | [32] | |||||
| 410 | T | K | 0.0 | Buried; no room for Lys, Ala introduces hole | Buried | [38] |
| A | [65] | |||||
| 411 | G | D** | 0.0 | Buried; no room for side chain | Buried | [48] |
| (double mutant with D313Y) | [74] | |||||
| 414 | L | S | 0.0 | Buried | Buried | [37] |
ASA: Accessible Surface Area as defined in Materials and Methods
Phi/psi: Main chain dihedral angles as seen in a Ramachandran plot
This mutation was described in the original citation as G85N, which is not possible from the point mutation identified.
This mutation was described in the original citation as G120Y.
Mutation database
The database of mutations were compiled from the following sources: Online Mendelian Inheritance in Man at Johns Hopkins University (http://www.ncbi.nlm.nih.gov/omim) (McKusick 1998), SWISS-PROT (http://www.expasy.org/sprot) [10], Human Gene Mutation Database (http://www.hgmd.org) [11], and the references listed in Table II. Residue numbering begins with the translation initiator Methionine numbered as +1, where the mature α-galactosidase A protein spans from L32 to L429. Four trinucleotide deletions are included in the α-galactosidase A point mutation list, as they result in deletions of single amino acids from the protein.
Solvent accessibility
Solvent accessible surfaces were calculated from the human α-galactosidase A structure with crystallography and NMR Suite [12] using a probe radius of 1.4 Å. Buried residues are defined as those with less than 2 Å2 solvent accessible surface area (ASA) per side chain atom, except for glycines (which have only one side chain atom) are defined as buried when all four atoms in the residue average less than 2Å 2 solvent accessible surface area.
Figure preparation
Molecular figures were drawn using the program MOLSCRIPT [13].
Results
We have assembled a database of 331 mutations in the exons of the GLA gene, including 278 missense mutations, 49 nonsense mutations and 4 single amino acid deletion mutations [10,11,14]. The mutated residues are found in 204 different locations in the protein sequence. We describe two different mutation sets in this paper: the combined set of α-galactosidase A missense and nonsense mutations, and the smaller set of missense mutations in the sequence that codes for the mature α-galactosidase A glycoprotein. The first set of mutations includes missense mutations, nonsense mutations and single amino acid deletions in the exons coding for the full-length α-galactosidase A protein, 331 mutations in total. These affect 204 of the 429 amino acids in the α-galactosidase A protein, or nearly half of the residues in the mature polypeptide. Of the 204 residues, 152 are missense mutants, 28 are nonsense mutants, and 21 are both missense and nonsense mutants. The second set of mutations, a subset of the first, includes only the missense mutations in the exons coding for the mature 398-residue polypeptide (after removal of the signal sequence). This database contains 270 distinct missense mutations affecting 167 residues. Because this set contains only point mutations affecting the mature polypeptide, it reveals properties of the folded protein. Table I summarizes the database of Fabry disease-causing mutations.
Table II lists the point and stop mutations found in patients with Fabry disease, along with their effects on the protein. Figure 2 plots the effects of Fabry disease-causing mutations on the structure of human α-galactosidase A. Surprisingly, the affected residues are not restricted to the region around the active site, but distribute throughout the structure, with some as far as 50 Å from the substrate-binding site. A single mutation in the GLA gene causes Fabry disease in all cases except for in two individuals with double mutations (corresponding to E66Q/R112C and L45R/H46S in the protein sequence). In general, mutations leading to a complete loss of enzyme activity affect residues in the interior of α-galactosidase A, while mutations leading to an α-galactosidase A protein with some residual enzyme activity tend to be found in more surface exposed residues.
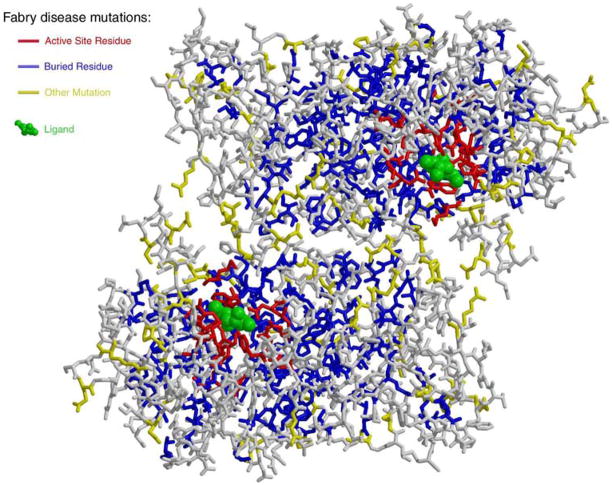
Figure 2.
Changes in α-galactosidase A associated with Fabry disease. The structure of human α-galactosidase A is shown in stick format in the same orientation as in Figure 1. For each missense mutation found in the Fabry disease mutation database, the affected residue is coloured by the resulting change in the glycoprotein. Perturbations of the active site are in red, changes to the buried core of the protein are shown in blue and other changes are shown in yellow. The ligand is shown in green to indicate the active site.
The database of Fabry disease mutations indicates the importance of different amino acids to the protein’s fold and function. A synopsis of the Fabry disease-causing mutations by amino acid affected appears in Table III. Based upon their frequency of mutation, Trp, Cys, Gln, Tyr, and Gly are the residue types most commonly altered by Fabry disease mutations. For example, 15 of the 16 Trp residues (94%) in the mature α-galactosidase A protein have been found to be affected by Fabry disease-causing mutations. Five of those Trp residues are affected exclusively by nonsense mutations, so 10 of the 16 Trp residues (63%) in α-galactosidase A are changed by missense mutations in patients with Fabry disease. Similarly, 10 of the 12 Cys residues in the protein are found as mutants in patients with Fabry disease, highlighting the importance of this residue to the structural integrity of the folded protein. In total, 77% of the 22 Gln residues in the structure are found altered in patients with Fabry disease, but most of these mutations are nonsense mutations that convert the Gln codon to a stop codon. Over two-thirds of the 31 Gly residues are altered in patients with Fabry disease, and all of them appear in missense mutations. Glycine, unique among the amino acids for its lack of a side chain, can appear in more main chain dihedral conformations than the other residues. As expected, the unique chemical properties of glycine make it sensitive to perturbation, and it is over-represented in the database. The basic residues Arg and Lys, although chemically quite similar, appear in the Fabry disease mutation database with markedly different frequencies: more than half of the 19 arginines in the structure are affected by mutations in patients with Fabry disease, but only two of the 17 lysines are affected. Arginine is more commonly found buried or partially buried in the structure compared with lysine, which is nearly always surface exposed, so arginine plays a more critical role in the folding of the protein. Surprisingly, Phe is less likely to be found altered in patients with Fabry disease compared with the average residue, with 5 of the 15 Phe residues (33%) affected by Fabry disease-causing mutations. The dearth of Phe residues in the database is unexpected for a large hydrophobic residue typically found in the interior of a protein.
Table III.
Synopsis of residues in the mature α-galactosidase A protein that are affected by Fabry disease-causing mutations.
| Residue type | Total no. residues in protein | % of total (and no. of) residues affected by mutations | No. residues affected by AA substitution only | No. residues affected by AA substitution or nonsense | No. residues affected by nonsense only | Percentage of total residues affected by AA substitution |
|---|---|---|---|---|---|---|
| Trp | 16 | 94% (15) | 1 | 9 | 5 | 63 |
| Cys | 12 | 83% (10) | 7 | 3 | 0 | 83 |
| Gln | 22 | 77% (17) | 5 | 2 | 10 | 32 |
| Gly | 31 | 68% (21) | 21 | 0 | 0 | 68 |
| Tyr | 15 | 67% (10) | 3 | 2 | 5 | 33 |
| Glu | 18 | 56% (10) | 6 | 2 | 2 | 44 |
| Met | 15 | 53% (8) | 8 | N/A | N/A | 53 |
| Arg | 19 | 53% (10) | 6 | 3 | 1 | 47 |
| Ile | 21 | 52% (11) | 11 | N/A | N/A | 52 |
| Ala | 28 | 46% (13) | 13 | N/A | N/A | 46 |
| Asp | 29 | 45% (13) | 13 | N/A | N/A | 45 |
| Ser | 23 | 43% (10) | 8 | 0 | 2 | 35 |
| Thr | 14 | 43% (6) | 6 | N/A | N/A | 43 |
| Pro | 19 | 42% (8) | 8 | N/A | N/A | 42 |
| Leu | 41 | 41% (17) | 15 | 0 | 2 | 37 |
| Asn | 20 | 40% (8) | 8 | N/A | N/A | 40 |
| Phe | 15 | 33% (5) | 5 | N/A | N/A | 33 |
| Val | 16 | 19% (3) | 3 | N/A | N/A | 19 |
| His | 7 | 14% (1) | 1 | N/A | N/A | 14 |
| Lys | 17 | 12% (2) | 1 | 0 | 1 | 6 |
| Total | 398 | 50% (198) | 149 | 21 | 28 | 43 |
AA, amino acid
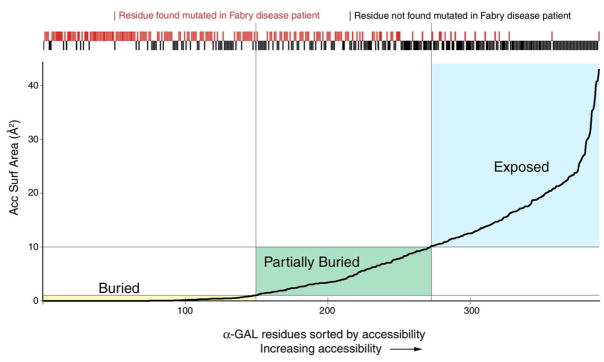
We have used the amount of buried surface area per side chain atom as a measure the degree of accessibility of a given residue. This correlates with the importance of a residue in the folded conformation of the protein. If each residue in α-galactosidase A is ranked by increasing surface accessibility, the residues found altered by Fabry disease mutations cluster among the buried residues (Figure 3). Conversely, residues that are exposed to solvent are rarely found to be affected by point mutation in Fabry disease. Thus, there is a correlation between the extent of burial of a residue in the core of the α-galactosidase A protein and Fabry disease mutations associated with that residue. As a metric for the importance of a given residue, other researchers have used the degree of conservation of a residue in α-galactosidase A orthologues [15] and the number atoms with bad contact distances in a hypothetical model of the mutant protein [16].
Figure 3.
Fabry disease-causing mutations and surface accessibility. The accessible surface area for each residue in the human α-galactosidase A structure is shown, with each residue ranked by its accessibility. The plot shows the 398 residues in the mature human α-galactosidase A sequence ordered by increasing accessibility (along the X axis) and the average accessibility of each atom in the residue (on the Y axis). Boxes identify the buried (yellow), partially buried (green), and exposed (blue) residues. Above the plot, red tick marks represent residues found altered in patients with Fabry disease and black marks represent residues not (yet) found altered in Fabry disease. The tick marks show that Fabry disease is most often caused by defects in the folding of the α-galactosidase A molecule, as nearly all of the buried residues have been found altered in patients with Fabry disease (left side and red tick marks). Conversely, exposed residues are rarely found substituted in patients with Fabry disease (right side and black tick marks). This plot shows that Fabry disease is most often caused by changes to the hydrophobic core of the α-galactosidase A protein.
Discussion
Half of residues in the protein have been found altered in the ensemble of Fabry disease patients (Table II and Figure 2). As might be expected, the active site is exquisitely sensitive to change, as all of the residues that make contact with the ligand have been affected by mutation in patients with Fabry disease. Surprisingly, most of the residues affected by Fabry disease point mutations do not cluster around the active site, but are found distributed throughout the hydrophobic core of the protein. Most of the missense mutations that cause Fabry disease produce changes in the hydrophobic core of the α-galactosidase A protein, as 65% of the missense mutations code for buried amino acid residues.
The structure of the human α-galactosidase A glycoprotein raises the possibility of tailoring the treatment of Fabry disease to the individual’s specific defect in the GLA gene. One group of patients can in principle respond to pharmacological chaperone therapy, whereas a second group will not respond to small molecule therapy and must be treated with recombinant ERT. In the case of patients with a defective α-galactosidase A active site or with a grossly altered α-galactosidase A polypeptide (i.e. those with changes in the protein’s active site, nonsense mutations, frameshift mutations or splice defect mutations), recombinant ERT seems more appropriate. In these patients, if the folded protein does not have catalytic capacity or if the full-length polypeptide is not even synthesized, small molecule chaperones to assist in the folding of the protein are unlikely to show efficacy, and ERT might be a preferred treatment. In the case of patients with a defect in the hydrophobic core of the molecule, small molecule chaperones might assist in the correct folding of the enzyme in the endoplasmic reticulum. This might be an appropriate treatment choice for patients who show some residual enzyme activity and for those who have amino acid changes that fall a long way from the active site of the enzyme.
Conclusion
At the amino acid level, Fabry disease is most often caused by a perturbation of the hydrophobic core of the protein. The literature on GLA mutations and Fabry disease prior to the availability of the three-dimensional structure presumed that the disease-causing missense mutations would produce changes in the enzyme near the active site. Using the crystal structure and an expanded database of Fabry disease mutations, we show that alterations in residues near the active site of the enzyme represent only 10% of the total. The majority of the amino acid changes responsible for Fabry disease fall in the hydrophobic core of the protein, thus Fabry disease is primarily a protein folding disease. Fabry disease, one of the best characterized members of the LSD family, might prove to be a model for other protein folding diseases, including Alzheimer’s disease, Parkinson’s disease, prion diseases and polyglutamine diseases such as Huntington’s disease.
Acknowledgments
We are grateful for financial support from the Charles H. Hood Foundation, Inc., Boston, MA.
References
- 1.Desnick RJ, Ioannou YA, Eng CM. α-Galactosidase A deficiency: Fabry disease. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The metabolic and molecular bases of inherited disease. 8. New York: McGraw-Hill; 2001. pp. 3733–74. [Google Scholar]
- 2.Fabry J. Ein Beitrag zur Kenntnis der Purpura haemorrhagica nodularis (Purpura papulosa haemorrhagica Hebrae) Arch Dermatol Syph. 1898;43:187–200. [Google Scholar]
- 3.Brady RO, Gal AE, Bradley RM, Martensson E, Warshaw AL, Laster L. Enzymatic defect in Fabry’s disease. Ceramidetrihexosidase deficiency. N Engl J Med. 1967;276:1163–7. doi: 10.1056/NEJM196705252762101. [DOI] [PubMed] [Google Scholar]
- 4.Brady RO. Enzyme replacement therapy: conception, chaos and culmination. Philos Trans R Soc Lond B Biol Sci. 2003;358:915–9. doi: 10.1098/rstb.2003.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brady RO. Gaucher and Fabry diseases: from understanding pathophysiology to rational therapies. Acta Paediatr Suppl. 2003;92:19–24. doi: 10.1111/j.1651-2227.2003.tb00215.x. [DOI] [PubMed] [Google Scholar]
- 6.Garman SC, Garboczi DN. The molecular defect leading to Fabry disease: structure of human α-galactosidase. J Mol Biol. 2004;337:319–35. doi: 10.1016/j.jmb.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 7.Bourne Y, Henrissat B. Glycoside hydrolases and glycosyltransferases: families and functional modules. Curr Opin Struct Biol. 2001;11:593–600. doi: 10.1016/s0959-440x(00)00253-0. [DOI] [PubMed] [Google Scholar]
- 8.Lee K, Jin X, Zhang K, Copertino L, Andrews L, Baker-Malcolm J, et al. A biochemical and pharmacological comparison of enzyme replacement therapies for the glycolipid storage disorder Fabry disease. Glycobiology. 2003;13:305–13. doi: 10.1093/glycob/cwg034. [DOI] [PubMed] [Google Scholar]
- 9.Ioannou YA, Zeidner KM, Gordon RE, Desnick RJ. Fabry disease: preclinical studies demonstrate the effectiveness of alpha-galactosidase A replacement in enzyme-deficient mice. Am J Hum Genet. 2001;68:14–25. doi: 10.1086/316953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000;28:45–8. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krawczak M, Cooper DN. The human gene mutation database. Trends Genet. 1997;13:121–2. doi: 10.1016/s0168-9525(97)01068-8. [DOI] [PubMed] [Google Scholar]
- 12.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 13.Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–50. [Google Scholar]
- 14.McKusick VA. Mendelian inheritance in man: a catalog of human genes and genetic disorders. John Hopkins University Press; Baltimore: 1998. [Google Scholar]
- 15.Shabbeer J, Yasuda M, Benson SD, Desnick RJ. Fabry disease: identification of 50 novel α-galactosidase A mutations causing the classic phenotype and three-dimensional structural analysis of 29 missense mutations. Hum Genomics. 2006;2:297–309. doi: 10.1186/1479-7364-2-5-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuzawa F, Aikawa S, Doi H, Okumiya T, Sakuraba H. Fabry disease: correlation between structural changes in alpha-galactosidase, and clinical and biochemical phenotypes. Hum Genet. 2005;117:317–28. doi: 10.1007/s00439-005-1300-5. [DOI] [PubMed] [Google Scholar]
- 17.Blanch LC, Meaney C, Morris CP. A sensitive mutation screening strategy for Fabry disease: detection of nine mutations in the alpha-galactosidase A gene. Hum Mutat. 1996;8:38–43. doi: 10.1002/(SICI)1098-1004(1996)8:1<38::AID-HUMU5>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 18.Eng CM, Ashley GA, Burgert TS, Enriquez AL, D’Souza M, Desnick RJ. Fabry disease: thirty-five mutations in the α-galactosidase A gene in patients with classic and variant phenotypes. Mol Med. 1997;3:174–82. [PMC free article] [PubMed] [Google Scholar]
- 19.Shabbeer J, Yasuda M, Luca E, Desnick RJ. Fabry disease: 45 novel mutations in the α-galactosidase A gene causing the classical phenotype. Mol Genet Metab. 2002;76:23–30. doi: 10.1016/s1096-7192(02)00012-4. [DOI] [PubMed] [Google Scholar]
- 20.Tse KC, Chan KW, Tin VP, Yip PS, Tang S, Li FK, et al. Clinical features and genetic analysis of a Chinese kindred with Fabry’s disease. Nephrol Dial Transplant. 2003;18:182–6. doi: 10.1093/ndt/18.1.182. [DOI] [PubMed] [Google Scholar]
- 21.Garzuly F, Marodi L, Erdos M, Grubits J, Varga Z, Gelpi E, et al. Megadolichobasilar anomaly with thrombosis in a family with Fabry’s disease and a novel mutation in the alpha-galactosidase A gene. Brain. 2005;128:2078–83. doi: 10.1093/brain/awh546. [DOI] [PubMed] [Google Scholar]
- 22.Teragaki M, Tanaka A, Akioka K, Lan HT, Nishi Y, Yamano T, et al. Fabry disease female proband with clinical manifestations similar to hypertrophic cardiomyopathy. Jpn Heart J. 2004;45:685–9. doi: 10.1536/jhj.45.685. [DOI] [PubMed] [Google Scholar]
- 23.Nakao S, Takenaka T, Maeda M, Kodama C, Tanaka A, Tahara M, et al. An atypical variant of Fabry’s disease in men with left ventricular hypertrophy. N Engl J Med. 1995;333:288–93. doi: 10.1056/NEJM199508033330504. [DOI] [PubMed] [Google Scholar]
- 24.Madsen KM, Hasholt L, Sorensen SA, Fermer ML, Dahl N. Two novel mutations (L32P) and (G85N) among five different missense mutations in six Danish families with Fabry’s disease. Hum Mutat. 1995;5:277–8. doi: 10.1002/humu.1380050316. [DOI] [PubMed] [Google Scholar]
- 25.Eng CM, Resnick-Silverman LA, Niehaus DJ, Astrin KH, Desnick RJ. Nature and frequency of mutations in the α-galactosidase A gene that cause Fabry disease. Am J Hum Genet. 1993;53:1186–97. [PMC free article] [PubMed] [Google Scholar]
- 26.Davies J, Christomanou H, Winchester B, Malcolm S. Detection of 8 new mutations in the alpha-galactosidase A gene in Fabry disease. Hum Mol Genet. 1994;3:667–9. doi: 10.1093/hmg/3.4.667. [DOI] [PubMed] [Google Scholar]
- 27.Koide T, Ishiura M, Iwai K, Inoue M, Kaneda Y, Okada Y, et al. A case of Fabry’s disease in a patient with no α-galactosidase A activity caused by a single amino acid substitution of Pro-40 by Ser. FEBS Lett. 1990;259:353–6. doi: 10.1016/0014-5793(90)80046-l. [DOI] [PubMed] [Google Scholar]
- 28.Ashton-Prolla P, Tong B, Shabbeer J, Astrin KH, Eng CM, Desnick RJ. Fabry disease: twenty-two novel mutations in the alpha-galactosidase A gene and genotype/phenotype correlations in severely and mildly affected hemizygotes and heterozygotes. J Investig Med. 2000;48:227–35. [PubMed] [Google Scholar]
- 29.Davies JP, Eng CM, Hill JA, Malcolm S, MacDermot K, Winchester B, et al. Fabry disease: fourteen alpha-galactosidase A mutations in unrelated families from the United Kingdom and other European countries. Eur J Hum Genet. 1996;4:219–24. doi: 10.1159/000472202. [DOI] [PubMed] [Google Scholar]
- 30.Rosenthal D, Lien YH, Lager D, Lai LW, Shang S, Leung N, et al. A novel alpha-galactosidase a mutant (M42L) identified in a renal variant of Fabry disease. Am J Kidney Dis. 2004;44:e85–9. [PubMed] [Google Scholar]
- 31.Iga MI, Okayama A, Matsuyama M, Sasaki T, Murai K, Hashida S, et al. Human gene mutations in GLA. Hum Genet. 2001;109:126. [Google Scholar]
- 32.Germain DP, Shabbeer J, Cotigny S, Desnick RJ. Fabry disease: twenty novel α-galactosidase A mutations and genotype–phenotype correlations in classical and variant phenotypes. Mol Med. 2002;8:306–12. [PMC free article] [PubMed] [Google Scholar]
- 33.Sakuraba H, Oshima A, Fukuhara Y, Shimmoto M, Nagao Y, Bishop DF, et al. Identification of point mutations in the alpha-galactosidase A gene in classical and atypical hemizygotes with Fabry disease. Am J Hum Genet. 1990;47:784–9. [PMC free article] [PubMed] [Google Scholar]
- 34.Wang ZX, Zhang Y, Bu DF, Zhang W, Yuan Y. Novel GLA gene mutations in two Chinese families with classic Fabry disease. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2005;22:489–92. [PubMed] [Google Scholar]
- 35.Blaydon D, Hill J, Winchester B. Fabry disease: 20 novel GLA mutations in 35 families. Hum Mutat. 2001;18:459. doi: 10.1002/humu.1219. [DOI] [PubMed] [Google Scholar]
- 36.Cooper A, Cooper JA, Wraith JE. Human gene mutations in GLA. Hum Genet. 2000;107:535–6. [Google Scholar]
- 37.Rodriguez-Mari A, Coll MJ, Chabas A. Molecular analysis in Fabry disease in Spain: fifteen novel GLA mutations and identification of a homozygous female. Hum Mutat. 2003;22:258. doi: 10.1002/humu.9172. [DOI] [PubMed] [Google Scholar]
- 38.Ashley GA, Shabbeer J, Yasuda M, Eng CM, Desnick RJ. Fabry disease: twenty novel alpha-galactosidase A mutations causing the classical phenotype. J Hum Genet. 2001;46:192–6. doi: 10.1007/s100380170088. [DOI] [PubMed] [Google Scholar]
- 39.Eng CM, Niehaus DJ, Enriquez AL, Burgert TS, Ludman MD, Desnick RJ. Fabry disease: twenty-three mutations including sense and antisense CpG alterations and identification of a deletional hot-spot in the α-galactosidase A gene. Hum Mol Genet. 1994;3:1795–9. doi: 10.1093/hmg/3.10.1795. [DOI] [PubMed] [Google Scholar]
- 40.Topaloglu AK, Ashley GA, Tong B, Shabbeer J, Astrin KH, Eng CM, et al. Twenty novel mutations in the α-galactosidase A gene causing Fabry disease. Mol Med. 1999;5:806–11. [PMC free article] [PubMed] [Google Scholar]
- 41.Shabbeer J, Robinson M, Desnick RJ. Detection of alpha-galactosidase a mutations causing Fabry disease by denaturing high performance liquid chromatography. Hum Mutat. 2005;25:299–305. doi: 10.1002/humu.20144. [DOI] [PubMed] [Google Scholar]
- 42.Schäfer E, Baron K, Widmer U, Deegan P, Neumann HP, Sunder-Plassmann G, et al. Thirty-four novel mutations of the GLA gene in 121 patients with Fabry disease. Hum Mutat. 2005;25:412. doi: 10.1002/humu.9327. [DOI] [PubMed] [Google Scholar]
- 43.Chen CH, Shyu PW, Wu SJ, Sheu SS, Desnick RJ, Hsiao KJ. Identification of a novel point mutation (S65T) in alpha-galactosidase A gene in Chinese patients with Fabry disease. Mutations in brief no. 169. Online Hum Mutat. 1998;11:328–30. doi: 10.1002/(SICI)1098-1004(1998)11:4<328::AID-HUMU11>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 44.Lai LW, Whitehair O, Wu MJ, O’Meara M, Lien YH. Analysis of splice-site mutations of the α-galactosidase A gene in Fabry disease. Clin Genet. 2003;63:476–82. doi: 10.1034/j.1399-0004.2003.00077.x. [DOI] [PubMed] [Google Scholar]
- 45.Ishii S, Sakuraba H, Suzuki Y. Point mutations in the upstream region of the alpha-galactosidase A gene exon 6 in an atypical variant of Fabry disease. Hum Genet. 1992;89:29–32. doi: 10.1007/BF00207037. [DOI] [PubMed] [Google Scholar]
- 46.Okumiya T, Kawamura O, Itoh K, Kase R, Ishii S, Kamei S, et al. Novel missense mutation (M72V) of α-galactosidase gene and its expression product in an atypical Fabry hemizygote. Hum Mutat. 1998;(Suppl 1):S213–6. doi: 10.1002/humu.1380110169. [DOI] [PubMed] [Google Scholar]
- 47.Slee PH, van Boven LJ, Slee DS. Fabry disease: data from four families. Ned Tijdschr Geneeskd. 2000;144:2412–5. [PubMed] [Google Scholar]
- 48.Guffon N, Froissart R, Chevalier-Porst F, Maire I. Mutation analysis in 11 French patients with Fabry disease. Hum Mutat. 1998;(Suppl 1):S288–90. doi: 10.1002/humu.1380110190. [DOI] [PubMed] [Google Scholar]
- 49.Takata T, Okumiya T, Hayashibe H, Shimmoto M, Kase R, Itoh K, et al. Screening and detection of gene mutations in Japanese patients with Fabry disease by non-radioactive single-stranded conformation polymorphism analysis. Brain Dev. 1997;19:111–6. doi: 10.1016/s0387-7604(96)00486-x. [DOI] [PubMed] [Google Scholar]
- 50.Lee JK, Kim GH, Kim JS, Kim KK, Lee MC, Yoo HW. Identification of four novel mutations in five unrelated Korean families with Fabry disease. Clin Genet. 2000;58:228–33. doi: 10.1034/j.1399-0004.2000.580311.x. [DOI] [PubMed] [Google Scholar]
- 51.Dobrovolny R, Dvorakova L, Ledvinova J, Magage S, Bultas J, Lubanda JC, et al. Relationship between X-inactivation and clinical involvement in Fabry heterozygotes. Eleven novel mutations in the alpha-galactosidase A gene in the Czech and Slovak population. J Mol Med. 2005;83:647–54. doi: 10.1007/s00109-005-0656-2. [DOI] [PubMed] [Google Scholar]
- 52.Kimura K, Sato-Matsumura KC, Nakamura H, Onodera Y, Morita K, Enami N, et al. A novel A97P amino acid substitution in alpha-galactosidase A leads to a classical Fabry disease with cardiac manifestations. Br J Dermatol. 2002;147:545–8. doi: 10.1046/j.1365-2133.2002.04902.x. [DOI] [PubMed] [Google Scholar]
- 53.Kotanko P, Kramar R, Devrnja D, Paschke E, Voigtlander T, Auinger M, et al. Results of a nationwide screening for Anderson-Fabry disease among dialysis patients. J Am Soc Nephrol. 2004;15:1323–9. doi: 10.1097/01.asn.0000124671.61963.1e. [DOI] [PubMed] [Google Scholar]
- 54.Whybra C, Kampmann C, Willers I, Davies J, Winchester B, Kriegsmann J, et al. Anderson-Fabry disease: clinical manifestations of disease in female heterozygotes. J Inherit Metab Dis. 2001;24:715–24. doi: 10.1023/a:1012993305223. [DOI] [PubMed] [Google Scholar]
- 55.Okumiya T, Ishii S, Kase R, Kamei S, Sakuraba H, Suzuki Y. α-galactosidase gene mutations in Fabry disease: heterogeneous expressions of mutant enzyme proteins. Hum Genet. 1995;95:557–61. doi: 10.1007/BF00223869. [DOI] [PubMed] [Google Scholar]
- 56.Ploos van Amstel JK, Jansen RP, de Jong JG, Hamel BC, Wevers RA. Six novel mutations in the α-galactosidase A gene in families with Fabry disease. Hum Mol Genet. 1994;3:503–5. doi: 10.1093/hmg/3.3.503. [DOI] [PubMed] [Google Scholar]
- 57.Eng CM, Desnick RJ. Molecular basis of Fabry disease: mutations and polymorphisms in the human α-galactosidase A gene. Hum Mutat. 1994;3:103–11. doi: 10.1002/humu.1380030204. [DOI] [PubMed] [Google Scholar]
- 58.Germain D, Biasotto M, Tosi M, Meo T, Kahn A, Poenaru L. Fluorescence-assisted mismatch analysis (FAMA) for exhaustive screening of the α-galactosidase A gene and detection of carriers in Fabry disease. Hum Genet. 1996;98:719–26. doi: 10.1007/s004390050292. [DOI] [PubMed] [Google Scholar]
- 59.Rosenberg KM, Schiffmann R, Kaneski C, Brady RO, Sorensen SA, Hasholt L. Five novel mutations in fourteen patients with Fabry disease. Hum Mutat. 2000;15:207–8. doi: 10.1002/(SICI)1098-1004(200002)15:2<207::AID-HUMU16>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 60.Morrone A, Cavicchi C, Bardelli T, Antuzzi D, Parini R, Di Rocco M, et al. Fabry disease: molecular studies in Italian patients and X inactivation analysis in manifesting carriers. J Med Genet. 2003;40:e103. doi: 10.1136/jmg.40.8.e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yasuda M, Shabbeer J, Benson SD, Desnick RJ. Fabry Disease: Identification/Characterization of Novel Double Mutations in the α-Galactosidase A Gene. ASHG Ann Meet Abs. 2002:926. [Google Scholar]
- 62.Ries M, Gupta S, Moore DF, Sachdev V, Quirk JM, Murray GJ, et al. Pediatric Fabry disease. Pediatrics. 2005;115:e344–55. doi: 10.1542/peds.2004-1678. [DOI] [PubMed] [Google Scholar]
- 63.Davies JP, Winchester BG, Malcolm S. Mutation analysis in patients with the typical form of Anderson-Fabry disease. Hum Mol Genet. 1993;2:1051–3. doi: 10.1093/hmg/2.7.1051. [DOI] [PubMed] [Google Scholar]
- 64.Meaney C, Blanch LC, Morris CP. A nonsense mutation (R220X) in the alpha-galactosidase A gene detected in a female carrier of Fabry disease. Hum Mol Genet. 1994;3:1019–20. doi: 10.1093/hmg/3.6.1019. [DOI] [PubMed] [Google Scholar]
- 65.Yang CC, Lai LW, Whitehair O, Hwu WL, Chiang SC, Lien YH. Two novel mutations in the α-galactosidase A gene in Chinese patients with Fabry disease. Clin Genet. 2003;63:205–9. doi: 10.1034/j.1399-0004.2003.00050.x. [DOI] [PubMed] [Google Scholar]
- 66.Germain DP, Poenaru L. Fabry disease: identification of novel α-galactosidase A mutations and molecular carrier detection by use of fluorescent chemical cleavage of mismatches. Biochem Biophys Res Commun. 1999;257:708–13. doi: 10.1006/bbrc.1999.0310. [DOI] [PubMed] [Google Scholar]
- 67.Knol IE, Ausems MG, Lindhout D, van Diggelen OP, Verwey H, Davies J, et al. Different phenotypic expression in relatives with Fabry disease caused by a W226X mutation. Am J Med Genet. 1999;82:436–9. doi: 10.1002/(sici)1096-8628(19990219)82:5<436::aid-ajmg14>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 68.Redonnet-Vernhet I, Ploos van Amstel JK, Jansen RP, Wevers RA, Salvayre R, Levade T. Uneven X inactivation in a female monozygotic twin pair with Fabry disease and discordant expression of a novel mutation in the α-galactosidase A gene. J Med Genet. 1996;33:682–8. doi: 10.1136/jmg.33.8.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Altarescu GM, Goldfarb LG, Park KY, Kaneski C, Jeffries N, Litvak S, et al. Identification of fifteen novel mutations and genotype–phenotype relationship in Fabry disease. Clin Genet. 2001;60:46–51. doi: 10.1034/j.1399-0004.2001.600107.x. [DOI] [PubMed] [Google Scholar]
- 70.Azibi K, Caillaud C, Manicom J, Puech JP, Kahn A, Poenaru L. Fabry disease: identification of ten novel mutations. ASHG Ann Meet Abs. 2001:2647. [Google Scholar]
- 71.Verovnik F, Benko D, Vujkovac B, Linthorst GE. Remarkable variability in renal disease in a large Slovenian family with Fabry disease. Eur J Hum Genet. 2004;12:678–81. doi: 10.1038/sj.ejhg.5201184. [DOI] [PubMed] [Google Scholar]
- 72.Dominissini S, Cariati R, Nevyjel M, Guerci V, Ciana G, Bembi B, et al. Comparative in vitro expression study of four Fabry disease causing mutations at glutamine 279 of the alpha-galactosidase A protein. Hum Hered. 2004;57:138–41. doi: 10.1159/000079244. [DOI] [PubMed] [Google Scholar]
- 73.von Scheidt W, Eng CM, Fitzmaurice TF, Erdmann E, Hubner G, Olsen EG, et al. An atypical variant of Fabry’s disease with manifestations confined to the myocardium. N Engl J Med. 1991;324:395–9. doi: 10.1056/NEJM199102073240607. [DOI] [PubMed] [Google Scholar]
- 74.Yasuda M, Shabbeer J, Benson SD, Maire I, Burnett RM, Desnick RJ. Fabry disease: characterization of alpha-galactosidase A double mutations and the D313Y plasma enzyme pseudodeficiency allele. Hum Mutat. 2003;22:486–92. doi: 10.1002/humu.10275. [DOI] [PubMed] [Google Scholar]
- 75.Sachdev B, Takenaka T, Teraguchi H, Tei C, Lee P, McKenna WJ, et al. Prevalence of Anderson-Fabry disease in male patients with late onset hypertrophic cardiomyopathy. Circulation. 2002;105:1407–11. doi: 10.1161/01.cir.0000012626.81324.38. [DOI] [PubMed] [Google Scholar]
- 76.Okumiya T, Ishii S, Takenaka T, Kase R, Kamei S, Sakuraba H, et al. Galactose stabilizes various missense mutants of α-galactosidase in Fabry disease. Biochem Biophys Res Commun. 1995;214:1219–24. doi: 10.1006/bbrc.1995.2416. [DOI] [PubMed] [Google Scholar]
- 77.Beyer E, Djatlovitskaya E, Zairatyants O, Berestova A, Mendelson M, Brook E, et al. Identification of Fabry disease in two brothers. J Inherit Metab Dis. 1990;13:230–1. doi: 10.1007/BF01799692. [DOI] [PubMed] [Google Scholar]
- 78.Azibi K, Caillaud C, Heltianu C, Dussau J, Puech JP, Poenaru L. Novel mutations and genetic markers in Fabry disease. ASHG Ann Meet Abs. 2002:2170. [Google Scholar]
- 79.Bernstein HS, Bishop DF, Astrin KH, Kornreich R, Eng CM, Sakuraba H, et al. Fabry disease: six gene rearrangements and an exonic point mutation in the α-galactosidase gene. J Clin Invest. 1989;83:1390–9. doi: 10.1172/JCI114027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miyazaki T, Kajita M, Ohmori S, Mizutani N, Niwa T, Murata Y, et al. A novel mutation (E358K) in the α-galactosidase A gene detected in a Japanese family with Fabry disease. Hum Mutat. 1998;(Suppl 1):S139–40. doi: 10.1002/humu.1380110147. [DOI] [PubMed] [Google Scholar]
- 81.Miyamura N, Araki E, Matsuda K, Yoshimura R, Furukawa N, Tsuruzoe K, et al. A carboxy-terminal truncation of human alpha-galactosidase A in a heterozygous female with Fabry disease and modification of the enzymatic activity by the carboxy-terminal domain. Increased, reduced, or absent enzyme activity depending on number of amino acid residues deleted. J Clin Invest. 1996;98:1809–17. doi: 10.1172/JCI118981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Germain DP, Salard D, Fellmann F, Azibi K, Caillaud C, Bernard MC, et al. Identification of a novel de novo mutation (G373D) in the α-galactosidase A gene (GLA) in a patient affected with Fabry disease. Hum Mutat. 2001;17:353. doi: 10.1002/humu.41. [DOI] [PubMed] [Google Scholar]
- 83.Shin YS, Podskarbi T. A novel mutation of the alpha-galactosidase A gene in a family with juvenile-onset form of Fabry disease. ASHG Ann Meet Abs. 2001:1799. [Google Scholar]
- 84.Galanos J, Nicholls K, Grigg L, Kiers L, Crawford A, Becker G. Clinical features of Fabry’s disease in Australian patients. Intern Med J. 2002;32:575–84. doi: 10.1046/j.1445-5994.2002.00291.x. [DOI] [PubMed] [Google Scholar]
- 85.Cariolou MA, Christodoulides M, Manoli P, Kokkofitou A, Tsambaos D. Novel trinucleotide deletion in Fabry’s disease. Hum Genet. 1996;97:468–70. doi: 10.1007/BF02267068. [DOI] [PubMed] [Google Scholar]
- 86.Novo FJ, Kruszewski A, MacDermot KD, Goldspink G, Gorecki DC. Editing of human α-galactosidase RNA resulting in a pyrimidine to purine conversion. Nucleic Acids Res. 1995;23:2636–40. doi: 10.1093/nar/23.14.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]