Abstract
TIA-1 and TIAR are related proteins that bind to an AU-rich element (ARE) in the 3′ untranslated region of tumor necrosis factor alpha (TNF-α) transcripts. To determine the functional significance of this interaction, we used homologous recombination to produce mutant mice lacking TIA-1. Although lipopolysaccharide (LPS)-stimulated macrophages derived from wild-type and TIA-1–/– mice express similar amounts of TNF-α transcripts, macrophages lacking TIA-1 produce significantly more TNF-α protein than wild-type controls. The half-life of TNF-α transcripts is similar in wild-type and TIA-1–/– macrophages, indicating that TIA-1 does not regulate transcript stability. Rather, the absence of TIA-1 significantly increases the proportion of TNF-α transcripts that associate with polysomes, suggesting that TIA-1 normally functions as a translational silencer. TIA-1 does not appear to regulate the production of interleukin 1β, granulocyte–macrophage colony-stimulating factor or interferon γ, indicating that its effects are, at least partially, transcript specific. Mice lacking TIA-1 are hypersensitive to the toxic effects of LPS, indicating that this translational control pathway may regulate the organismal response to microbial stress.
Keywords: AU-rich element/TIA-1/TNF-α/translation
Introduction
Tumor necrosis factor alpha (TNF-α) is a proinflammatory cytokine produced by activated macrophages and lymphocytes (Beutler, 1999; Feldmann and Maini, 1999; Kollias et al., 1999). The expression of TNF-α is regulated by both transcriptional and post-transcriptional mechanisms (Biragyn and Nedospasov, 1995; Crawford et al., 1997; Han and Beutler, 1990; Han et al., 1990; Prichett et al., 1995; Wang et al., 1997; Raabe et al., 1998). Post-transcriptional control of TNF-α expression is achieved by regulating mRNA stability, polyadenylation and translational initiation (Han and Beutler, 1990; Han et al., 1990; Wang et al., 1997). An adenine and uridine (AU)-rich element (ARE) in the 3′ untranslated region (3′ UTR) of TNF-α transcripts (Caput et al., 1986; Kruys et al., 1989; Lewis et al., 1998) is an important determinant of post-transcriptional control. Transgenic mice expressing TNF-α transcripts lacking the ARE develop chronic inflammatory polyarthritis and inflammatory bowel disease as a consequence of overexpressing TNF-α (Keffer et al., 1991; Kontoyiannis et al., 1999).
Trans-acting factors that bind to the TNF-α ARE are essential for post-transcriptional control of TNF-α expression. For example, tristetraprolin (TTP) binds to the TNF-α ARE and promotes the degradation of TNF-α transcripts (Taylor et al., 1996; Carballo et al., 1998; Lai et al., 1999). Mutant mice lacking TTP develop cachexia, arthritis and autoimmunity as a consequence of overexpressed TNF-α mRNA and protein (Taylor et al., 1996). In addition to TTP, Hel-N1 (Levine et al., 1993), HuR (Myer et al., 1997; Fan and Steitz, 1998; Peng et al., 1998; Sokolowski et al., 1999), AUF1 [heterogeneous nuclear ribonucleoprotein (hnRNP) D] (Zhang et al., 1993) and TIAR (Gueydan et al., 1999) are ARE-binding proteins that have been proposed to regulate the expression of TNF-α (Sakai et al., 1999). Hel-N1 (Levine et al., 1993; Gao et al., 1994; Ford et al., 1999) and HuR (Myer et al., 1997; Fan and Steitz, 1998; Peng et al., 1998; Sokolowski et al., 1999) stabilize ARE-containing transcripts, whereas AUF1 destabilizes these transcripts (Loflin et al., 1999). It is likely, therefore, that the stability of ARE-containing transcripts is determined by the relative expression of functionally antagonistic ARE-binding proteins. Although TIAR is a component of an ARE-binding complex (Gueydan et al., 1999), the functional consequences of TIAR binding have not been described.
TIAR and its closely related homologue, TIA-1, are members of the RNA-recognition motif (RRM) family of RNA-binding proteins (Tian et al., 1991; Kawakami et al., 1992, 1994; Beck et al., 1996; Dember et al., 1996). Both proteins have three RRM domains in their N-termini that confer high-affinity binding to uridine-rich motifs (Dember et al., 1996). Like components of the general hnRNP complex, TIA-1 and TIAR continuously shuttle between the nucleus and the cytoplasm (N.Kedersha and P.Anderson, in preparation), suggesting that they might participate in the nucleocytoplasmic transport of selected mRNAs. These proteins also regulate the general translational arrest that accompanies environmental stress. Following the stress-induced phosphorylation of translation initiation factor eIF-2α, TIA-1 and TIAR recruit most cytoplasmic mRNAs to discrete foci known as stress granules (Kedersha et al., 1999). The TIA-1/TIAR-dependent sequestration of these mRNAs prevents their translational initiation. In this capacity, TIA-1 and TIAR function as translational silencers that appear to influence the duration of stress-induced translational arrest. Mutant mice lacking TIAR exhibit partial embryonic lethality and defective germ-cell maturation, implicating this protein in certain aspects of vertebrate development (Beck et al., 1998).
The discovery of TIAR as a component of the ARE-associated complex that assembles on the 3′ UTR of TNF-α transcripts (Gueydan et al., 1999) provided the first clue that TIA-1 and TIAR might specifically regulate the expression of TNF-α. To test this hypothesis, we produced mutant mice lacking TIA-1 and compared the lipopolysaccharide (LPS)-induced expression of TNF-α in wild-type and TIA-1–/– macrophages. Our results indicate that LPS-induced expression of TNF-α is significantly increased in macrophages lacking TIA-1. The functional effects of TIA-1 appear to result from translational silencing rather than regulation of mRNA stability. Thus, the ARE-binding protein TIA-1 represses the expression of TNF-α by a mechanism that differs from that used by other known ARE-binding proteins.
Results
Targeted disruption of the tia-1 gene
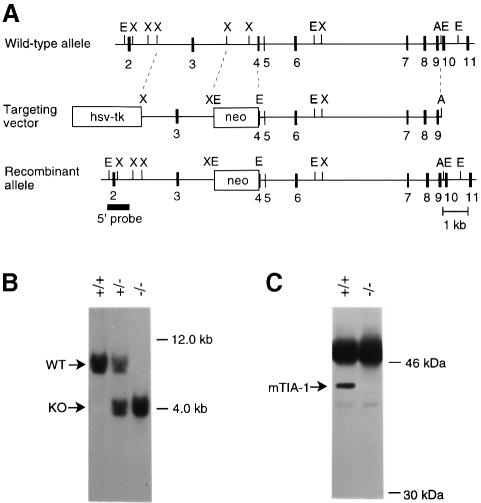
We have previously described the targeted disruption of the tiar gene in mice (Beck et al., 1998). We used a similar approach to disrupt the tia-1 gene using homologous recombination in embryonic stem (ES) cells (Figure 1A). The rearranged tia-1 locus (termed tia-1–) was tested by Southern blotting of genomic DNA from ES cells for proper integration of the targeting vector by using probes 5′ and 3′ of the gene fragments used in the targeting vector. Digestion of ES-cell DNA with two different restriction enzymes and Southern blot analysis using a probe for the neomycin resistance (neor) cassette detected the presence of only one copy of the targeting vector in the rearranged genome (data not shown). Transmission of the tia-1– allele through the mouse germline was demonstrated by Southern blot analysis of DNA from offspring of mice derived from the tia-1– ES cells (Figure 1B). Immunoblot analysis of cell lysates of embryonic fibroblasts from tia-1+/– × tia-1+/– breedings using an antibody against TIA-1 confirmed lack of expression of TIA-1 and inactivation of the tia-1 locus (Figure 1C).
Fig. 1. Construction of TIA-1–/– mouse and MEFs. (A) The expected gene replacement at the mouse tia-1 locus. Correct gene targeting should result in replacement of parts of intron 3 and exon 4 by a marker gene for positive selection (PGK–neor). The herpes simplex virus thymidine kinase expression cassette was used for negative selection. The 5′ probe used for Southern blot analysis is indicated. A, AatII; E, EcoRI, X, XbaI. (B) Southern blot analysis of DNA from offspring derived from heterozygous matings. Genomic DNA was digested with EcoRI and analyzed using the 5′ probe, yielding the 7.5 and 4.0 kb fragments expected for the wild-type and mutant allele, respectively. Southern blot analysis of ES-cell DNA using a 3′ probe and a neo probe showed proper targeting and single insertion of the transfected vector (data not shown). (C) Immunoprecipitation of lysates from tia-1+/+ and tia-1–/– embryonic fibroblasts using ML29 monoclonal antibody (mAb) and subsequent protein immunoblot with mAb 2G9 confirms the absence of TIA-1 protein in tia-1–/– cells.
Crosses of tia-1+/– × tia-1+/– mice suggested a ∼50% lethality among tia-1–/– adult offspring (Table I). Genotype analysis of embryos from tia-1+/– × tia-1+/– and tia-1+/– × tia-1–/– breedings did not reveal any significant lethality among tia-1–/– embryos of different stages. Since genotype analysis was generally done ∼3 weeks post partum, this indicates that tia-1–/– mice die between embryonic day 16.5 and 3 weeks of age. The surviving mice appear normal and live for ≤2 years. Gross and histological analysis of heart, lung, testes, thymus, liver and kidney did not reveal any morphological abnormalities. Flow cytometric analysis of splenocytes [CD45RA, immunoglobulin M, CD3, CD4 and CD8] and counting of blood smears confirmed the presence of all the major hematopoietic lineages in normal proportions (data not shown).
Table I. Embryonic lethality in mice lacking TIA-1a.
| +/+ | +/– | –/– | (Observed –/–)/(expected –/–) | |
|---|---|---|---|---|
| A Offspring of tia-1+/– × tia-1+/– cross | ||||
| Adult mice | 38 | 67 | 16 | 16/35 (46%) |
| Embryos (E12.5–E16.5) | 18 | 30 | 14 | 14/16 (89%) |
| B Offspring of tia-1+/– × tia-1–/– cross | ||||
| Embryos (E9.5–E11.5) | 31 | 32 | 32/31 (103%) |
aTIA-1 deficiency is partially lethal. Genotype distribution of (A) tia-1+/– × tia-1+/– progeny and (B) tia-1+/– × tia-1–/– progeny of adult mice and embryos of different developmental stages.
TIA-1, like TIAR, is an ARE-binding protein
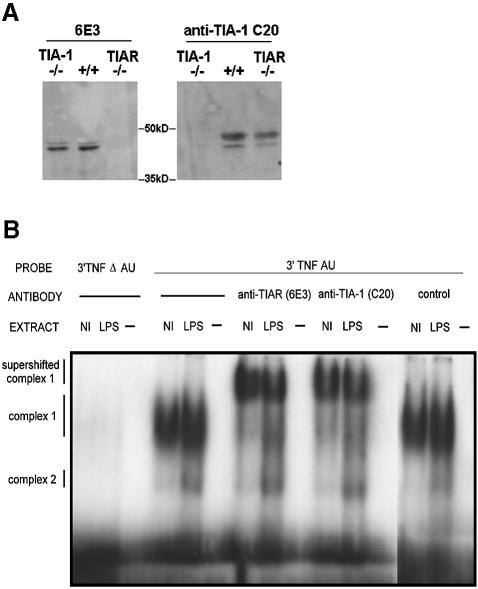
The identification of TIAR as a component of the ARE-binding protein complex that assembles on the 3′ UTR of TNF-α transcripts provided the first clue that TIA-1 and TIAR might regulate the expression of specific mRNAs (Gueydan et al., 1999). We used mouse embryonic fibroblasts (MEFs) derived from wild-type, TIA-1–/– and TIAR–/– mice to confirm the specificity of antibodies reactive with TIAR (6E3) and TIA-1 (anti-TIA-1 C20) (Figure 2A). Thus, 6E3 recognizes TIAR in wild-type and TIA-1–/– MEFs, but not in TIAR–/– MEFs (left panel). Conversely, anti-TIA-1 C20 recognizes TIA-1 in wild-type and TIAR–/– MEFs, but not in TIA-1–/– MEFs (right panel). TIAR has been reported previously to bind to the TNF-α ARE (Gueydan et al., 1999). It is also a component of the ARE-binding complex 1, which binds to the 3′ UTR of TNF-α transcripts in vivo (Gueydan et al., 1999). We used antibodies specific for TIA-1 and TIAR to show that both TIA-1 and TIAR are components of complex 1. As shown in Figure 2B, extracts from normal (Nl) or LPS-treated RAW 264.7 cells were incubated with radiolabeled probes corresponding to the 3′ UTR of TNF-α with (3′TNF AU) or without (3′TNF ΔAU) the ARE, separated by SDS–PAGE and visualized using autoradiography. Although LPS extracts assemble two distinct protein complexes (complexes 1 and 2) under these conditions, complex 1 is selectively shifted in the presence of antibodies reactive with TIAR (6E3) or TIA-1 (C20). A control goat antiserum (anti-MAD2; Santa Cruz) did not supershift complex 1, confirming the specificity of this effect and indicating that both TIA-1 and TIAR are ARE-binding proteins that interact with the TNF-α ARE in vitro.
Fig. 2. (A) Mouse embryonic fibroblasts derived from wild-type, TIA-1–/– and TIAR–/– mice. 6E3 recognizes TIAR (left panel), while anti-TIA-1 C20 recognizes TIA-1 (right panel). (B) EMSA using Raw 264.7 cells. Extracts from control or LPS-treated RAW cells were incubated with radiolabeled probes from the 3′ UTR of TNF-α with or without the ARE before separation by SDS–PAGE. Anti-TIA-1 (ML29) and anti-TIAR (6E3) supershifted complex 1, whereas a goat anti-MAD2 (Santa Cruz) control did not, indicating that both TIA-1 and TIAR are TNF-α ARE-binding proteins. The relative migration of complex 1, complex 2 and supershifted complex 1 are indicated on the left.
TIA-1 selectively suppresses the expression of TNF-α
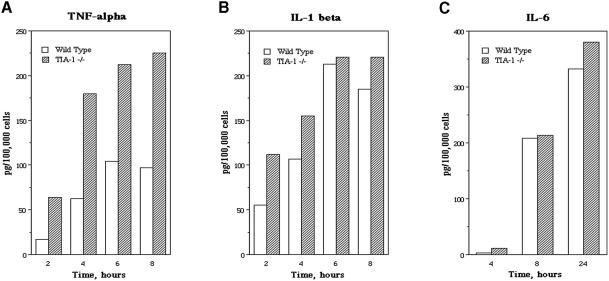
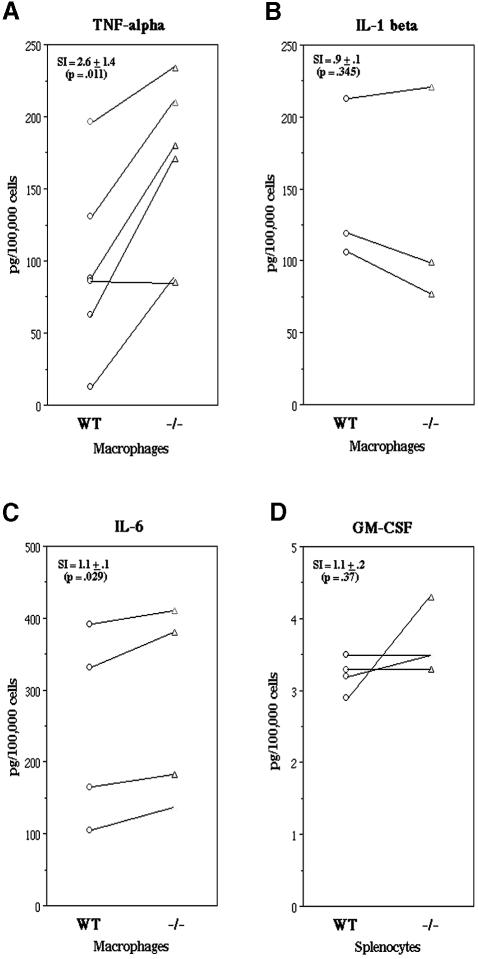
The effect of TIA-1 on the expression of TNF-α was examined by comparing the LPS-induced expression of TNF-α in wild-type (BALB/c) and TIA-1–/– (backcrossed for 10 generations in the BALB/c background) peritoneal macrophages. Thioglycolate-elicited macrophages were removed from the peritoneal cavities of age and gender-matched wild-type and mutant mice, and cultured in the absence or presence of LPS (1 µg/ml) for the times indicated before assaying for the production of TNF-α, interleukin (IL) 1β and IL-6 by enzyme-linked immunosorbent assay (ELISA) (Figure 3). None of these monocyte-derived cytokines were detected in supernatants harvested from wild-type or mutant macrophages cultured in the absence of LPS (data not shown). In contrast, LPS induced the secretion of TNF-α (Figure 3A), IL-1β (Figure 3B) and IL-6 (Figure 3C) in both wild-type and mutant macrophages. At every time-point examined, the amount of inflammatory cytokines produced by TIA-1–/– macrophages exceeded that produced by wild-type macrophages (Figure 3). The significance of these differences was determined by applying the paired Student’s t-test to the levels of cytokine produced by wild-type and mutant macrophages in several independent experiments (Figure 4). This analysis reveals that TIA-1–/– macrophages produce significantly more TNF-α than wild-type macrophages [stimulation index (SI) 2.6 ± 1.4; p = 0.011, paired Student’s t-test; n = 6; Figure 4A]. The production of IL-1β in TIA-1–/– macrophages was not significantly different from that in wild-type macrophages (SI 0.9 ± 0.1; p = 0.345; n = 3; Figure 4B). TIA-1–/– macrophages produced slightly more IL-6 than wild-type macrophages (SI 1.1 ± 0.1; p = 0.029, n = 4; Figure 4C), but this may have been an indirect consequence of TNF-α production since TNF-α can induce the expression of IL-6 (Feldmann and Maini, 1999). We also compared the expression of granulocyte–macrophage colony-stimulating factor (GM-CSF), another cytokine that is subject to ARE-dependent post-transcriptional control, in wild-type and TIA-1–/– mice (Figure 4D). Because LPS did not induce the production of GM-CSF in peritoneal macrophages, we compared the production of GM-CSF in splenocytes stimulated with LPS and anti-CD3/anti-CD28. Under these conditions, the secretion of GM-CSF was not significantly different in wild-type mice and TIA-1–/– mice (SI 1.1 ± 0.2; p = 0.37; n = 4; Figure 4D). In three separate experiments using age- and gender-matched mice, LPS- and anti-CD3/CD28-induced expression of interferon γ (IFN-γ) did not differ between wild-type and TIA-1–/– splenocytes (mean supernatant concentrations: wild type, 80.6 ± 28.3 pg/1 × 105 cells; TIA-1–/–, 94.6 ± 39.6 pg/1 × 105 cells; p = 0.169, paired Student’s t-test; n = 3). These data indicate that TIA-1 selectively regulates the production of TNF-α.
Fig. 3. Thioglycolate-elicited peritoneal macrophages collected from wild-type and TIA-1–/– BALB/c mice were treated with LPS (1 µg/ml) for the times indicated. Supernatant TNF-α and IL-6 concentrations (A and C) and combined cell lysate and supernatant IL-1β concentrations (B) were measured by ELISA. Each experiment was repeated at least three times; one representative experiment is depicted in each panel. Open bars, wild type; closed bars, TIA-1–/–.
Fig. 4. LPS-induced cytokine production by BALB/c mouse peritoneal macrophages. Cytokine concentrations were measured by ELISA. Each set of connected data points represents a separate experiment using age- and gender-matched mice. (A) Thioglycolate-elicited peritoneal macrophages collected from wild-type and TIA-1–/– mice were stimulated with LPS for 4 h and supernatant TNF-α concentrations were measured. (B) IL-1β concentrations in cell lysate and supernatant (combined values) of wild-type and TIA-1–/– mouse thioglycolate-elicited peritoneal macrophages after LPS stimulation for 6 h. (C) IL-6 concentrations in supernatant of wild-type and TIA-1–/– mouse thioglycolate-elicited peritoneal macrophages after stimulation with LPS for 24 h. (D) GM-CSF production by wild-type and TIA-1–/– splenocytes after LPS and anti-CD3/CD28 stimulation for 24 h. SI, stimulation index (average fold increase in cytokine production by TIA-1–/– mice compared with wild-type mice).
Because BALB/c mice lacking TIAR die in utero (our unpublished results), we were unable to compare directly the effects of TIA-1 and TIAR on the expression of TNF-α. In preliminary experiments, peritoneal macrophages derived from TIAR nullizygotes that had been backcrossed for four or five generations into the C57BL/6 background (∼10% of TIAR nullizygotes survive to birth in the C57BL/6 background) produced significantly more TNF-α than wild-type C57BL/6 mice (data not shown), suggesting that the effects of TIA-1 and TIAR may be similar. Attempts to breed double knockouts have been unsuccessful due to the high rate of embryonic lethality. It remains to be determined whether the effects of TIA-1 and TIAR on the production of TNF-α are additive or synergistic.
TIA-1 is a translational silencer
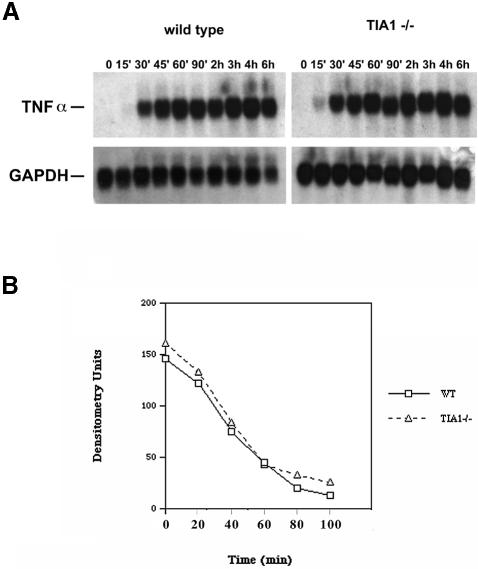
In order to determine whether the altered expression of TNF-α results from increased expression of TNF-α transcripts, we compared the expression of TNF-α mRNA in LPS-treated wild-type and TIA-1–/– peritoneal macrophages. As shown in Figure 5A, TNF-α mRNA was undetectable in macrophages cultured in the absence of LPS (t = 0). LPS-induced expression of TNF-α mRNA is similar in wild-type and TIA-1–/– macrophages harvested between 30 min and 8 h after the addition of LPS, suggesting that the increased secretion of TNF-α by TIA-1–/– macrophages is not a consequence of increased levels of TNF-α transcripts. Because the quantity and size of the TNF-α transcripts are similar in wild-type and TIA-1–/– macrophages, it is unlikely that TIA-1 alters the stability of TNF-α transcripts. This was confirmed by comparing the half-life of TNF-α transcripts in LPS-activated peritoneal macrophages derived from wild-type and TIA-1–/– mice. In these experiments, LPS-activated macrophages were treated with 5,6-dichlorobenzimidazole riboside (DRB) to inhibit new RNA synthesis, harvested at the times indicated, and processed for northern blotting using a TNF-α-specific probe. TNF-α mRNA concentrations were measured densitometrically and plotted as a function of time in Figure 5B. It is clear from this analysis that the stability of TNF-α transcripts is not significantly different in wild-type and TIA-1–/– macrophages.
Fig. 5. (A) TNF-α mRNA expression in wild-type and TIA-1–/– peritoneal macrophages. Cells were stimulated with LPS for the times indicated and cell lysates were analyzed for TNF-α and GAPDH mRNA by northern blotting. (B) Stability of TNF-α transcripts in wild-type and TIA-1–/– peritoneal macrophages. Cells were stimulated with LPS for 2 h and then treated with 50 µM DRB to inhibit new RNA synthesis. At the times indicated, cell lysates were analyzed for TNF-α mRNA by northern blotting. Expression levels were measured by densitometry.
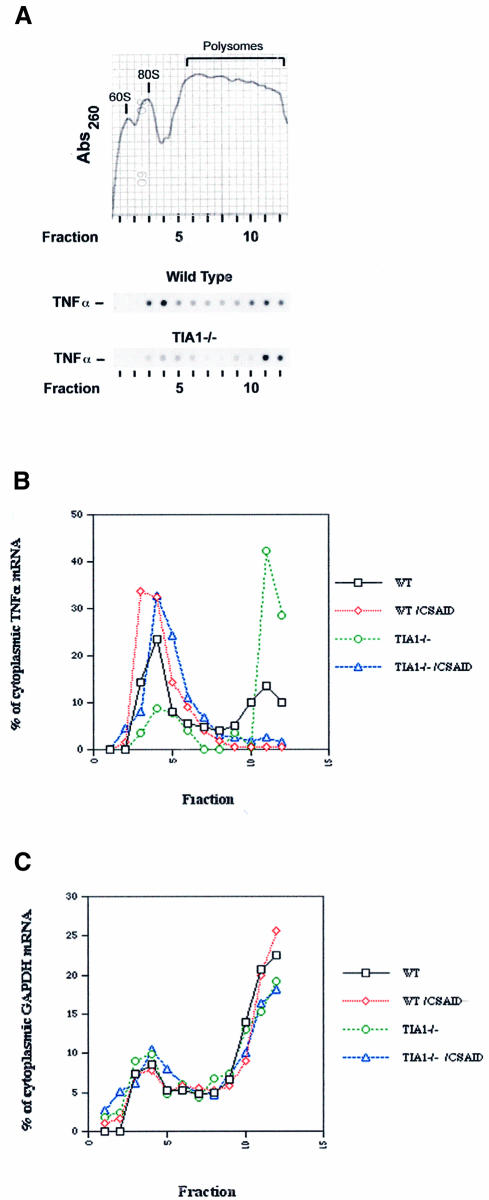
TIA-1 and TIAR function as translational silencers that sequester non-heat shock mRNAs in cells subjected to environmental stress (Kedersha et al., 1999). To determine whether a similar mechanism is used to repress the expression of TNF-α, we fractionated cytoplasmic extracts from wild-type and TIA-1–/– macrophages over sucrose gradients to compare the polysome profiles of TNF-α and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNAs. The relative migration of monosomes (80S) and polysomes was determined by continuously monitoring the absorbance at 260 nm during gradient elution (Figure 6A; although this profile is from the wild-type cells, the absorbance profiles from TIA-1–/– cells are superimposable). Individual fractions were processed for northern blotting, and probed to identify transcripts encoding TNF-α and GAPDH. Representative autoradiograms from wild-type and TIA-1–/– mice are shown in Figure 6A. Densitometric analysis was used to determine the proportion of each transcript found in polysomal (fractions 7–12) and non-polysomal (fractions 1–6) regions of the gradient (Figure 6B and C). Although TNF-α transcripts are distributed between polysomal and non-polysomal fractions in wild-type macrophages (Figure 6B, black squares), there is a clear shift towards the polysomal fractions in macrophages lacking TIA-1 (Figure 6B, green circles). Averaged over five independent experiments, the fraction of TNF-α transcripts associated with polysomes was 0.57 ± 0.06 (n = 5) in wild-type and 0.71 ± 0.06 (n = 5) in TIA-1–/– macrophages (p = 0.007, unpaired Student’s t-test). In contrast, the fraction of GAPDH transcripts associated with polysomes was not significantly different in wild-type (0.69 ± 0.1; n = 5) and TIA-1–/– (0.65 ± 0.09; n = 5) macrophages (Figure 6C).
Fig. 6. Sucrose gradient analysis. Peritoneal macrophages were activated with LPS in the absence or presence of the CSAID SB202190 (5 µM) for 2 h. Cell lysates were layered over a 20–47% sucrose gradient and centrifuged at 40 000 r.p.m. for 3 h. (A) Representative profile of the 260 nm UV absorption across the gradients. The profile shown was obtained from wild-type cell lysates in the absence of CSAID; the absorption peaks corresponding to the 60S, 80S and polysome-containing fractions are indicated. Analyses from TIA-1–/– lysates were virtually identical. TNF-α mRNA levels in individual fractions from wild-type and TIA-1–/– lysates were determined by hybridization and autoradiography. Quantitation of TNF-α (B) and GAPDH (C) transcripts in individual fractions was determined by densitometry. Black squares, wild-type macrophages; red diamonds, wild-type macrophages plus CSAID; green circles, TIA-1–/– macrophages; blue triangles, TIA-1–/– macrophages plus CSAID.
Cytokine-suppressive anti-inflammatory drugs (CSAIDs) inhibit the stress-induced p38–MAPKAP kinase 2 (MK2) signaling cascade and block the LPS-induced release of translational suppression (Prichett et al., 1995; Lee and Young, 1996; Salituro et al., 1999). These drugs function at the level of translational initiation, suggesting that they might directly or indirectly target TIA-1 and/or TIAR. As shown in Figure 6B, the CSAID SB202190 shifts TNF-α transcripts (but not GAPDH transcripts, Figure 6C) from polysomal to non-polysomal fractions in both wild-type and TIA-1–/– macrophages, indicating that the absence of TIA-1 does not abrogate the response to CSAIDs.
Endotoxin lethality
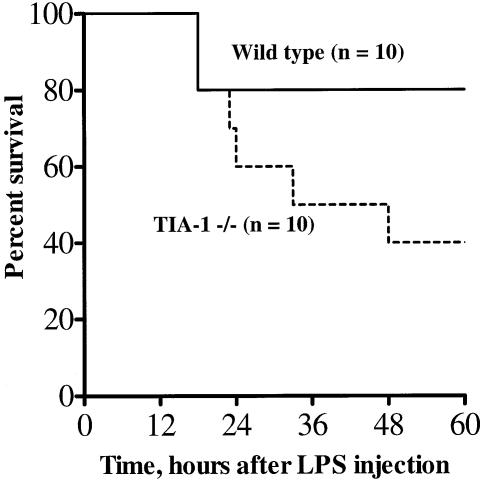
The physiological significance of TIA-1 was confirmed by comparing the susceptibility of wild-type and TIA-1–/– mice to endotoxin. As shown in Figure 7, TIA-1–/– mice are more susceptible to LPS-induced endotoxic shock than are wild-type mice. By 48 h after intraperitoneal LPS injection, 60% of TIA-1–/– mice had died, compared with 20% of wild-type mice. Although we have not yet shown that this is a consequence of increased expression of TNF-α, it strongly suggests that TIA-1 can regulate the production of one or more of the mediators of endotoxic shock.
Fig. 7. Increased susceptibility of TIA-1–/– mice to endotoxic shock. Kaplan–Meier survival plots for wild-type and TIA-1–/– BALB/c mice after intraperitoneal injection with 100 µg LPS.
Discussion
TIA-1 and TIAR are structurally related RNA-binding proteins that are essential for normal vertebrate development (Beck et al., 1998). Targeted disruption of TIAR results in embryonic lethality (Beck et al., 1998), the penetrance of which is strain dependent. The rate of embryonic lethality is 100% in the BALB/c background and 90% in the C57BL/6 background (our unpublished data). Mice that survive to birth are sterile due to defective germ-cell maturation (Beck et al., 1998). Targeted disruption of TIA-1 also results in embryonic lethality, but the penetrance is <50% in both BALB/c and C57BL/6 backgrounds. TIA-1 nullizygotes are fully fertile. Breeding experiments reveal that mice lacking both TIA-1 and TIAR die before embryonic day 7, indicating that one or the other of these proteins must be present for normal embryonic development. These results indicate that the functions of these related proteins are partially redundant. At the same time, TIAR is uniquely required for normal germ-cell maturation.
Consistent with their functional redundancy, the tissue distribution and subcellular localization of TIA-1 and TIAR are quite similar (Kawakami et al., 1992; Beck et al., 1996). Although both proteins are concentrated in the nucleus at steady state, heterokaryon analysis reveals that both proteins continuously shuttle between the nucleus and the cytoplasm (N.Kedersha and P.Anderson, manuscript in preparation). In this respect, TIA-1 and TIAR resemble the hnRNPs that assemble around nascent RNA transcripts and facilitate transport from the nucleus to the cytoplasm (Krecic and Swanson, 1999). Like the hnRNPs, TIA-1 and TIAR can function as general RNA-binding proteins, which interact with many, if not most, mRNAs in vitro (Dember et al., 1996). At the same time, these proteins can selectively interact with RNAs possessing uridine-rich motifs (Dember et al., 1996). In their ability to function as both general and specific RNA-binding proteins, TIA-1 and TIAR resemble hnRNPs K and E1, proteins that participate in general RNA export and also bind to the 3′ UTRs of 5-lipoxygenase transcripts to repress translational initiation (Ostareck-Lederer et al., 1994; Ostareck et al., 1997).
The general RNA-binding activity of TIA-1 and TIAR allows these proteins to regulate the translational arrest that accompanies environmental stress. Stress-induced phosphorylation of eIF-2α, a regulatory component of the heterotrimeric G-protein complex that loads initiator tRNAs on to 40S ribosomal subunits, inhibits translation by preventing the assembly of preinitiation complexes (Berlanga et al., 1998; Gray and Wickens, 1998; Srivastava et al., 1998). TIA-1 and TIAR function downstream of this event to sequester untranslated mRNAs at cytoplasmic RNP particles known as stress granules (Kedersha et al., 1999). By preventing the translational reinitiation of these mRNAs, TIA-1 and TIAR are likely to influence the duration of stress-induced translational arrest. The association of TIA-1 and TIAR with the TNF-α ARE suggests that these proteins might also regulate the metabolism of specific transcripts to which they bind with high affinity. The increased production of TNF-α in LPS-stimulated macrophages lacking either TIA-1 or TIAR is consistent with this possibility. Although the increased production of TNF-α observed in macrophages lacking either TIA-1 or TIAR is relatively modest, the ability of these proteins to complement one another functionally suggests that cells lacking both TIA-1 and TIAR might produce significantly more TNF-α. It is also likely that additional ARE-binding proteins, including TTP (Taylor et al., 1996), cooperate with TIA-1 and TIAR in the regulation of TNF-α production. The functional redundancy between TIA-1 and TIAR might also explain why mice lacking one or the other of these proteins do not spontaneously develop inflammatory arthritis as has been reported in mice lacking TTP (Taylor et al., 1996).
Two distinct regulatory complexes are known to bind to the TNF-α ARE (Hel et al., 1996; Gueydan et al., 1999). TIA-1 and TIAR are components of complex 1, which assembles on the AUUUAUUUA nonamer repeats (also known as class II AREs) encoded in the 3′ UTR of the TNF-α transcript (Gueydan et al., 1999). Class II AREs regulate the stability of cytokine mRNAs such as TNF-α and GM-CSF (Caput et al., 1986; Shaw and Kamen, 1986; Wilson and Treisman, 1988; Kruys et al., 1989; Han and Beutler, 1990; Han et al., 1990; Chen and Shyu, 1994; Chen et al., 1995; Wang et al., 1997; Xu et al., 1997). Trans-acting ARE-binding proteins known to participate in this process include HuR (Vakalopoulou et al., 1991; Ma et al., 1996; Myer et al., 1997; Fan and Steitz, 1998; Peng et al., 1998), Hel-N1 (Levine et al., 1993; Gao et al., 1994) and AUF1 (hnRNP D) (Zhang et al., 1993; De Maria and Brewer, 1996; Laroia et al., 1999; Loflin et al., 1999). Whereas the ELAV proteins HuR and Hel-N1 stabilize transcripts bearing class II AREs (Fan and Steitz, 1998; Peng et al., 1998), AUF1 destabilizes these transcripts (Zhang et al., 1993; De Maria and Brewer, 1996; Laroia et al., 1999; Loflin et al., 1999). Trans-acting factors that influence mRNA stability have similar effects on transcripts encoding TNF-α or GM-CSF (Loflin et al., 1999). In contrast, the stability of TNF-α transcripts does not appear to be altered in macrophages lacking TIA-1. The increased production of TNF-α in LPS-stimulated macrophages lacking TIA-1 appears to result from an increased rate of translational initiation since a greater fraction of transcripts are found in association with polysomes. This suggests that TIA-1 and TIAR normally function as translational silencers, which is consistent with their function in stressed cells (Kedersha et al., 1999). Whereas ARE-binding proteins that regulate transcript stability coordinately influence the production of several ARE-containing cytokines (Loflin et al., 1999), TIA-1 appears to regulate selectively the production of TNF-α.
Our results introduce TIA-1 and TIAR as translational silencers that can independently and selectively regulate the production of TNF-α. Previous studies using macrophage cell lines have clearly shown that translational silencing is important in the post-transcriptional control of TNF-α production (Han and Beutler, 1990; Han et al., 1990; Biragyn and Nedospasov, 1995; Crawford et al., 1997; Wang et al., 1997). In the unstimulated macrophage cell line, RAW 264.7, TNF-α transcripts are expressed but excluded from polysomes and not translated (Han et al., 1990; Crawford et al., 1997). Comparison of TNF-α mRNA distribution into polysomes in wild-type and TIA-1–/– macrophages indicates that TIA-1 controls the association of TNF-α mRNA with polysomes. It remains to be determined whether TIA-1/TIAR-induced translational silencing is achieved by regulation of translational initiation. In any case, the ability of TIA-1 and TIAR to inhibit TNF-α mRNA translation suggests that these proteins might be targets of the stress kinase signaling cascade that is blocked by CSAIDs. CSAIDs block the LPS-induced production of TNF-α by preventing translational derepression (Lee et al., 1994; Prichett et al., 1995). This is accomplished by inhibiting the p38–MK2 signaling cascade (Han and Ulevitch, 1999; Kotlyarov et al., 1999; Winzen et al., 1999), suggesting that these kinases phosphorylate a translational silencer that associates with TNF-α transcripts. The ability of CSAIDs to repress the expression of TNF-α similarly in wild-type and TIA-1–/– macrophages indicates that TIA-1 is not an essential target of these drugs. TIA-1 might, thus, act as a constitutive translational suppressor controlling excessive TNF-α production. Alternatively, the functional redundance of TIA-1 and TIAR leaves open the possibility that cells lacking both TIA-1 and TIAR might be resistant to the suppressive effects of CSAIDs.
Taken together, our results suggest that TIA-1 and TIAR are translational silencers that regulate the cellular and organismal response to stress. At the cellular level, these proteins contribute to the general translational arrest that accompanies environmental stress. By controlling the duration of translational arrest, TIA-1 and TIAR might determine whether stressed cells live to repair the stress-induced damage or die by apoptosis. At the organismal level, these proteins regulate the expression of at least one inflammatory mediator that serves as a sentinel to signal the presence of microbial infection. It remains to be determined whether the translational control exerted by these proteins is limited to the stress response or is a general feature of normal cellular metabolism.
Materials and methods
Generation of mutant mice
A phosphoglycerate kinase (PGK)–thymidine cassette followed by a 2.8 kb XbaI–XbaI fragment containing exon 3 of the TIA-1 gene (Beck et al., 1998), a PGK–promoter neor cassette and a 6.5 kb TIA-1 gene fragment comprising the 3′-half of exon 4 (generated by exonuclease III digestion) through exon 9 and ending at an AatI site, were inserted into the pSK+ plasmid. D3 ES cells were electroporated with the XhoI-linearized targeting construct and selected in G418 (150 mg/ml) and gancylovir (2 mM) and expanded for Southern blot analysis. Of ∼1000 clones screened, one yielded fragments of the expected size and was used for injection into BALB/c blastocytes, which were transferred to the uterus of pseudopregnant Swiss Webster mice. Germline transmission was obtained on further crossing of male chimeras with BALB/c females. These mice were then backcrossed for 10 generations on the BALB/c strain before use in the functional assays described. Mice lacking TIAR have been described previously (Beck et al., 1998). Because TIAR nullizygosity is uniformly lethal in the BALB/c background, these mice were backcrossed for five generations into the C57BL/6 strain for use in these experiments.
Southern blotting
Tail biopsies were recovered from ∼3 week old mice and subjected to proteinase K digestion for 10–16 h at 55°C in a buffer containing 50 mM Tris–HCl (pH 8.5), 20 mM EDTA, 10 mM NaCl, 0.5% SDS and 0.5 mg/ml proteinase K (Boehringer). Genomic DNA was recovered by phenol extraction and precipitation according to standard procedures. About 20 µg of genomic DNA digested with EcoRI was used for Southern blotting using standard methods.
Primary embryonic fibroblasts
Female pregnant mice from tia-1+/– × tia-1+/– breedings were killed on embryonic day 14.5 (where the time when a copulation plug was found is defined as embryonic day 0.5). Embryos were recovered and the visceral yolk sac was used for genotyping. The head and organs of each embryo were removed, the remaining carcass was trypsinized and the resulting cells were grown in tissue culture plates at 37°C/5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum.
Protein analysis
Primary embryonic fibroblasts (∼7 × 106 cells) were washed once with phosphate-buffered saline (PBS); cells were then recovered with a cell lifter. Cells were lysed for 20 min on ice in a buffer containing 150 mM NaCl, 1% Nonidet P-40 (NP-40), 50 mM Tris–HCl pH 8.0, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride (PMSF), 10 mg/ml aprotinin and 10 mg/ml leupeptin. Insoluble material was removed by centrifugation (10 000 g); lysates were precleared with Sepharose 4B beads and then incubated with 5 µg of anti-TIA-1 (ML29) and 25 µl protein G–Sepharose slurry (Pharmacia Biotech, Inc.) for 3–5 h. Immunoprecipitates were washed three times in a buffer containing 150 mM NaCl, 50 mM Tris–HCl pH 8.0 and 1% NP-40 and washed immunoprecipitated proteins were resolved by SDS–PAGE (10% gels) using reducing conditions. Proteins were then transferred to Immobilon P membrane (Millipore) and probed with anti-TIA-1 (2G9, 5 µg/ml). Immunoblots were developed with protein A/G-conjugated horseradish peroxidase (Pierce) and the chemiluminescence reagent, luminol, essentially as described by the supplier (DuPont–NEN).
Electrophoretic mobility shift assays (EMSAs)
S100 extracts from RAW 264.7 mouse macrophages were incubated with a 3′ TNF probe as described previously (Gueydan et al., 1996). Supershifts with anti-TIAR, anti-TIA-1 or control (goat anti-MAD2; Santa Cruz) antibodies were performed by incubating 15 µg of S100 extract with 0.2 µg antibody for 25 min on ice in a total volume of 15 µl before the EMSA. The EMSAs were electrophoresed on non-denaturing 3.5% polyacrylamide gels.
Isolation of peritoneal macrophages and splenocytes
Age- and gender-matched mice were each injected intraperitoneally with 2 ml sterile thioglycolate broth (Benton Dickinson) in order to increase the yield of peritoneal macrophages. Three days later, the mice were killed in a carbon dioxide chamber and peritoneal cells were collected by peritoneal lavage with Hanks’ balanced salt solution (HBSS). The spleens were removed and splenocytes were separated by crushing with forceps and aspiration through a 21 gauge needle. The cells were washed with HBSS and red blood cells were lysed by the addition of 0.9% ammonium chloride (Fisher) for 3 min. Following repeat washing with HBSS, the cells were plated at a concentration of 1 × 106 (peritoneal cells) or 2 × 106 (splenocytes) cells/ml medium; cells were treated as indicated within 24 h of cell collection. The cell medium consisted of DMEM supplemented with 10% heat-inactivated fetal bovine serum, 1% penicillin/streptomycin, 3.5 µl/l β-mercaptoethanol and insulin– transferrin–sodium selenite medium supplement (Sigma).
Cell treatment with LPS and/or anti-CD3/CD28
LPS, Escherichia coli serotype O111:B4 (Sigma) at a concentration of 1 µg/ml was used to stimulate mouse-derived peritoneal macrophages or splenocytes for the durations indicated in each experiment. Anti-murine CD3 and anti-murine CD28 antibodies were added at a 1:100 dilution from hybridoma supernatants, a concentration determined to induce maximal proliferation of splenocytes in culture (antibodies were provided by Dr Frank Borriello, Brigham and Women’s Hospital). After treatment as indicated, the cell supernatants were collected and centrifuged at 13 600 g for 3 min and the supernatants were frozen and stored at –80°C. For IL-1β experiments, combined cell lysates and supernatants were collected. At the times indicated, the cells were frozen on dry ice, then thawed, then frozen and thawed once more before centrifugation and subsequent storage at –80°C.
Cytokine detection
Sandwich ELISAs were used to determine cytokine concentrations in each cell supernatant. Nunc-Immuno plates were used for the ELISAs. TNF-α, GM-CSF, IFN-γ and IL-6 capture and biotinylated detection antibodies were obtained from Pharmingen. IL-1β capture and biotinylated detection antibodies were obtained from R&D Systems. Commercially available streptavidin-conjugated horseradish peroxidase (Southern Biotechnology Associates, Inc.) and ABTS (Boehringer Mannheim) were used as the final steps in detection. The optical density of each sample was measured on a Titertek Multiscan MCC/340 MK 11 plate reader at the recommended wavelength and the cytokine concentration in each sample was calculated from a cytokine recombinant protein standard curve. Each cytokine concentration was expressed as amount per 1 × 105 cells.
Northern blot analysis
Total RNA was isolated using Trizol (Gibco-BRL) according to the manufacturer’s protocol with the addition of 40 µg glycogen (Boehringer Mannheim) before isopropanol precipitation. Five micrograms of RNA was loaded into each well of a denaturing 1.3% agarose gel (containing 0.5× MOPS, 3% formaldehyde and 100 ng/ml ethidium bromide) and transferred to NytranN (Schleicher and Schuell). 32P-labelled random-primed probes were generated from the mouse TNF-α 3′ UTR and the mouse full-length GAPDH cDNA using a kit (Stratagene). Hybridization (9× SSC, 50% formamide, 0.1 mg/ml calf thymus DNA, 0.05 M sodium phosphate pH 6.4, 1× Denhardt’s solution, 0.1% SDS) and autoradiography were done first with the TNF-α probe and then with the GAPDH probe. Filters were washed in 0.5× SSC and 0.1% SDS at 65°C.
TNF-α transcript stability
Peritoneal macrophages were cultured from wild-type and TIA-1–/– mice and treated with LPS (1 µg/ml) for 2 h. Total RNA was isolated from parallel cultures before and at various times after transcriptional arrest with DRB (final concentration 50 µM). The relative levels of TNF-α transcripts in 5 µg of total RNA from each sample were determined by northern blot analysis, autoradiography and densitometry, as described above.
Cell fractionation and polysome analysis
Peritoneal macrophages from TIA-1–/– and wild-type mice were cultured as described above using 60 mm plates. LPS (1 µg/ml) in the absence or presence of the CSAID SB202190 (5 µM) was added for 2 h. Cells were then washed twice in ice-cold PBS containing 10 mg/ml cycloheximide. Cells were scraped into 1.0 ml of ice-cold lysis buffer (140 mM KCl, 1 mM dithiothreitol, 20 mM Tris pH 8, 5 mM MgCl2, 0.5% NP-40, 0.5 U/µl RNAsin (Promega), 10 mM cycloheximide, 10 µM PMSF, 5 nM leupeptin, 5 µm benzamidine, 5 ng/ml aprotinin) and mechanically disrupted by douncing. Nuclei and debris were removed by microcentrifugation for 10 min. The supernatant was layered on to 11 ml of a 20–47% continuous sucrose gradient. Centrifugation was performed at 40 000 r.p.m. for 3 h 15 min using a SW40Ti rotor. One milliliter fractions were collected, starting from the top of the gradient. UV absorption at 260 nm was monitored continuously to identify fractions containing monosomes and polysomes. Five hundred microliters of each fraction was digested with 0.5 mg/ml proteinase K after the addition of SDS (0.2% final concentration) and EDTA (5 mM final concentration) at 37°C for 6 min, phenol extracted, ethanol precipitated and resuspended in 20 µl of DEPC-treated water. Five microliters was added to 5 µl water and 30 µl formamide, heated to 65°C for 5 min, mixed with 400 µl 6× SSC and applied to a well of a dot-blot apparatus. Hybridization and washing were done with TNF-α and GAPDH probes as described above. The relative amounts of TNF-α and GADPH mRNA in each fraction were determined by densitometry.
Endotoxin lethality
Age- and gender-matched wild-type and TIA-1–/– BALB/c mice were each injected intraperitoneally with 100 µg LPS (E.coli serotype O111:B4, Sigma) diluted in 1 ml sterile PBS. Subsequent survival was monitored.
Acknowledgments
Acknowledgements
We thank Lana Tsao and Nadia Rosenthal for providing GAPDH cDNA and Stephen O’Brien for technical assistance. This work was supported by grants from the National Institutes of Health (P.A.), the Arthritis Foundation (P.A., M.S.) and the EC biotech Program (BIO4-CT95-0045). P.A. and M.S. are scholars of the Leukemia and Lymphoma Society.
References
- Beck A.R.P., Medley,Q.G., O’Brien,S., Anderson,P. and Streuli,M. (1996) Structure, tissue distribution and genomic organization of the murine RRM-type RNA binding proteins TIA-1 and TIAR. Nucleic Acids Res., 24, 3829–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck A.R.P., Miller,I.J., Anderson,P. and Streuli,M. (1998) RNA-binding protein TIAR is essential for primordial germ cell development. Proc. Natl Acad. Sci. USA, 95, 2331–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlanga J., Herrero,S. and DeHaro,C. (1998) Characterization of the hemin-sensitive eukaryotic initiation factor 2α kinase from mouse nonerythroid cells. J. Cell Biol., 273, 32340–32346. [DOI] [PubMed] [Google Scholar]
- Beutler B. (1999) The role of tumor necrosis factor in health and disease. J. Rheum., 26, 16–21. [PubMed] [Google Scholar]
- Biragyn A. and Nedospasov,S.A. (1995) Lipopolysaccharide-induced expression of TNF-α gene in the macrophage cell line ANA-1 is regulated at the level of transcription processivity. J. Immunol., 155, 674–683. [PubMed] [Google Scholar]
- Caput D., Beutler,B., Hartog,K., Thayer,R., Brown-Shimer,S. and Cerami,A. (1986) Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc. Natl Acad. Sci. USA, 83, 1670–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo E., Lai,W.S. and Blackshear,P.J. (1998) Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science, 281, 1001–1005. [DOI] [PubMed] [Google Scholar]
- Chen C.Y.A. and Shyu,A.B. (1994) Selective degradation of early-response-gene mRNAs: functional analyses of sequence features of the AU-rich elements. Mol. Cell. Biol., 14, 8471–8482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y.A., Xu,N. and Shyu,A.B. (1995) mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte–macro phage colony-stimulating factor transcripts: different deadenylation kinetics and uncoupling from translation. Mol. Cell. Biol., 15, 5777–5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford E.K., Ensor,J.E., Kalvakolanu,I. and Hasday,J.D. (1997) The role of 3′ poly(A) tail metabolism in tumor necrosis factor-α regulation. J. Biol. Chem., 272, 21120–21127. [DOI] [PubMed] [Google Scholar]
- De Maria C.T. and Brewer,G. (1996) AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J. Biol. Chem., 271, 12179–12184. [DOI] [PubMed] [Google Scholar]
- Dember L.M., Kim,N.D., Liu,K.Q. and Anderson,P. (1996) Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J. Biol. Chem., 271, 2783–2788. [DOI] [PubMed] [Google Scholar]
- Fan X.C. and Steitz,J.A. (1998) Overexpression of HuR, a nuclear–cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J., 17, 3448–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann M. and Maini,R. (1999) The role of cytokines in the pathogenesis of rheumatoid arthritis. Rheumatology, 38, 3–7. [PubMed] [Google Scholar]
- Ford L.P., Watson,J., Keene,J.D. and Wilusz,J. (1999) ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes Dev., 13, 188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F.B., Carson,C.C., Levine,T.D. and Keene,J.D. (1994) Selection of a subset of mRNAs from combinatorial 3′ untranslated region libraries using neuronal RNA-binding protein Hel-N1. Proc. Natl Acad. Sci. USA, 91, 11207–11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray N. and Wickens,M. (1998) Control of translation initiation in animals. Annu. Rev. Cell. Dev. Biol., 14, 399–458. [DOI] [PubMed] [Google Scholar]
- Gueydan C., Houzet,L., Marchant,A., Sels,A., Huez,G. and Kruys,M. (1996) Engagement of tumor necrosis factor mRNA by an endotoxin-inducible cytoplasmic protein. Mol. Med., 2, 479–488. [PMC free article] [PubMed] [Google Scholar]
- Gueydan C., Droogmans,L., Chalon,P., Huez,G., Caput,D. and Kruys,V. (1999) Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor α mRNA. J. Biol. Chem., 274, 2322–2326. [DOI] [PubMed] [Google Scholar]
- Han J. and Beutler,B. (1990) The essential role of the UA-rich sequence in endotoxin-induced cachectin/TNF synthesis. Eur. Cytokine Network, 1, 71–75. [PubMed] [Google Scholar]
- Han J. and Ulevitch,R.J. (1999) Emerging targets for anti-inflammatory therapy. Nature Cell Biol., 1, E39–40. [DOI] [PubMed] [Google Scholar]
- Han J., Brown,T. and Beutler,B. (1990) Endotoxin-responsive sequences control cachectin/tumor necrosis factor biosynthesis at the translational level. J. Exp. Med., 171, 465–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hel Z., Skamene,E. and Radzioch,D. (1996) Two distinct regions in the 3′ untranslated region of tumor necrosis factor α mRNA form complexes with macrophage proteins. Mol. Cell. Biol., 16, 5579–5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami A., Tian,Q., Duan,X., Streuli,M., Schlossman,S.F. and Anderson,P. (1992) Identification and functional characterization of a TIA-1-related nucleolysin. Proc. Natl Acad. Sci. USA, 89, 8681–8685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami A., Tian,Q., Streuli,M., Poe,M., Edelhoff,S., Disteche,C.M. and Anderson,P. (1994) Intron–exon organization and chromosomal localization of the human TIA-1 gene. J. Immunol., 152, 4937–4945. [PubMed] [Google Scholar]
- Kedersha N.L., Gupta,M., Li,W., Miller,I. and Anderson,P. (1999) RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J. Cell Biol., 147, 1431–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keffer J., Probert,L., Cazlaris,H., Georgopoulos,S., Kaslaris,E., Kioussis,D. and Kollias,G. (1991) Transgenic mice expressing human tumour necrosis factor: a predictive genetic model of arthritis. EMBO J., 10, 4025–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollias G., Douni,E., Kassiotis,G. and Kontoyiannis,D. (1999) The function of TNF and receptors in models of multi-organ inflammation, rheumatoid arthritis, multiple scleroisis and inflammatory bowel disease. Ann. Rheum. Dis., 58, I32–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontoyiannis D., Pasparakis,M., Pizarro,T.T., Cominelli,F. and Kollias,G. (1999) Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint- and gut-associated immunopathologies. Immunity, 10, 387–398. [DOI] [PubMed] [Google Scholar]
- Kotlyarov A., Neininger,A., Schubert,C., Eckert,R., Birchmeier,C., Volk,H.D. and Gaestel,M. (1999) MAPKAP kinase 2 is essential for LPS-induced TNF-α biosynthesis. Nature Cell Biol., 1, 94–97. [DOI] [PubMed] [Google Scholar]
- Krecic A. and Swanson,M. (1999) hnRNP complexes: composition, structure and function. Curr. Opin. Cell Biol., 11, 363–371. [DOI] [PubMed] [Google Scholar]
- Kruys V., Marinz,O., Shaw,G., Deschamps,J. and Huez,G. (1989) Translational blockade imposed by cytokine-derived UA-rich sequences. Science, 245, 852–855. [DOI] [PubMed] [Google Scholar]
- Lai W.S., Carballo,E., Strum,J.R., Kennington,E.A., Phillips,R.S. and Blackshear,P.J. (1999) Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor α mRNA. Mol. Cell. Biol., 19, 4311–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroia G., Cuesta,R., Brewer,G. and Schneider,R.J. (1999) Control of mRNA decay by heat shock–ubiquitin–proteasome pathway. Science, 284, 499–502. [DOI] [PubMed] [Google Scholar]
- Lee J.C. and Young,P.R. (1996) Role of CSBP/p38/RK stress response kinase in LPS and cytokine signaling mechanisms. J. Leukoc. Biol., 59, 152–157. [DOI] [PubMed] [Google Scholar]
- Lee J.C. et al. (1994) A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature, 372, 739–746. [DOI] [PubMed] [Google Scholar]
- Levine T.D., Gao,F., King,P.H., Andrews,L.G. and Keene,J.D. (1993) Hel-N1: an autoimmune RNA-binding protein with specificity for 3′ uridylate-rich untranslated regions of growth factor mRNAs. Mol. Cell. Biol., 13, 3494–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis T., Gueydan,C., Huez,G., Toulme,J.J. and Kruys,V. (1998) Mapping of a minimal AU-rich sequence required for lipopoly saccharide-induced binding of a 55-kDa protein on tumor necrosis factor-α mRNA. J. Biol. Chem., 273, 13781–13786. [DOI] [PubMed] [Google Scholar]
- Loflin P., Chen,C.Y.A. and Shyu,A.B. (1999) Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev., 13, 1884–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W.J., Cheng,S., Campbell,C., Wright,A. and Furneaux,H. (1996) Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem., 271, 8144–8151. [DOI] [PubMed] [Google Scholar]
- Myer V.E., Fan,X.C. and Steitz,J.A. (1997) Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J., 16, 2130–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostareck D.H., Ostareck-Lederer,A., Wilm,M., Thiele,B.J., Mann,M. and Hentze,M.W. (1997) mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell, 89, 597–606. [DOI] [PubMed] [Google Scholar]
- Ostareck-Lederer A., Ostareck,D.H., Standart,N. and Thiele,B.J. (1994) Translation of 15-lipoxygenase mRNA is inhibited by a protein that binds to a repeated sequence in the 3′ untranslated region. EMBO J., 13, 1476–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S.S.Y., Chen,C.Y.A., Xu,N. and Shyu,A.B. (1998) RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J., 17, 3461–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prichett W., Hand,A., Sheilds,J. and Dunnington,D. (1995) Mechanism of action of bicyclic imidazoles defines a translational regulatory pathway for tumor necrosis factor α. J. Inflamm., 45, 97–105. [PubMed] [Google Scholar]
- Raabe T., Bukrinsky,M. and Currie,R.A. (1998) Relative contribution of transcription and translation to the induction of tumor necrosis factor-α by lipopolysaccharide. J. Biol. Chem., 273, 974–980. [DOI] [PubMed] [Google Scholar]
- Sakai K., Kitagawa,Y. and Hirose,G. (1999) Binding of neuronal ELAV-like proteins to the uridine-rich sequence in the 3′-untranslated region of tumor necrosis factor-α messenger RNA. FEBS Lett., 446, 157–162. [DOI] [PubMed] [Google Scholar]
- Salituro F., Germann,U., Wilson,K., Bemis,G., Fox,T. and Su,M. (1999) Inhibitors of p38 MAP kinase: therapeutic intervention in cytokine-mediated diseases. Curr. Med. Chem., 6, 807–823. [PubMed] [Google Scholar]
- Shaw G. and Kamen,R. (1986) A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell, 46, 659–667. [DOI] [PubMed] [Google Scholar]
- Sokolowski M., Furneaux,H. and Schwartz,S. (1999) The inhibitory activity of the AU-rich RNA element in the human papillomavirus type 1 late 3′ untranslated region correlates with its affinity for the Elav-like HuR protein. J. Virol., 73, 1080–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S., Kumar,K. and Kaufman,R. (1998) Phosphorylation of eIF2α mediates apoptosis in response to activation of the double stranded RNA-dependent protein kinase. J. Biol. Chem., 273, 2416–2423. [DOI] [PubMed] [Google Scholar]
- Taylor G.A. et al. (1996) A pathogenetic role for TNFα in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity, 4, 445–454. [DOI] [PubMed] [Google Scholar]
- Tian Q., Streuli,M., Saito,H., Schlossman,S.F. and Anderson,P. (1991) A polyadenylate binding protein localized to the granules of cytolytic lymphocytes induces DNA fragmentation in target cells. Cell, 67, 629–639. [DOI] [PubMed] [Google Scholar]
- Vakalopoulou E., Schaack,J. and Shenk,T. (1991) A 32-kilodalton protein binds to AU-rich domains in the 3′ untranslated regions of rapidly degraded mRNAs. Mol. Cell. Biol., 11, 3355–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E., Ma,W.J., Aghajanian,C. and Spriggs,D.R. (1997) Post transcriptional regulation of protein expression in human epithelial carcinoma cells by adenine-uridine-rich elements in the 3′-untranslated region of tumor necrosis factor-α messenger RNA. Cancer Res., 57, 5426–5433. [PubMed] [Google Scholar]
- Wilson T. and Treisman,R. (1988) Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3′ AU-rich sequences. Nature, 336, 396–399. [DOI] [PubMed] [Google Scholar]
- Winzen R. et al. (1999) The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J., 18, 4969–4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Chen,C.Y.A. and Shyu,A.B. (1997) Modulation of the fate of cytoplasmic mRNA by AU-rich elements: key sequence features controlling mRNA deadenylation and decay. Mol. Cell. Biol., 17, 4611–4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Wagner,B.J., Ehrenman,K., Schaefer,A.W., De Maria,C.T., Crater,D., De Haven,D., Long,L. and Brewer,G. (1993) Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell. Biol., 13, 7652–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]