Abstract
The eukaryotic DNA recombination repair protein BRCA2 is functional in the parasitic protozoan Trypanosoma brucei. The mechanism of the involvement of BRCA2 in homologous recombination includes its interaction with the DNA recombinase proteins of the RAD51 family. BRCA2 is known to interact with RAD51 through its unique and essential BRC sequence motifs. T. brucei BRCA2 homolog (TbBRCA2) has fifteen repeating BRC motifs as compared to mammalian BRCA2 that has only eight. We report here our yeast 2-hybrid analysis studies on the interactions of TbBRCA2 BRC motifs with five different RAD51 paralogues of T. brucei. Our study revealed that a single BRC motif is sufficient to bind to these RAD51 paralogues. To test the possibility whether a single 44 amino acid long repeating unit of the TbBRCA2 BRC motif may be exploited as an inhibitor of T. brucei growth, we ectopically expressed this peptide segment in the procyclic form of the parasite and evaluated its effects on cell survival as well as the sensitivity of these cells to the DNA damaging agent methyl methane sulfonate (MMS). Expression of a single BRC motif led to MMS sensitivity and inhibited cellular proliferation in T. brucei.
Keywords: Trypanosoma brucei, BRCA2, RAD51, homologous recombination, DNA repair, Methyl methane sulfonate, yeast 2-hybrid analysis, aptamer
1. Introduction
Homologous recombination (HR) of DNA is an essential component of any form of life [1-3]. It is particularly important to the parasitic protozoan Trypanosoma brucei because through HR this extracellular parasite can switch its cell surface antigen coat and survives from humoral immunity of its mammalian hosts [4-11]. Parasites of the T. brucei group, often known as African trypanosomes, can successfully proliferate in the mammalian bloodstream due to a sophisticated strategy of antigenic variation of a homogeneous Variant Surface Glycoprotein (VSG) coat [7-11]. Once an infected mammalian host mounts the appropriate antibody response, it can effectively clear a given VSG variant via antibody mediated lysis [11]. However, VSG switch variants are continuously being generated causing temporary escape from immuno-destruction [4-7]. Individual trypanosomes have many hundreds of VSG genes and thus cyclical waves of parasitemia make up a chronic infection which can persist for years [4-11]. One of the major pathways that T. brucei accomplish these antigenic variations is through gene conversion which uses the HR machineries of the cell [11, 12].
Another potential involvement of HR in trypanosome digenetic life cycle is the stress management phase when the parasite switches hosts from the insect vector to mammal and vice versa during transmission [13-15]. The heat and oxygen level shocks experienced by the parasite cells during these transmission stages may induce DNA damage and understandably the parasite must have fully functional HR mechanisms to cope with such stresses to survive [13-15].
One of the critical proteins in the HR pathway in many eukaryotes is the multifunctional scaffolding protein BRCA2 [1-3]. BRCA2 is widely expressed in eukaryotes with the exception of yeast and few other cells. It encodes a large nuclear protein localized to the nucleus of S-phase cells. BRCA2 has been implicated in processes fundamental to all cells including RAD51-mediated recombination repair [16, 17]. Mouse and human BRCA2-deficient cells accumulate spontaneous chromosomal aberrations during cell division in culture, implicating BRCA2 in the maintenance of genome stability [16]. BRCA2-deficient cells are hypersensitive to genotoxic agents that have the potential to cause DNA double-strand breaks (DSBs), implicating BRCA2 in cell cycle signaling and/or DSB repair [16, 17].
Mitotic cells can repair DNA DSBs by two major recombination mechanisms, nonhomologous end joining (NHEJ) and homologous recombination [18]. In NHEJ, DNA ends are joined with little or no base pairing at the joining site and the end-joining product can suffer insertion or deletion mutations [19]. In contrast, DSB repair by homologous recombination requires the presence of an intact DNA duplex with extensive homology to the region flanking the break to serve as a repair template. The preferred template for homologous recombination repair is the sister chromatid [20]. A key step in DSB repair by homologous recombination is the invasion of a 3′ single-strand DNA (ssDNA) end into the intact template. RAD51 protein carries out this reaction. RAD51 functions as a polymer, made up of hundreds of monomers that coat ssDNA and form a nucleoprotein filament that catalyzes the strand invasion reaction, which is followed by new DNA synthesis [21]. The resulting intermediate can either disassemble (i.e., the newly synthesized strand can be displaced and anneal with the non-invading 3′-ssDNA end to elicit non-crossover gene conversion only) or be processed to a Holliday junction intermediate to yield gene conversion with or without crossover [22, 23]. Homologous recombination is considered to be error free when it involves sister chromatids [20], but it can also be deleterious when it takes place between repetitive sequences, and in excess, it can promote genome instability and cause diseases [24-26]. The first evidence linking BRCA2 to homologous recombination was its direct interaction with RAD51. The interaction is mediated by six of eight internal BRC repeats (BRC1-BRC4, BRC7, and BRC8) that are encoded by BRCA2 exon 11 and are highly conserved among mammals [21]. BRCA2 and RAD51 colocalize to subnuclear foci following DNA damage and during the S and G2 phases of the cell cycle [27]. Structural, cell biological, and biochemical evidence indicates that BRCA2 regulates RAD51 assembly at sites of DNA damage [28]. Direct evidence of a role for BRCA2 in assisting RAD51-mediated chromosomal repair was provided by the demonstration that with a chromosomal DSB in direct repeats, gene conversion was decreased by >100-fold in the BRCA2-deficient human cancer cell line CAPAN-1 and 4-to 6-fold in BRCA2-deficient mouse cells compared with wild-type cells [29, 30]. The 4-fold decrease in gene conversion was accompanied by a 2- to 3-fold increase in deletion events, suggesting that DSB repair by error-prone mechanisms predominates in BRCA2-deficient cells [30]. It also indicates that chromosomal instability provoked by BRCA2 deficiency is the result of incorrect routing of DSB processing down error-prone pathways because error-free processing by homologous recombination is unavailable [30, 31]. Unlike its mammalian hosts, T. brucei apparently lacks any NHEJ mechanisms [32, 33]. Thus, any DNA damage events requiring double-stranded DNA break repair should be lethal to the cells if BRCA2 is impaired in these cells.
Homologous recombination is largely driven by RAD51 [2]. T. brucei was shown to encode six RAD51-related proteins (TbRAD51.1, TbRAD51.2, TbRAD51.3, TbRAD51.4, TbRAD51.5 and TbRAD51.6) [34]. Two of these RAD51-related proteins, TbRAD51.3 and TbRAD51.5 were shown to contribute to DNA repair, homologous recombination and RAD51 function in the cell [34]. Surprisingly, however, only TbRAD51.3 contributes to VSG switching [34]. TbRAD51.2, a homolog of mammalian meiosis-specific RAD51 DMC1 [35], also has been studied in detail in T. brucei and was found not to act in DNA recombination, repair or antigenic variation in bloodstream stage cells [36].
T. brucei BRCA2 (TbBRCA2) is essential for HR and VSG switching and structurally distinct from mammalian BRCA2 [37]. Thus, it could be a target for pharmacological intervention. To explore this possibility, we studied the molecular biology of TbBRCA2 BRC repeats and their interactions with different major RAD51 paralogues of the parasite. Here we report that a single BRC motif is sufficient to bind to these proteins. We show evidence that a single 44 amino acid long repeat unit of the TbBRCA2 BRC motif may be exploited as an inhibitor of T. brucei growth as the ectopically expressed peptide segment shunts the proliferation of the parasite cells in vitro and increased the sensitivity of these cells to the DNA damaging agent methyl methane sulfonate (MMS).
2. Materials and methods
2.1. Trypanosome strains, media, and transfection
The procyclic form of T. brucei 427 double resistant cell line (29-13) expressing the tetracycline repressor gene (TetR) and T7RNA polymerase (T7RNAP) were grown in SDM-79 medium (JRH Biosciences, Lenexa, KS) containing 10% heat-inactivated fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA) and appropriate antibiotics (hygromycin: 50 μg/ml; G418: 15 μg/ml) at 27 °C [38, 39]. Bloodstream form cells were maintained in HMI-9 medium with G418 (2.5 μg/ml) at 37 °C. Procyclic cells were transfected by electroporating 3 × 107 cells with 20 μg of Not I linearized plasmid DNA construct that had been phenol/chloroform treated and ethanol precipitated. Electroporation was performed in 0.5 ml of Zimmerman Post-Fusion medium (132 mM NaCl, 8 mM Na2HPO4, 1.5 mM KH2PO4, 0.5 mM magnesium acetate, 0.09 mM calcium acetate at pH 7.0), with a BTX Electro Cell Manipulator 600 set at 1.6 kV and 25 μF capacitance [35, 36]. Cells were allowed to recover for 24 h before transfectants were selected for with 2.5 μg/ml phleomycin. Expression of constructs was induced with 1 μg/ml doxycycline.
2.2. RNA isolation, DNase treatment, and real-time RT-PCR
Total RNA was isolated from the bloodstream and procyclic trypanosomes using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. All primers used for amplification and real-time PCR are listed in Table 1. Total RNA was DNase I treated using RQ1 (Promega, Madison, WI) following the protocol provided by the supplier. cDNA was synthesized from total RNA using the iScript cDNA synthesis Kit (Bio-Rad Laboratories, Hercules, CA). Real-time PCR was done using the IQ SYBR Green Supermix Kit (Bio-Rad Laboratories). Beta-actin was used as the endogenous control for normalization of the real-time PCR data.
Table 1. Oligonucleotides used in this study.
| No | Primer name | Nucleotide sequence (5′-3′) | Purpose |
|---|---|---|---|
| 1 | RAD51.1RTF | GATATCAAAAAGTTGATG | qRT-PCR for RAD51.1 |
| 2 | RAD51.1RTR | CCAACTTCAATGCCACCTCC | |
| 3 | RAD51.2RTF | GGGTTTGTCTGATGCC | qRT-PCR for RAD51.2 |
| 4 | RAD51.2RTR | GCCTCTGTGTCAACATAAATAGC | |
| 5 | RAD51.3RTF | GAGGCGATTGAGACGGCTCGAGTC | qRT-PCR for RAD51.3 |
| 6 | RAD51.3RTR | CGTTGTAGTACGATTTCCCTC | |
| 7 | RAD51.4RTF | GGGAACGGCACGCGTCTTG | qRT-PCR for RAD51.4 |
| 8 | RAD51.4RTR | GGATGCTGATTGCTGCTGATCC | |
| 9 | RAD51.5RTF | CCCGCCTTGTGCGCCGCATGATATC | qRT-PCR for RAD51.5 |
| 10 | RAD51.5RTR | CGACGGATACTCCTCGTGGTACAG | |
| 11 | RAD51.6RTF | CGTTCTGTCGAGAATACACATCG | qRT-PCR for RAD51.6 |
| 12 | RAD51.6RTR | GTCCAGCGCAAAACCCGCCTTC | |
| 13 | BRCA2RTF | CGCGTGCTTCCGCCCTTTTTCCTTG | qRT-PCR for TbBRCA2 |
| 14 | BRCA2RTR | GGGGTGACCAGGTCGGAAGCGTG | |
| 15 | TbRAD51.1F | ATGAACACTCGCACCAAAAATAAGAAACGC | ORF PCR for RAD51.1 |
| 16 | TbRAD51.1R | CTAGTCCCTAACGTCTCCCACACCG | |
| 17 | TbRAD51.2F | ATGCAGCACGTGGGAACGCG | ORF PCR for RAD51.2 |
| 18 | TbRAD51.2R | CTACTCACGTGCGTCAAC | |
| 19 | TbRAD51.3F | ATGTCCGTGGAGCAATGCTCC | ORF PCR for RAD51.3 |
| 20 | TbRAD51.3R | TCAAAGAGTGGGTCGGAAAACATCGCG | |
| 21 | TbRAD51.4F | ATGGATGCACTCTGGCCAGTG | ORF PCR for RAD51.4 |
| 22 | TbRAD51.4R | CTAGCAGTGAACAAATGTTGG | |
| 23 | TbRAD51.5F | ATGACGGCGGAGGAGATTTCACAG | ORF PCR for RAD51.5 |
| 24 | TbRAD51.5R | TCAGGGTAAAAAGATGTTTCC | |
| 25 | TbBRC2HF1 | GACCATATGCGTACGAAGTGCCGCTCCGATG | PCR for BRC repeats 1-15 |
| 26 | TbBRC2HR1 | GACCTCGAGCCCTACTCCTTTCGCAACCGCTC | |
| 27 | TbBRCVARYF | GACCATATGACTGAAAATGGC | BRC repeats variants PCR |
| 28 | TbBRCVARYR | GACCTCGAGATTCATTCGTGC | |
| 29 | TbBRCFR1/2LEWF | GACAAGCTTCGTACGAAGTGCCGCTCCG | PCR of single BRC repeat |
| 30 | TbBRCFR1/2LEWR | GACTCTAGACCCTACTCCTTTCGCAACCGC | |
| 31 | pLEW79TAPR | AAGCGGTTGGCTGCTGAGAC | TAP tagged repeat (RT-PCR) |
2.3. Southern blot analysis
Southern blot analysis was performed to determine the gene copy number for BRCA2 in T. brucei. Genomic DNA (5-10 μg) was digested with different restriction endonucleases. The restriction endonucleases chosen do not have a site in the DNA used as probe. The digests were resolved on a 0.8% agarose gel and transferred to nylon membrane in denaturing buffer (Schleicher and Schuell, Keene, New Hampshire) following standard protocols. The blots were probed with a 32P-labeled BRCA2 ORF fragment (+3705 to +4305 of TbBRCA2 ORF with the first nucleotide of the ORF as +1) amplified from genomic DNA with the primer set 5′-CACCATGGCCATCGATTTTGCTG GCTTGTTCG-3′ and 5′-CTATACCTGTTCTTCTTCACTGCTTAAGG-3′ following standard protocols.
2.4. DNA cloning and sequence analysis
All primers used for amplification are listed in Table 1. For the evaluation of protein-protein interactions by yeast two hybrid analysis, the open reading frame (ORF) of TbRAD51.1 (XP_828893), TbRAD51.2 (XP_827266), TbRAD51.3 (XP_828338), TbRAD51.4 (XP_828775), and TbRAD51.5 (EAN78580) and the BRC repeat region of TbBRCA2 (XM_001218766; TbBRC, 2271bp) were PCR amplified from T. brucei genomic DNA using Pfu DNA polymerase (Stratagene, LaJolla, CA). The PCR products were purified and cloned into pCR4-TOPO (Invitrogen) and then sequence verified. Purified plasmid DNAs with RAD51 ORFs were digested with EcoR I to isolate the insert, purified and cloned into the EcoR I site of pGBKT7 (BD Clontech, Mountain View, CA). Purified plasmid with the TbBRCA2 BRC repeat region (TbBRC) was digested with Nde I and Xho I to isolate the insert, purified and cloned into pGADT7 (BD Clontech). The inserts of these constructs were again sequenced to verify that the ORF's were in frame with their respective fusion partners.
To construct plasmids with varying lengths of BRC repeats, deletion mutagenesis was performed. Varying lengths of BRC repeats were PCR amplified from the pGADTbBRC construct using Pfu DNA polymerase. The primers TbBRCVARYF and TbBRCVARYR (Table 1) were used to generate one repeat (R1), two repeats (R2) and so on. Primers TbBRC2HF1 and VARYR (Table 1) were used to generate a single repeat with a flank at the N-terminus of the peptide with putative nuclear localization sequences (NLS), (FLR1). Primers VARYF, TbBRC2HR1 (Table 1) were used to amplify the 14th and 15th repeats with a flank containing NLS3 (14/15FL) and 15FL respectively. Prior to cloning into the pGADT7 vector the PCR products were purified and cloned into pCR4-TOPO and sequence verified, as before. Purified plasmid DNA constructs with varying lengths of BRC repeats were digested with Nde I and Xho I, purified, and cloned into pGADT7. The inserts of these constructs were sequence verified to confirm that the ORF's were in frame with their respective fusion partners.
To construct a plasmid to over-express a single BRC motif the DNA sequence encoding a single BRC repeat flanked by the putative NLSs (see Supplemental Fig. 1S) was amplified from the BRC repeat region of TbBRCA2 (TbBRC) using the primers TbBRCFR1/2LEWF and TbBRCFR1/2LEWR (Table 1). The primer sequence included Hind III (forward) and Xba I (reverse) restriction sites. The PCR product was purified and cloned into pLEW79MHTAP previously digested with Hind III and Xba I and purified. The resulting plasmid (pLEW79BRCTAP) was linearized by Not I and tranfected into cells as previously described [38, 39].
2.5. Reverse transcription (RT)-PCR
Reverse transcription (RT)-PCR was performed on total RNA isolated from procyclic and bloodstream trypanosomes. RT-PCR was performed using a Qiagen (Valencia, CA) OneStep RT-PCR kit following the protocol outlined by the supplier. For each RT-PCR reaction, 4.5 μg of RQI (DNase I, Promega)-treated total RNA from trypanosomes was used. Primers, detailed in Table 1, were used for PCR amplifications.
2.6. Yeast two hybrid analysis
Yeast cells, Saccharomyces cerevisiae strain AH109 (BD Biosciences Clontech), were co-transfected with pair wise combinations of bait (pGBKT7) and prey vectors (pGADT7) with lithium acetate (Matchmaker System 3; BD Biosciences Clontech), as described in the manufacturer's protocol, and nutritionally selected. The yeast cells were co-transfected individually with pGADT7-TbBRC fusion construct and each pGBKT7-RAD51 homologue (pGBKT7RAD51.1-RAD51.5). Yeast cells were plated onto synthetic dropout medium lacking leucine, tryptophan, and histidine in the presence of 5-bromo-4-chloro-3-indolyl-ft-D-galactopyranoside (X-α-Gal; Clontech) to select for yeast containing weaker interacting proteins. Yeast cells were also plated onto synthetic dropout medium lacking leucine, tryptophan, histidine, and adenine in the presence of 5-bromo-4-chloro-3-indolyl-ft-D-galactopyranoside (X-α-Gal; Clontech) to select for yeast containing stronger interacting proteins. The positive control (supplied with the reagent kit) used was SV40 T-antigen and p53, known to interact very strongly. The negative controls used were cells co-transfected with empty pGADT7/pGBKT7 vector, untransformed AH109 cells and singly transformed (e.g., with Gal4BD-RAD51.1 and other variants) yeast cells. We did not find any evidence for auto-activation when the negative controls were used (see results).
2.7. Western blot analysis
Western blot analysis was performed using protein extracts made from procyclic trypanosomes. Cell lysates containing equal amounts of protein were resolved by 4–12% SDS–PAGE, transferred to PVDF membrane and probed with a secondary rabbit IgG (anti-goat) antibody conjugated to HRP (1:1000) and detected by enhanced chemiluminescent technology (GE Healthcare, Brentwood, TN). Western blot analysis was performed using protein extracts made from yeast cells co-transfected with yeast two hybrid constructs expressing the BRC repeat region of TbBRCA2 (pGADT7TbBRC 1-15) and individual RAD51 homologues (pGBKT7RAD51.1-RAD51.5). Cell lysates containing equal amounts of protein were resolved by 4-12% SDS–PAGE, transferred to PVDF membrane and probed with primary mouse monoclonal antibodies, anti-HA (1:200 for HA-tagged TbBRC) and anti-Myc (1:1000 for Myc-tagged RAD51.1-RAD51.5). A secondary anti-mouse antibody conjugated to HRP (1:3000) was used and detected by enhanced chemiluminescent technology (GE Healthcare). TAP expression at the protein level was determined by Western blot analysis using a secondary rabbit IgG (anti-goat) antibody conjugated to HRP (1:1000) procured from Santa Cruz Biotechnology (Santa Cruz, CA). Antibody against Tet repressor protein was purchased from Clontech Laboratories (Mountain View, CA) and used in a 1:1000 dilution. Antibody against α-tubulin of T. brucei was obtained from Prof. K. Gull of University of Oxford and was used in a dilution of 1:10,000.
2.8. α-Galactosidase assay
α-Galactosidase assays were done using X-α-Gal (BD Clontech) as a substrate to detect α-Galactosidase activity. X-α-Gal was dissolved at 20 mg/ml in dimethylformamide (DMF), and then diluted further to 4 mg/ml in DMF. The appropriate dropout plates were made at room temperature; 200μl of X-α-Gal was spread onto the plate and allowed to dry for 15 min at room temperature. The transfected yeast cells were then streaked and incubated at 30 °C until blue colonies formed (∼3 days).
2.9. β-Galactosidase assay
β-Galactosidase assays were done using the Beta Glo Assay reagents and protocols (Promega) to detect β-Galactosidase activity. Yeast cells as well as the Beta-Glo reagent were brought to room temperature. Equal volumes of the reagent and the yeast cell culture were added together. The sample contents were mixed for 30 seconds. Samples were then incubated for 30 min at room temperature and the luciferase activity was measured using a luminometer.
2.10. DNA damage sensitivity assay
Sensitivities of the procyclic trypanosomes to methyl methane sulfonate (MMS) were assayed. Cells were grown to a maximum density of 1×106 cells/ml, and plated in 24 well plates with increasing concentrations of MMS, plus a no drug control. For each cell line, at each drug concentration, six repetitions were performed. Cells were counted by hemocytometer.
3. Results and discussion
3.1. T. brucei BRCA2 has several distinct features in comparison to its mammalian homolog
Although BRCA2 was originally discovered as breast cancer susceptibility gene #2 in humans [16, 17], later on it was found to be a ubiquitous protein in many eukaryotic organisms [37, 40-44]. A few years back we discovered this gene in the Kinetoplastid protozoan Leishmania [45]. Recently, a BRCA2 homolog was characterized in T. brucei [37]. We have retrieved the nucleotide sequences of the BRCA2 homolog of T. brucei from the GeneDB (http://www.genedb.org/genedb/tryp/, systematic name: Tb927.1.640), designed primers and amplified the cDNA for this gene. The gene is located at the chromosome 1 and the 4947 bp ORF codes for a protein of 1648 amino acids (see Supplementary Tables 1S and 2S).
The BRC-repeat region in human BRCA2 is followed by an ∼1000-residue COOH-terminal region that corresponds to the best-conserved portion of BRCA2 across dog, mouse, rat, and chicken [46] orthologs [68% average identity for this region, compared with 42% for the entire protein], as well as in putative orthologs found in Arabidopsis and rice [47]. The COOH-terminal region also contains 27% of the tumor-derived missense mutations in the breast cancer information core (BIC) database [48]; this indicates that it has an important role in the tumor-suppressor function of BRCA2. This region binds the 70–amino acid DSS1 (deleted in split-hand/split foot syndrome) protein [49], which was originally identified as one of three genes that map to a 1.5-Mb locus deleted in an inherited developmental malformation syndrome [50]. DSS1 ortholog in the trypanosomatids is not yet identified. The human BRC repeats are degenerate but those of TbBRCA2 are identical (except the TbBRC15) [37] (see also Supplementary Fig. 1S).
3.2. TbBRCA2 and all six isoforms of TbRAD51 are expressed in the procyclic and the bloodstream forms of T. brucei
While there is only one isoform of TbBRCA2 so far identified through genome sequencing, six different isoforms (paralogues) of TbRAD51 are reported in T. brucei [34]. Figs. 1A and 1B show the expression of the TbBRCA2 gene in the procyclic and the bloodstream forms. This gene is expressed in both of these forms at a significant level as was revealed by end-point PCR (Fig. 1A) as well as real-time RT-PCR (Fig. 1B). While protein level data are not available due to lack of specific antibody against TbBRCA2, expression of TbBRCA2 in these cells may suggest importance of this protein in the life cycle of this parasite. We also found similar levels of different TbRAD51 isoforms in the procyclic and bloodstream forms of T. brucei (Fig. 2). Although the functions of each of the isoforms of TbRAD51 in the biology of T. brucei life cycle are not known, the expressions of all these six isoforms in relatively high levels in both the procyclics and bloodstream forms suggest potential roles of these proteins in these parasites. Due to the lack of available antibodies against the RAD51 paralogues, we could not evaluate the levels of the corresponding proteins in these cells.
Fig. 1. Expression of TbBRCA2 in T. brucei procyclics and bloodstream forms.
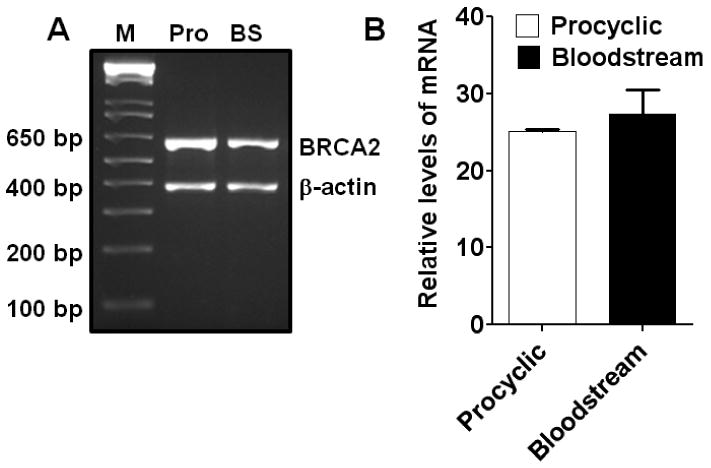
(A) Photograph of ethidium bromide-stained agarose gel showing the products of end-point RT-PCR with RNAs isolated from the procyclics (Pro) and bloodstream (BS) forms of T. brucei for TbBRCA2 mRNA. (B) Real-time RT-PCR data showing the relative levels of TbBRCA2 mRNAs present in the procyclic and bloodstream forms of T. brucei. β-Actin mRNA was used as a normalization control for both the experiments in A and B. Results are mean ± SEM (n=3).
Fig. 2. Expressions of different isoforms of RAD51 in T. brucei procyclics and bloodstream forms.
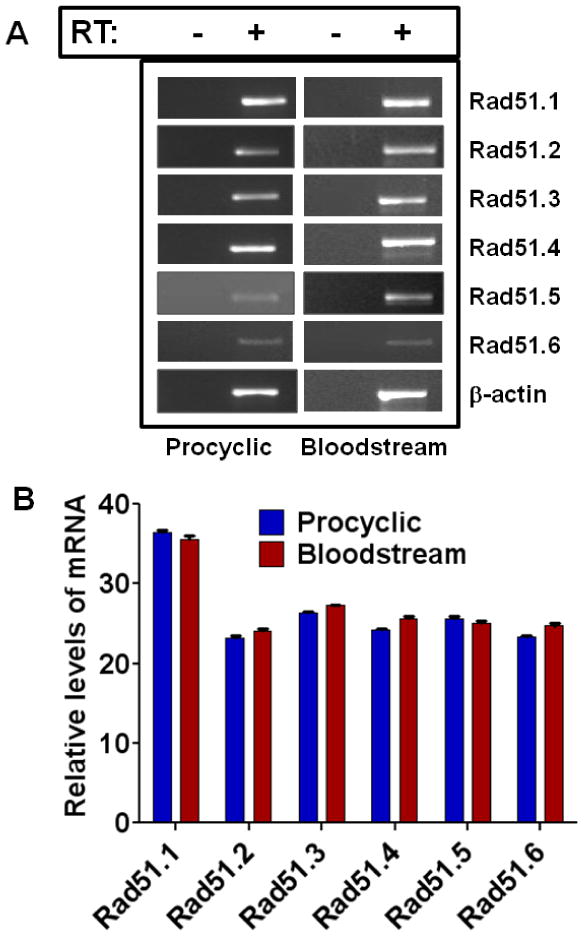
(A) Photographs of ethidium bromide-stained agarose gels showing the products of end-point RT-PCR with RNAs isolated from the procyclics and bloodstream forms of T. brucei for different isoforms of RAD51. PCR performed with RNAs without prior treatment with reverse transcriptase did not yield any product, showing the lack of genomic DNA contaminations of the RNA preparations. RT: reverse transcriptase. (B) Real-time RT-PCR data showing the relative levels of mRNAs of different isoforms of TbRAD51 recombinases present in the procyclics and bloodstream forms of T. brucei. β-Actin mRNA was used as a normalization control for both the experiments in A and B. Results are mean ± SEM (n=3).
3.3. Different RAD51 isoforms of T. brucei differentially interact with the BRC repeats of TbBRCA2
As mentioned above, BRCA2 interacts with RAD51 through its BRC repeats [16]. We evaluated whether TbBRCA2 BRC repeats can physically interact with all the TbRAD51 isoforms by yeast 2-hybrid analysis. TbBRCA2 has 15 BRC repeats. We amplified the DNA segment containing all 15 BRC repeats along with a 42 amino acid flank at the N-terminus of the repeat #1 and a 55 amino acid flank at the C-terminus of repeat #15 (Fig. 3A). The flanks included contain all three putative nuclear localization signals (NLSs) of TbBRCA2 (see Supplemental Fig. 1S [37]). We cloned this fragment in the MCS of pGAD-T7 plasmid. When expressed in S. cerevisiae the mRNA and the hybrid protein (∼99 kDa) containing Gal4AD, HA epitope and the TbBRCA2 BRC repeats were expressed, as was verified by Western blot analysis using HA-antibody (Fig. 3B). We also amplified the full length ORFs of TbRAD51.1, -51.2, -51.3, -51.4, -51.5 and cloned them in the MCS of pGBK-T7 plasmid (Fig. 3A). The expressions of the hybrid c-myc-tagged proteins were also verified by Western blot analysis with myc-antibody (Fig. 3B). We do not know whether all the paralogues of TbRAD51 have recombinase activity. Since least amount of data is available for TbRAD51.6 [34], we did not include this isoform of TbRAD51 for BRC interaction in this study.
Fig. 3. Differential interactions between TbBRCA2 BRC-cluster and individual RAD51 protein in yeast 2-hybrid analysis.
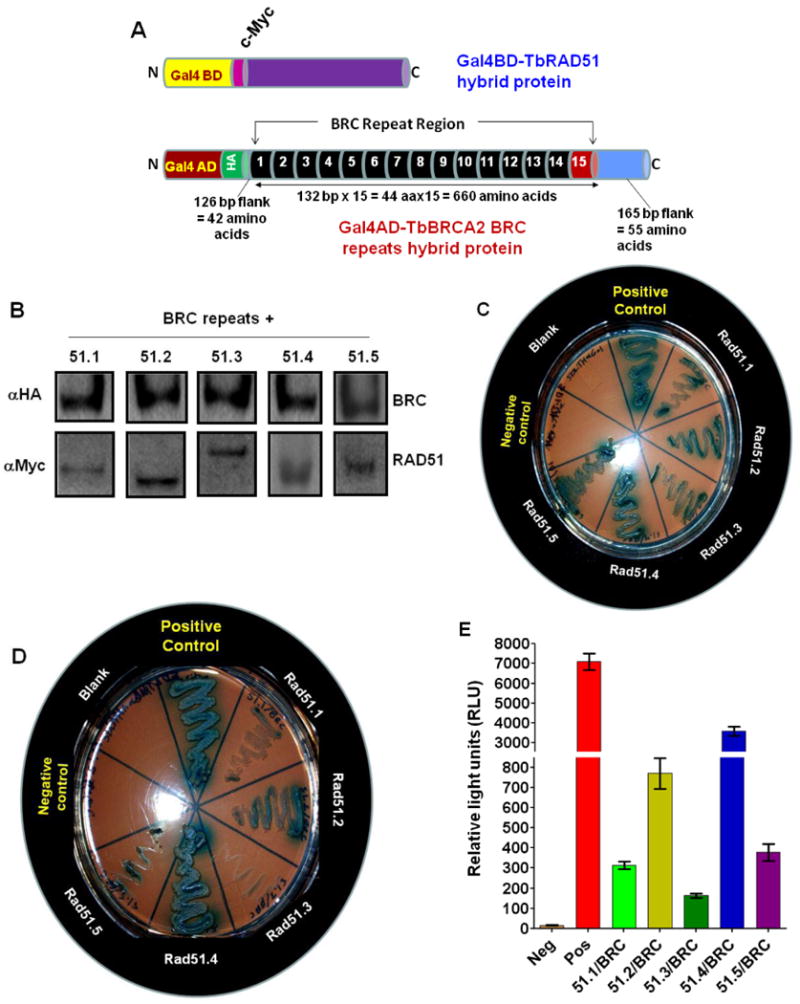
(A) Maps of different recombinant hybrid proteins generated with five TbRAD51 proteins and the BRC repeat region (TbBRC) clones tested in yeast two hybrid systems. TbRAD51s are expressed as fusion proteins with yeast GAL4 DNA binding domain (GAL4BD). The fusion proteins also have a c-Myc epitope in between the fusion partners. TbBRC repeats are expressed as a fusion protein with yeast GAL4 activation domain (GAL4AD). This fusion protein also has a HA-epitope in between the fusion partners. The sizes of the repeats and the flanks of the BRC repeat construct are shown. (B) Evaluation of the expressions of BRC repeat and specific TbRAD51 hybrid proteins in the transfected S. cerevisiae cells by Western blotting analysis. Hybrid GAL4AD-BRC repeat protein was detected with HA antibody whereas the hybrid GAL4BD-TbRAD51 proteins were detected with c-Myc antibody. The molecular sizes of the hybrid proteins are: GAL4AD-BRC: ∼99 kDa; GAL4BD-TbRAD51.1: ∼61 kDa; GAL4BD-TbRAD51.2: ∼58 kDa; GAL4BD-TbRAD51.3: ∼76 kDa; GAL4BD-TbRAD51.4: ∼66 kDa; and GAL4BD-TbRAD51.5: ∼61 kDa. (C) Evaluation of specific interactions between individual TbRAD51 and TbBRC repeats by yeast 2-hybrid analyses under moderate stringency in selective dropout medium: SD medium without leucine, tryptophan, and histidine plus X-α-gal. Controls: SV40 T-antigen/P53 (positive control); empty vectors pGBKT7/pGADT7 (negative control). Experimental yeast cells were co-transfected with pGADT7-TbBRC and one of the pGBKT7-TbRAD51 constructs (as indicated). (D) Verification of specific interactions between individual TbRAD51 and TbBRC repeats by yeast 2-hybrid analyses under higher stringency in selective dropout medium: SD medium without leucine, tryptophan, histidine, and adenine plus X-α-gal. The bait and prey protein pairs in different segments in the photograph are as in ‘C’. (E) Further verification of specific interactions between individual TbRAD51 and TbBRC repeats by Beta Glo Assay. Results are mean ± SEM (n=6).
We did not find any significant growth and beta-galactosidase activity of yeast cells transfected with individual Gal4BD-RAD51 constructs (negative controls) in yeast 2-hybrid analysis (Supplementary Fig. 2S). Yeast cells were transfected with pGBKT7-TbRAD51.1, pGBKT7-TbRAD51.2, pGBKT7-TbRAD51.3, pGBKT7-TbRAD51.4 and pGBKT7-TbRAD51.5. Since the pGBKT7 plasmid has TRP1 reporter gene, yeast cells transfected with these plasmid constructs were able to grow in the tryptophan-drop-out SD medium (Supplementary Fig. 2SA). When evaluated for growth in higher stringency selective dropout medium i.e., SD medium without leucine, tryptophan, histidine, and adenine, the yeast cells transfected with individual Gal4BD-RAD51 constructs failed to grow (Supplementary Fig. 2SB). We verified further the lack of reporter gene expression in yeast cells transfected with individual Gal4BD-RAD51 constructs by Beta Glo Assay (Supplementary Fig. 2SC). All these control data verified the validity of the yeast 2-hybrid system we used for this study.
We then evaluated the physical interactions between the BRC repeat clusters of TbBRCA2 with different isoforms of TbRAD51 under two conditions of stringency using the Matchmaker 3-reporter genes system (HIS3, ADE2 and LacZ; BD Clontech). Under low stringent conditions (adenine is present in the growth medium) weaker protein-protein interactions along with stronger interacting pairs are identified. We found that under low stringent conditions, all the five isoforms of TbRAD51 bind to TbBRCA2 BRC repeats (Fig. 3C). On the other hand, when stringency was increased by dropping out adenine from the growth medium, only TbRAD51.2 and TbRAD51.4 appeared to be binding to the TbBRCA2 BRC repeats tightly (Fig. 3D). We quantitated the relative strength of interaction between the proteins by evaluating the expression of β-galactosidase using β-Glo assays. This assay revealed that TbRAD51.4 has the highest affinity for the BRC-repeats of TbBRCA2 in comparison to the other isoforms tested (Fig. 3E). TbRAD51.2 (DMC1) also showed significant affinity of the BRC repeats (Fig. 3E). The C-terminal domain of human BRCA2 protein showed some stabilization effects on the binding of RAD51 to BRCA2 [35]. Since we do not have the C-terminal domain of BRCA2 in our construct, that may have affected the differential affinity of the binding observed in our study. Although TbRAD51.2 (DMC1) was found not to participate in DNA recombination, repair or antigenic variation in bloodstream form of T. brucei [39], its potential role in meiotic recombination [38] during the sexual life cycle of the parasite in its insect vector host [52-54] may be important. Thus, further understanding the interaction of TbRAD51.2 with BRC repeats may be critical to understand the sexual life cycle of T. brucei and related parasites [54, 55]. The significance, if any, of these differential affinities of TbBRCA2 BRC repeats for different TbRAD51 isoforms in the biology of T. brucei is currently not understood.
3.4. A single BRC motif is sufficient to bind to RAD51 of T. brucei
Since TbBRCA2 BRC repeats preferentially bind with TbRAD51.2 and TbRAD51.4, we focused on these two isoforms of TbRAD51 for further studies. The minimum number of BRC repeat motifs that are necessary to establish physical interactions with these TbRAD51 proteins was determined by yeast 2-hybrid analysis. We also determined whether the non-BRC flanks used in the large repeat construct have any contribution in the establishment of binding between BRC repeats and TbRAD51. We designed primers to amplify different multimers of the BRC repeats from the original 15-mer construct as a template (Fig. 4A). Each of the amplified BRC repeat multimers was gel purified and subcloned in the MCS of pGAD-T7 plasmid. Similarly, we amplified the repeat #1 with the 42 amino acid N-terminal flank, repeat #14 and #15 with the 55 amino acid C-terminal flank as well as the repeat #15 with C-terminal flank and cloned them in the MCS of pGAD-T7 vector (Fig. 4B). All these constructs tested were efficient to bind tightly with both TbRAD51.2 (Fig. 4) and TbRAD51.4 (Fig. 5) in the yeast 2-hybrid system analysis. They established tight interactions both at the low (Figs. 4C and 5A) as well as at the high (Figs. 4D and 5B) stringent conditions. Although only one BRC motif was sufficient for the establishment of tight binding with TbRAD51 isoforms tested (Figs. 4E and 5C), there was some decrease with some of the combination constructs. For example, the BRC repeat #15 with the C-terminal 55 amino acid flank was significantly less efficient in binding with TbRAD51.2 under stringent conditions (Fig. 4E) whereas the dimer was less efficient in binding with TbRAD51.4 (Fig. 5C). The mechanisms of these differential efficiencies are not known. The conclusions from these studies were (i) a single BRC motif of 44 amino acids is sufficient for establishing physical interactions with specific paralogues of TbRAD51 tested; (ii) the conserved BRC repeats are little more efficient in the establishment of the binding than the non-conserved repeat #15; and, (iii) the non-BRC flanks do not have any positive contribution in the establishment of such binding.
Fig. 4. Determination of minimum numbers of BRC motifs required for its interaction with TbRAD51.2.
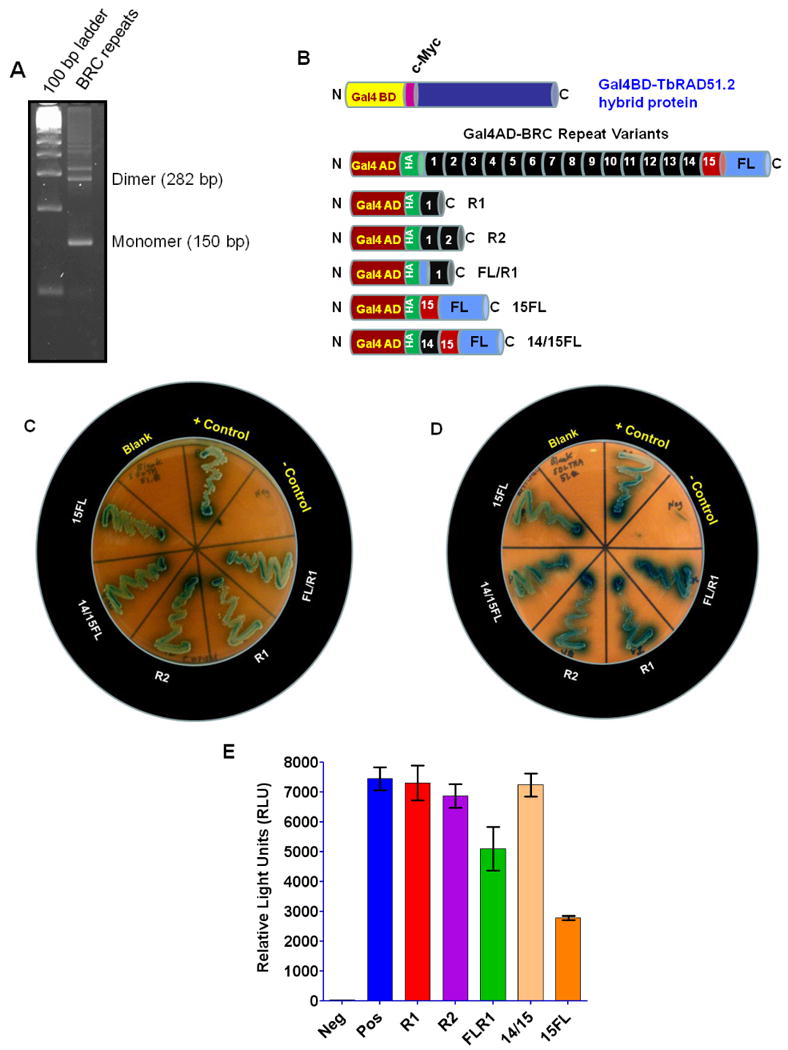
(A) Photograph of ethidium bromide stained agarose gel (4%) showing the products of PCR amplifications of the BRC repeat ladder. 100 bp DNA ladder was used as molecular size standards. The primers have flanking restriction endonucelase sites hence adding to the actual sizes of the monomer, dimer and so on. (B) Maps of hybrid proteins with TbRAD51.2 and the different BRC repeat variants used for yeast two hybrid analyses. TbRAD51.2 is expressed as a fusion protein with yeast GAL4 DNA binding domain (GAL4BD). TbBRC repeat variants are expressed as a fusion protein with yeast GAL4 activation domain (GAL4AD). (C) Evaluation of specific interactions between TbRAD51.2 and individual TbBRC repeat variants by yeast 2-hybrid analyses under moderate stringency in selective dropout medium: SD medium without leucine, tryptophan, and histidine plus X-α-gal. Controls: SV40 T-antigen/P53 (positive control); empty vectors pGBKT7/pGADT7 (negative control). Experimental yeast cells were co-transfected with pGBKT7-TbRAD51.2 and one of the pGADT7-TbBRC repeat variant constructs (as indicated). (D) Verification of specific interactions between TbRAD51.2 and individual TbBRC repeat variants by yeast 2-hybrid analyses under higher stringency in selective dropout medium: SD medium without leucine, tryptophan, histidine, and adenine plus X-α-gal. The bait and prey protein pairs in different segments in the photograph are as in ‘C’. (E) Further verification of specific interactions between TbRAD51.2 and individual TbBRC repeat variants by Beta Glo Assay. Please refer to Fig. 4B for the identities of the samples in the X-axis. 14/15 indicates 14/15FL. Results are mean ± SEM (n=6).
Fig. 5. Evaluation of specific interactions between TbRAD51.4 and different numbers of BRC repeats by Yeast Two Hybrid System analyses.
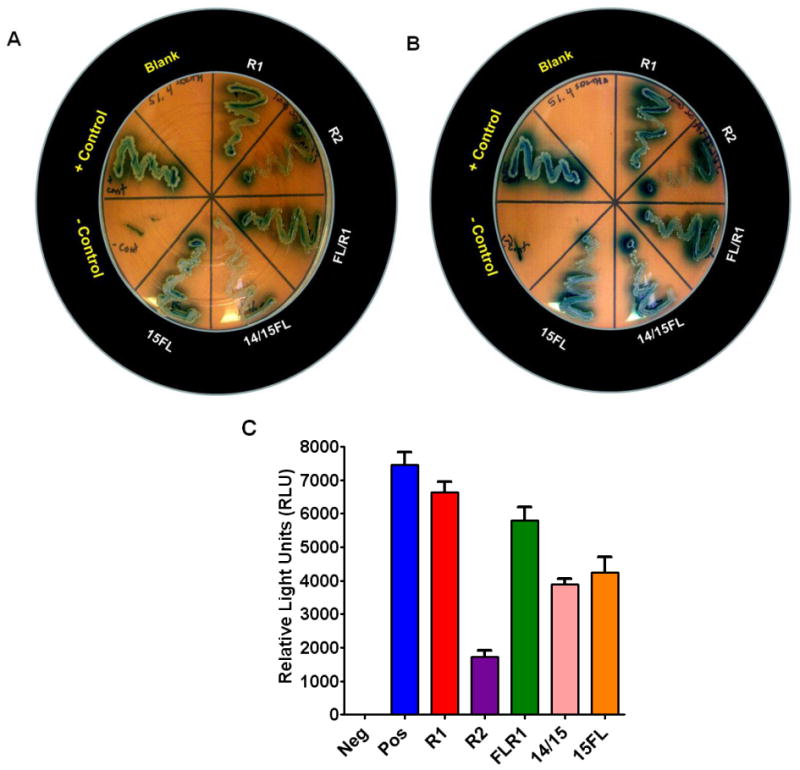
(A) Evaluation of specific interactions between TbRAD51.4 and individual TbBRC repeat variants by yeast 2-hybrid analyses under moderate stringency in selective dropout medium: SD medium without leucine, tryptophan, and histidine plus X-α-gal. Controls: SV40 T-antigen/P53 (positive control); empty vectors pGBKT7/pGADT7 (negative control). Experimental yeast cells were co-transfected with pGBKT7-TbRAD51.4 and one of the pGADT7-TbBRC repeat variant constructs (as indicated). (B) Verification of specific interactions between TbRAD51.4 and individual TbBRC repeat variants by yeast 2-hybrid analyses under higher stringency in selective dropout medium: SD medium without leucine, tryptophan, histidine, and adenine plus X-α-gal. The bait and prey protein pairs in different segments in the photograph are as in ‘A’. (C) Further verification of specific interactions between TbRAD51.4 and individual TbBRC repeat variants by Beta Glo Assay. Please refer to Fig. 4B for the identities of the samples in the X-axis. 14/15 indicates 14/15FL. Results are mean ± SEM (n=6).
3.5. Expression of a single BRC motif led to increased MMS sensitivity and inhibited cellular proliferation in T. brucei
TbBRCA2 knockout mutant of T. brucei has recently been shown to be significantly impaired in antigenic variation and to display genome instability in the bloodstream forms [37]. Whether this protein is critical in the survival of the procyclic forms of the parasite is not known. Our attempt to knockdown this gene by RNAi in the procyclic form of the parasite using RNAi constructs was not successful. Since BRCA2-RAD51 interactions are among the multiple cellular functions of the BRCA2 protein, we hypothesized that inhibition of such interactions using the 44 amino acid long BRC motif of TbBRCA2 as an aptamer will perturb the function of TbBRCA2 and kill the parasite cells. We tested our hypothesis by expressing a single BRC motif along with the 42 amino acid N-terminal flank that contains the two putative NLS of TbBRCA2 from a doxycycline-inducible system (pLEW79MHTAP; Fig. 6A). We evaluated the expression of the mRNA of the BRC motif construct by end-point RT-PCR. Although there was some leaky expression of some mRNA even in the absence of doxycycline, there was significant increase in the expression of the BRC motif mRNA (Fig. 6B) and protein (Fig. 6C) in the presence of doxycycline in the growth medium of the recombinant T. brucei procyclics. We used tetracycline repressor protein as well as α-tubulin as loading controls in the Fig. 6C. As we expected, the growth rate of the T. brucei procyclics were significantly slowed down in the presence of doxycycline when the expression of the BRC aptamer is induced (Fig. 6D). Doxycycline at the concentration tested, had no effect on the growth rate of the cells (data not shown). The cells transfected with the vector alone without the FLR1 insert proliferated similar to the wild-type T. brucei 29-30 procyclics (Fig. 6D). There was also some inhibition of growth even in the absence of doxycycline (Fig. 6D) which can be attributed to the leaky expression of the protein in the cells. These data show that the BRC aptamer is able to slow down the growth rates of T. brucei procyclics suggesting that TbBRCA2-TbRAD51 interaction may be essential for the normal growth of these cells in the culture medium.
Fig. 6. Expression and characterization of TAP-tagged BRC motif in T. brucei.
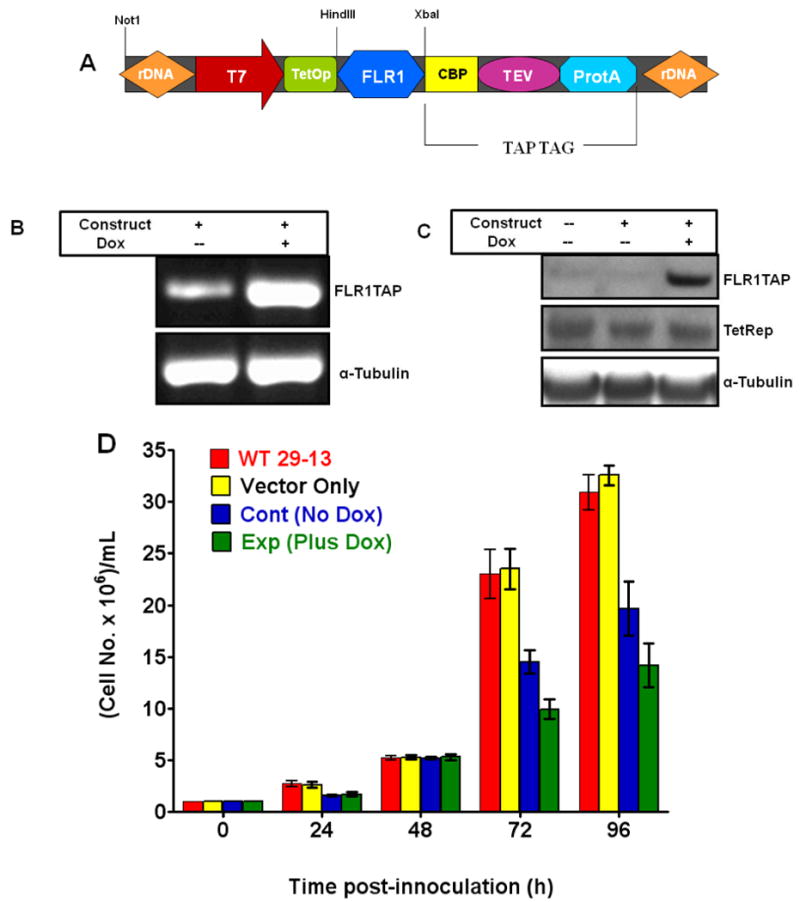
(A) Map of the pLEW79FLR1TAP construct used for the in vivo doxycycline-inducible expression of a TAP-tagged single BRC motif (44 amino acids) with a 42 amino acid N-terminal flank. The recombinant protein (∼30 kDa) also has domains from calmodulin binding peptide (CBP, 26 amino acids), recognition site for TEV protease (TEV, 9 amino acids) and IgG-binding domains of Protein A (ProtA, 116 amino acids). Evaluation of the in vivo expression of the BRC motif-containing aptamer mRNA by RT-PCR (B) and corresponding protein by Western blot analysis (C). Since the wild-type procyclic T brucei cells (29-13) have tetracycline repressor protein (TetRep), the expression of the aptamer protein was inducible with doxycycline (Dox). RT-PCR products were generated with primers specific to TbBRCA2 BRC repeat. β-actin was used as a loading control in this analysis. Western Blot analysis was performed using a HRP-conjugated rabbit anti-goat IgG antibody. α-tubulin and the tet repressor were used as loading controls in this analysis. (D) Effect of expression of the BRC aptamer on the growth rate of procyclic forms of T. brucei. The controls included wild-type cells (WT 29-13) alone and wild-type cells transfected with the empty vector (Vector only). The other control is the recombinant cells grown in medium without doxycycline (No Dox cont). Results are mean ± SEM (n=12).
Since one of the resultant consequences of interruption of TbBRCA2/TbRAD51 interactions would be perturbation of homologous recombination repair of DNA in the cells, we evaluated whether the expression of the BRC motif will make the cells more prone to DNA damage inducing agents like MMS. During the replication of the DNA, the replication fork is often stalled for various reasons [56-58] and this stalling is repaired by HR mechanisms in the cell [56-58]. MMS is known to inhibit the repair of the stalled replication forks during DNA replication through its inhibition of HR [59]. Our data show that over expression of the BRC motif in T. brucei procyclics significantly aggravates the lethal effects of MMS on these cells (Fig. 7). At any given concentration of MMS, in comparison to the control cells (wild-type 29-13 and the cells transfected with vector alone), the cells that are expressing the BRC aptamer are more sensitive to MMS toxicity (Fig. 7). The cells with the recombinant plasmid but growing in the absence of the inducer doxycycline were a little more sensitive to MMS than the control cells (Fig. 7). This is probably due to the leaky expression of the BRC aptamer. These data suggest that a combination of suitable DNA damaging agent and the optimized BRC motif peptide, if targeted to the T. brucei cells, should eliminate the parasites significantly and rapidly.
Fig. 7. Effect of expression of a single BRC motif in T. brucei on its tolerance of methyl methane sulfonate (MMS).
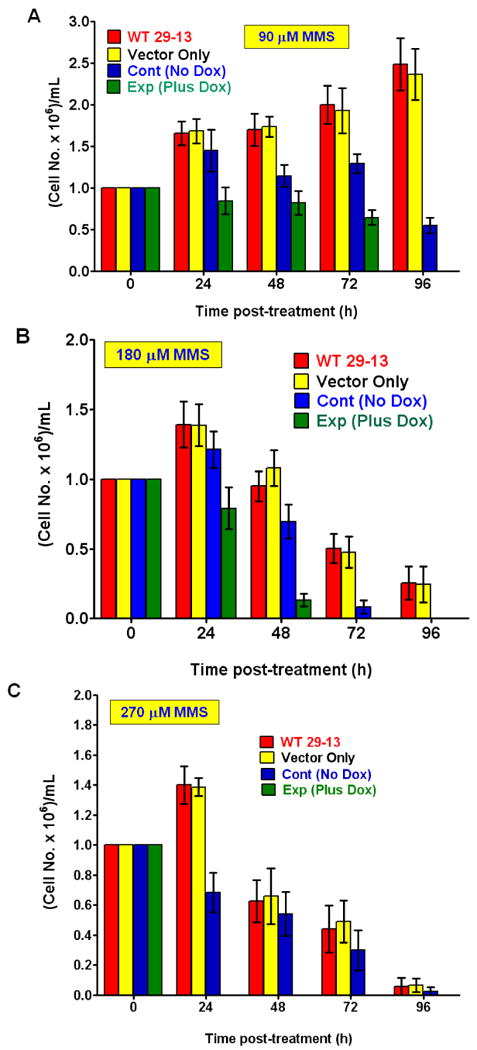
Growth of procyclic T. brucei cells were analyzed in the absence or in the presence of increasing concentrations of MMS (A: 90 μM; B: 180 μM and C: 270 μM) by counting the cells using a hemocytometer. The controls included wild-type cells (WT 29-13) alone and wild-type cells transfected with the empty vector (Vector only). The other control is the recombinant cells grown in medium without doxycycline (No Dox cont). Results are mean ± SEM (n=12).
3.6. Concluding remarks
Homologous recombination of DNA is a critical metabolic pathway in every organism. It is particularly important in the parasitic protozoa that have a digenetic life cycle involving an insect host as a transmitting organism between two vertebrates. When these parasites switch from warm blooded vertebrates to the blood-sucking insects, they are exposed to chemical and physico-chemical stress which may inflict DNA damage. The pathogens are seemingly well equipped with enhanced biochemical mechanisms to take care of such damages and must be able to repair the DNA to survive in the hosts, particularly during transitions. The error-free DNA recombination mechanisms are additionally very critical for the parasites of the T. brucei complex. One of the major survival mechanisms of these pathogens is the surface antigen switch (antigenic variation) inside the mammalian blood stream. This process requires robust HR mechanisms. Thus, any damage to the HR machinery in these parasites could be critical for the survival of the parasite under selected conditions during its life cycle. BRCA2 is recognized as a central protein in the HR mechanism in many organisms including T. brucei. In contrast to the earlier reported data that suggests multiple BRC repeats are important to TbBRCA2 functions [37], our study described here revealed that a single 44 amino acid long BRC motif of TbBRCA2 is sufficient to increase the sensitivity of the cells to MMS as well as to kill these cells. MMS induces blockage at the growing replication fork and this blockage is lethal to the growing cells if not repaired by HR [59]. TbBRCA2 are known to interact with other critical cellular molecules such as CDC45 [60]. Whether these non-RAD51 interactions potentially contribute to the aptamer-induced toxicity in T. brucei is currently not known. Our research provides the proof-of-concept for the efficacy of a peptide aptamer against a critical HR target in the parasitic protozoan T. brucei. Since other pathogens like T. cruzi and Leishmania also have similar machineries, further research in this area may develop an optimized peptide aptamer that will work against all tri-tryps.
Supplementary Material
Acknowledgments
We thank Drs. Mukul Mittal and Tanu Rana for reading this manuscript and for their valuable suggestions during the studies. We also thank Prof. George Cross for T. brucei 427 and 29-13 cells, Prof. Marilyn Parsons for the pLewMHTAP plasmid and Prof. Keith Gull for anti-tubulin antibody. This project is supported in parts by NIAID grants 5R01AI042327; 1R21AI076757-01A1 and NIGMS grant #3 S06 GM008037-34-S2 to GC. Graduate student stipend was provided to MH from the institutional training grants 2T32AI007281-21A1 and 5R25GM059994-10 obtained from NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kass EM, Jasin M. Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett. 2010;584:3703–3708. doi: 10.1016/j.febslet.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin SA, Lord CJ, Ashworth A. DNA repair deficiency as a therapeutic target in cancer. Curr Opin Genet Dev. 2008;18:80–6. doi: 10.1016/j.gde.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 4.Stockdale C, Swiderski MR, Barry JD, McCulloch R. Antigenic variation in Trypanosoma brucei: joining the DOTs. PLoS Biol. 2008;6:e185. doi: 10.1371/journal.pbio.0060185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palmer GH, Brayton KA. Gene conversion is a convergent strategy for pathogen antigenic variation. Trends Parasitol. 2007;23:408–13. doi: 10.1016/j.pt.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 6.Taylor JE, Rudenko G. Switching trypanosome coats: what's in the wardrobe? Trends Genet. 2006;22:614–20. doi: 10.1016/j.tig.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 7.Pays E. Regulation of antigen gene expression in Trypanosoma brucei. Trends Parasitol. 2005;21:517–20. doi: 10.1016/j.pt.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 8.Horn D. The molecular control of antigenic variation in Trypanosoma brucei. Curr Mol Med. 2004;4:563–76. doi: 10.2174/1566524043360078. [DOI] [PubMed] [Google Scholar]
- 9.Gull K. The biology of kinetoplastid parasites: insights and challenges from genomics and post-genomics. Int J Parasitol. 2001;31:443–52. doi: 10.1016/s0020-7519(01)00154-0. [DOI] [PubMed] [Google Scholar]
- 10.Vanhamme L, Lecordier L, Pays E. Control and function of the bloodstream variant surface glycoprotein expression sites in Trypanosoma brucei. Int J Parasitol. 2001;31:523–31. doi: 10.1016/s0020-7519(01)00143-6. [DOI] [PubMed] [Google Scholar]
- 11.Pays E, Vanhamme L, Pérez-Morga D. Antigenic variation in Trypanosoma brucei: facts, challenges and mysteries. Curr Opin Microbiol. 2004;7:369–374. doi: 10.1016/j.mib.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Navarro M, Gull K. A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature. 2001;414:759–63. doi: 10.1038/414759a. [DOI] [PubMed] [Google Scholar]
- 13.Folgueira C, Requena JM. A postgenomic view of the heat shock proteins in kinetoplastids. FEMS Microbiol Rev. 2007;31:359–77. doi: 10.1111/j.1574-6976.2007.00069.x. [DOI] [PubMed] [Google Scholar]
- 14.Turrens JF. Oxidative stress and antioxidant defenses: a target for the treatment of diseases caused by parasitic protozoa. Mol Aspects Med. 2004;25:211–20. doi: 10.1016/j.mam.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 15.Piacenza L, Alvarez MN, Peluffo G, Radi R. Fighting the oxidative assault: the Trypanosoma cruzi journey to infection. Curr Opin Microbiol. 2009;12:415–21. doi: 10.1016/j.mib.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 16.Rajendra E, Venkitaraman AR. Two modules in the BRC repeats of BRCA2 mediate structural and functional interactions with the RAD51 recombinase. Nucleic Acids Res. 2010;38:82–96. doi: 10.1093/nar/gkp873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thorslund T, West SC. BRCA2: a universal recombinase regulator. Oncogene. 2007;26:7720–30. doi: 10.1038/sj.onc.1210870. [DOI] [PubMed] [Google Scholar]
- 18.Liang F, Han M, Romanienko PJ, Jasin M. Homology directed repair is a major double-strand break repair pathway in mammalian cells. Proc Natl Acad Sci USA. 1998;95:5172–7. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–54. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 20.Johnson RD, Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 2000;19:3398–407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4:435–45. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 22.Belmaaza A, Chartrand P. One-sided invasion events in homologous recombination at double-strand breaks. Mutat Res. 1994;314:199–208. doi: 10.1016/0921-8777(94)90065-5. [DOI] [PubMed] [Google Scholar]
- 23.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bertrand P, Saintigny Y, Lopez BS. p53's double life: transactivation-independent repression of homologous recombination. Trends Genet. 2004;20:235–43. doi: 10.1016/j.tig.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 25.Kolomietz E, Meyn MS, Pandita A, Squire JA. The role of Alu repeat clusters as mediators of recurrent chromosomal aberrations in tumors. Genes Chromosomes Cancer. 2002;35:97–112. doi: 10.1002/gcc.10111. [DOI] [PubMed] [Google Scholar]
- 26.Meyn MS. Chromosome instability syndromes: lessons for carcinogenesis. Curr Top Microbiol Immunol. 1997;221:71–148. doi: 10.1007/978-3-642-60505-5_6. [DOI] [PubMed] [Google Scholar]
- 27.Scully R, Livingston DM. In search of the tumour suppressor functions of BRCA1 and BRCA2. Nature. 2000;408:429–32. doi: 10.1038/35044000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venkitaraman AR. Tracing the network connecting brca and fanconi anaemia proteins. Nat Rev Cancer. 2004;4:266–76. doi: 10.1038/nrc1321. [DOI] [PubMed] [Google Scholar]
- 29.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7:263–72. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 30.Tutt A, Bertwistle D, Valentine J, Gabriel A, Swift S, Ross G, Griffin C, Thacker J, Ashworth A. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 2001;20:4704–16. doi: 10.1093/emboj/20.17.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–82. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 32.Glover L, McCulloch R, Horn D. Sequence homology and microhomology dominate chromosomal double-strand break repair in African trypanosomes. Nucleic Acids Res. 2008;36:2608–18. doi: 10.1093/nar/gkn104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burton P, McBride DJ, Wilkes JM, Barry JD, McCulloch R. Ku heterodimer-independent end joining in Trypanosoma brucei cell extracts relies upon sequence microhomology. Eukaryot Cell. 2007;6:1773–81. doi: 10.1128/EC.00212-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Proudfoot C, McCulloch R. Distinct roles for two RAD51-related genes in Trypanosoma brucei antigenic variation. Nucleic Acids Res. 2005;33:6906–19. doi: 10.1093/nar/gki996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thorslund T, Esashi F, West SC. Interactions between human BRCA2 protein and the meiosis-specific recombinase DMC1. EMBO J. 2007;26:2915–22. doi: 10.1038/sj.emboj.7601739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Proudfoot C, McCulloch R. Trypanosoma brucei DMC1 does not act in DNA recombination, repair or antigenic variation in bloodstream stage cells. Mol Biochem Parasitol. 2006;145:245–53. doi: 10.1016/j.molbiopara.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 37.Hartley CL, McCulloch R. Trypanosoma brucei BRCA2 acts in antigenic variation and has undergone a recent expansion in BRC repeat number that is important during homologous recombination. Mol Microbiol. 2008;68:1237–51. doi: 10.1111/j.1365-2958.2008.06230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singha UK, Sharma S, Chaudhuri M. Down regulation of mitochondrial porin inhibits cell growth and alters respiratory phenotype in Trypanosoma brucei. Eukaryot Cell. 2009;8:1418–28. doi: 10.1128/EC.00132-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singha UK, Peprah E, Williams S, Walker R, Saha L, Chaudhuri M. Characterization of the mitochondrial inner membrane protein translocator Tim17 from Trypanosoma brucei. Mol Biochem Parasitol. 2008;159:30–43. doi: 10.1016/j.molbiopara.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lo T, Pellegrini L, Venkitaraman AR, Blundell TL. Sequence fingerprints in BRCA2 and RAD51: implications for DNA repair and cancer. DNA Repair (Amst) 2003;2:1015–28. doi: 10.1016/s1568-7864(03)00097-1. [DOI] [PubMed] [Google Scholar]
- 41.Warren M, Smith A, Partridge N, Masabanda J, Griffin D, Ashworth A. Structural analysis of the chicken BRCA2 gene facilitates identification of functional domains and disease causing mutations. Hum Mol Genet. 2002;11:841–51. doi: 10.1093/hmg/11.7.841. [DOI] [PubMed] [Google Scholar]
- 42.Petalcorin MI, Galkin VE, Yu X, Egelman EH, Boulton SJ. Stabilization of RAD-51-DNA filaments via an interaction domain in Caenorhabditis elegans BRCA2. Proc Natl Acad Sci USA. 2007;104:8299–8304. doi: 10.1073/pnas.0702805104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kojic M, Kostrub CF, Buchman AR, Holloman WK. BRCA2 homolog required for proficiency in DNA repair, recombination, and genome stability in Ustilago maydis. Mol Cell. 2002;10:683–91. doi: 10.1016/s1097-2765(02)00632-9. [DOI] [PubMed] [Google Scholar]
- 44.Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA–ssDNA junction. Nature. 2005;433:653–57. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 45.Misra S, Hall M, 3rd, Chaudhuri G. Molecular characterization of a human BRCA2 homolog in Leishmania donovani. J Parasitol. 2005;91:1492–5. doi: 10.1645/GE-579R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warren M, Smith A, Partridge N, Masabanda J, Griffin D, Ashworth A. Structural analysis of the chicken BRCA2 gene facilitates identification of functional domains and disease causing mutations. Hum Mol Genet. 2002;11:841–51. doi: 10.1093/hmg/11.7.841. [DOI] [PubMed] [Google Scholar]
- 47.Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, Zheng N, Chen PL, Lee WH, Pavletich NP. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–48. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 48.Szabo C, Masiello A, Ryan JF, Brody LC. The breast cancer information core: database design, structure, and scope. Hum Mutat. 2000;16:123–31. doi: 10.1002/1098-1004(200008)16:2<123::AID-HUMU4>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 49.Marston NJ, Richards WJ, Hughes D, Bertwistle D, Marshall CJ, Ashworth A. Interaction between the product of the breast cancer susceptibility gene BRCA2 and DSS1, a protein functionally conserved from yeast to mammals. Mol Cell Biol. 1999;19:4633–42. doi: 10.1128/mcb.19.7.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crackower MA, Scherer SW, Rommens JM, Hui CC, Poorkaj P, Soder S, Cobben JM, Hudgins L, Evans JP, Tsui LC. Characterization of the split hand/split foot malformation locus SHFM1 at 7q21.3-q22.1 and analysis of a candidate gene for its expression during limb development. Hum Mol Genet. 1996;5:571–79. doi: 10.1093/hmg/5.5.571. [DOI] [PubMed] [Google Scholar]
- 51.Esashi F, Galkin VE, Yu X, Egelman EH, West SC. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat Struct Mol Biol. 2007;14:468–74. doi: 10.1038/nsmb1245. [DOI] [PubMed] [Google Scholar]
- 52.Holzmuller P, Herder S, Cuny G, De Meeûs T. From clonal to sexual: a step in T. congolense evolution? Trends Parasitol. 2010;26:56–60. doi: 10.1016/j.pt.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 53.Gibson W, Stevens J. Genetic exchange in the trypanosomatidae. Adv Parasitol. 1999;43:1–46. doi: 10.1016/s0065-308x(08)60240-7. [DOI] [PubMed] [Google Scholar]
- 54.Gibson W, Bailey M. Genetic exchange in Trypanosoma brucei: evidence for meiosis from analysis of a cross between drug-resistant transformants. Mol Biochem Parasitol. 1994;64:241–52. doi: 10.1016/0166-6851(94)00017-4. [DOI] [PubMed] [Google Scholar]
- 55.Akopyants NS, Kimblin N, Secundino N, Patrick R, Peters N, Lawyer P, Dobson DE, Beverley SM, Sacks DL. Demonstration of genetic exchange during cyclical development of Leishmania in the sand fly vector. Science. 2009;324:265–8. doi: 10.1126/science.1169464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakano T, Katafuchi A, Matsubara M, Terato H, Tsuboi T, Masuda T, Tatsumoto T, Pack SP, Makino K, Croteau DL, Van Houten B, Iijima K, Tauchi H, Ide H. Homologous recombination but not nucleotide excision repair plays a pivotal role in tolerance of DNA-protein cross-links in mammalian cells. J Biol Chem. 2009;284:27065–76. doi: 10.1074/jbc.M109.019174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18:99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Daboussi F, Courbet S, Benhamou S, Kannouche P, Zdzienicka MZ, Debatisse M, Lopez BS. A homologous recombination defect affects replication-fork progression in mammalian cells. J Cell Sci. 2008;121:162–6. doi: 10.1242/jcs.010330. [DOI] [PubMed] [Google Scholar]
- 59.Lundin C, North M, Erixon K, Walters K, Jenssen D, Goldman ASH, Helleday T. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 2005;33:3799–3811. doi: 10.1093/nar/gki681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oyola SO, Bringaud F, Melville SE. A kinetoplastid BRCA2 interacts with DNA replication protein CDC45. Int J Parasitol. 2009;39:59–69. doi: 10.1016/j.ijpara.2008.07.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


