Abstract
Background
We sought to determine whether lower DICER1 expression is associated with recurrence in patients with endometrioid endometrial cancer. We also explored DNA methylation and haploinsufficiency as potential mechanisms related to altered DICER1 expression in these tumors.
Methods
DICER1 expression was assessed by qPCR in a selected cohort of endometrioid endometrial tumors (N=169). Loss of heterozygosity analyses were conducted using two single nucleotide polymorphisms and COBRA was used to assess methylation in the 5′-UTR region of DICER1 in representative tumors. The relationships between DICER1 expression and clinicopathologic variables, including overall and disease-free survival (OS and DFS), were assessed using non-parametric rank-sum tests and Cox proportional hazard models as appropriate. Survival distributions were described using the Kaplan-Meier method. A nested case-control analysis was conducted to confirm the association between transcript levels and recurrence.
Results
Lower DICER1 expression and advanced stage were associated with worse DFS (HR: 1.36, 95%CI: 1.05-1.75, P=0.02 and HR: 2.79, 95%CI: 1.59-4.90, P<0.001, respectively). Three variables were significantly associated with reduced OS: age (HR: 1.04, 95%CI: 1.02-1.06, P<0.0001), advanced stage (HR: 6.41, 95%CI: 3.57-11.52, P<0.0001) and high grade (HR: 2.96, 95%CI: 1.46-5.99, P=0.003). Nested-case control analysis confirmed lower DICER1 transcripts in recurrent cases (P=0.01). Deletion of DICER1 sequences was an infrequent event (5% of analyzed cases) and no methylation was observed in the 5′ DICER1 regulatory region.
Conclusions
Lower DICER1 transcripts correlate with recurrence and worse disease free survival in patients with endometrioid endometrial cancer. The factors influencing DICER1 levels in primary endometrial cancers remain unknown.
Keywords: Endometrial Cancer, DICER1, Expression, Recurrence, Survival, Outcomes
Introduction
Endometrial cancer is the fourth most common malignancy among women in the United States 1. Most endometrial cancer cases are diagnosed when the disease is still confined to the uterus. These patients have an excellent prognosis with a 5-year survival of >95% 2. Conversely, recurrent endometrial cancer carries a dismal prognosis with a median survival of <10 months and 5-year survival rates close to 15% 3. In 2009, more than 7,700 U.S. women died from endometrial cancer 1.
Despite a growing understanding of the pathophysiology and molecular biology of endometrial cancers, accurate prediction of which patients with early stage disease are destined to experience disease recurrence is still not possible. Multiple refinements to staging modalities and comprehensive risk assessment models that include a broad range of clinicopathologic and molecular variables have evolved over the last two decades, but continue to lack the desired prognostic precision 4, 5. Despite the considerable effort that has gone into developing approaches to better identify and treat endometrial cancer patients at risk for recurrence and progression, there has been little success in improving outcomes 5-7.
The relatively low incidence and paucity of events (recurrences and deaths) observed in patients with endometrial cancer make comparative efficacy and outcome research difficult. Discovery of novel molecular prognostic markers holds promise for improved risk prediction and personalized therapeutic approaches to the management of endometrial cancer.
As a post-transcriptional regulatory mechanism, RNA interference (RNAi) plays an important role in carcinogenesis 8. RNAi is orchestrated at least in part by small RNA molecules (e.g. microRNA – miRNA and short interfering RNA – siRNA) capable of targeting specific messenger RNAs ultimately resulting in gene silencing. Dicer is the enzyme responsible for the cleavage of miRNA precursors. Merritt et al demonstrated that low expression of Dicer and Drosha are associated with aggressive ovarian cancer phenotypes and poor clinical outcomes 9. Growing evidence suggests that Dicer′s expression levels are associated with worse clinical outcomes in various cancer types 9-14.
We hypothesized that lower DICER1 expression is associated with increased risk of recurrence and progression in patients with endometrioid endometrial cancer. We analyzed DICER1 expression in a series of recurrent pure endometrioid endometrial tumors as the first phase of a planned nested case-control design to test whether DICER1 transcript levels differ between recurrent and non-recurrent cases. DICER1 expression was also evaluated in a number of non-recurrent endometrioid endometrial tumors. We investigated tumors for DICER1 deletion (loss of heterozygosity - LOH) and promoter methylation as potential mechanisms leading to reduced DICER1 expression in primary endometrial cancers.
Materials and Methods
Study Population
Since 1991 the Division of Gynecologic Oncology at Washington University School of Medicine has prospectively collected matched tumor and normal tissue samples from endometrial cancer surgical specimens. None of these patients received preoperative radiation or chemotherapy. Surgical staging for cases included in this study was assigned on an ongoing basis based on International Federation of Gynecology and Obstetrics′ (FIGO) 1988 criteria 15. Clinical and pathologic information was prospectively collected and stored in a computerized database. Treatment was individualized at the discretion of the patient′s attending gynecologic oncologist. Follow-up typically occurred at 3-month intervals for the first 2 years and then at 6-month intervals for the following 3 years. Disease surveillance included physical examination and periodic pap smears. Diagnostic imaging and directed biopsies are performed as clinically indicated. All suspected recurrences were histologically confirmed. All participants have consented to enrollment in molecular studies approved by Washington University Human Research Protection Office (protocols 91-507 and 93-0828). Normal endometrial tissues were collected from volunteers undergoing hysterectomy for reasons other than a cancer diagnosis (protocol 04-0949).
Selection of Cases and Controls
Pure endometrioid endometrial tumors with available outcome data and high (≥ 70%) neoplastic cellularity (N=561) were considered for this study. First, we identified all women with documented recurrence (potential “cases”, N=81). Tissue for RNA preparation was available for 70 of these and DICER1 transcript levels were successfully determined in 57 cases. Potential “controls” were identified among contemporary non-recurrent women with available primary tumor RNA and follow-up longer than 3 years (most endometrial cancer recurrences can be expected to occur within 2-3 years of diagnosis). DICER1 levels could be assessed in 112 non-recurrent tumors. Thus, a total of 169 selected tumors (cohort enriched for recurrences) with available DICER1 expression and outcome data (recurrent N=57, non-recurrent N=112) were included and analyzed in this study.
For the planned nested case-control analysis, we successfully matched a sub-set of cases (N=29) to two non-recurrent controls (N=58) based on surgical stage, FIGO grade and age using the matching library from the Comprehensive R Archive Network (R Development Core Team, v4.7-6, http://CRAN.R-project.org/package=Matching).
RNA extraction and DICER1 expression analysis
RNA was extracted from primary tumors using the Trizol® reagent (Invitrogen, Carlsbad, CA) and further purified using RNEasy Mini Kit® reagents (Qiagen, Valencia, CA). RNA concentrations were determined using Nanodrop™ spectrophotometry (Thermo Fisher Scientific, Waltham, MA). cDNA for primary tumor RNAs, normal endometrium and the AN3CA cell line (normal and cell line serving as reference for our qPCR assays) was prepared using the Quantitect® Reverse Transcription Kit (Qiagen, Valencia, CA). We chose the AN3CA cell line because its RNA represents a “renewable resource” and by using it as our reference allows us and other investigators to more directly compare results from studies in additional primary tumors.
Transcript levels were determined using SYBR green methods 16. DICER1 and GAPDH were measured using SYBR® Green Mix (Bio-Rad, Hercules, CA) and an iCycler™ with optical module (Bio-Rad, Hercules, CA). All reactions were performed in triplicate. The DICER1 amplicon spans exons 23 and 24 (Forward primer: 5′-CTCCAGGCTTTTACACATGC-3′ and Reverse primer: 5′-CGATGCAAAGATGGTGTTGT-3′). Detection cycle counts for DICER1 transcripts were determined for each tumor sample (normalized based on GAPDH levels) and standardized relative to a pooled sample of 3 normal endometria. DICER1 transcript levels were expressed as units of AN3CA expression.
Loss of Heterozygosity (LOH) Analysis
Tumors with the highest (N=44) and lowest (N=44) DICER1 transcript levels were assessed for LOH using intragenic restriction fragment length polymorphisms (RFLPs). Sequences including rs1057035 and rs1209904 were PCR amplified in tumor and matched normal DNAs (rs1057035 Forward: 5′-AGTTAGGACTGCGGAAAGCA-3′ and Reverse: 5′-GCCGTTTCACTTTTTGTTGG-3′; rs1209904 Forward: 5′-TGCTGATCACACAGATCTTCAA-3′ and Reverse: 5′-GCTTTTGCCAAAGAAATTGG-3′). Amplification products were digested with MwoI and HpyCH4IV respectively (New England Biolabs, Ipswich, MA) and the resultant restriction digestion products resolved on 10% polyacrylamide gels.
Methylation Analysis
Methylation in the DICER1 5′ putative regulatory region was analyzed by combined bisulfate restriction analysis (COBRA) 17, 18. Tumor DNAs and a universally methylated control (Millipore, Billerica, MA) were bisulfite converted using the EZ DNA Methylation™ Gold Kit (Zymo Research, Orange, CA). Three COBRA assays were designed to assess methylation at 13 CpG pairs in the approximately 1200 nucleotide DICER1 CpG island. PCR reactions were carried out and products digested and resolved on 10% polyacrylamide gels (PCR reaction and digestion information provided in Table 1).
Table 1.
COBRA Assays
| Assay | Forward Primers * | Reverse Primers * | Amplicon Size (bp) |
Endonuclease Fragment Sizes (bp) |
|---|---|---|---|---|
| COBRA1 | Outer: ttTtTattatTtggggatT | Outer: AaAaatcccactAActcc | 528 | BssHII |
| Inner: gggaggtgTtTagagggaag | Inner: cccaAccactcaAAaAcaAA | 276 | 139 / 98 / 37 / 2 | |
|
| ||||
| COBRA2 | Outer: gTTaggTTTtgtTTaatTaTagg | Outer: tcacaAcccaAAcctc | 327 | BstUI |
| Inner: gggagTTagtgggattTtTTaag | Inner: caAAttactccattcacctAAAcc | 163 | 80 / 42 / 41 | |
|
| ||||
| COBRA3 | Outer: gggaggTTtgggTtgtg | Outer: cctAacaAtaatcacacaAAacc | 590 | Hpy99I |
| Inner: gaggTagTTTtgggagaTt | Inner: AAAccccaAAAtActcc | 179 | 147 / 32 | |
Uppercase T or A in primer sequence represent residues (cytosines) converted by bisulfite treatment in the reading or complementary strand respectively
COBRA: Combined bisulfite restriction analysis
Statistical Analysis
The relationship between DICER1 expression and clinicopathologic covariates was analyzed in the entire study cohort (N=169) using Spearman′s correlation and Kruskall-Wallis rank-sum test for continuous and categorical variables respectively. Overall survival (OS) was defined as the time from initial surgery to the date of death due to any cause. Survivors were censored at the date of last contact. Disease-free survival (DFS) was defined as the time from surgery to recurrence or progression. Survival distributions were described using the Kaplan-Meier product limit method and compared using log-rank test. Univariate and multivariate Cox proportional hazard models were fitted to assess the predictive effects of the covariates on OS and DFS. Factors with P-values <0.2 in the univariate analysis were included in the corresponding multivariate model. For the nested case-control the relative DICER1 expression level for cases (N=29) and 2:1 matched controls (N=58) were compared. All analyses were two-sided, and significance was set at a P value of 0.05. Statistical analyses were performed using SAS (SAS Institutes, Cary, NC).
Results
Reduced DICER1 expression in primary endometrial cancers that subsequently recurred
DICER1 expression was assessed by qPCR in 169 endometrioid endometrial tumors. The clinicopathologic characteristics of the study cohort are presented in Table 2. As previously noted, the cohort was enriched for cases that recurred. Therefore, the disease and clinical characteristics differ from what would be expected for a population based study (higher grade and stage and more patient receiving adjuvant therapies). Median follow-up was 59.2 months (0.7 – 162.2 months) and median DICER1 expression was 1.37 (0.13 – 12.28 arbitrary units). The relative expression of the AN3CA cell line relative to our sample of pooled normal endometria was 0.42 arbitrary units.
Table 2.
Demographic and Clinicopathologic Characteristics
| N = 169 | ||
|---|---|---|
| n | % | |
| Age (years) | 65.9 ± 11.5 * | |
| Race | ||
| Caucasian | 149 | 88.2 |
| Non-Caucasian | 20 | 11.8 |
| Stage (AJCC /FIGO 1988) | ||
| I | 81 | 47.9 |
| II | 24 | 14.2 |
| III | 49 | 29.0 |
| IV | 15 | 8.9 |
| FIGO grade | ||
| 1 | 50 | 29.6 |
| 2 | 69 | 40.8 |
| 3 | 50 | 29.6 |
| Adjuvant treatment | ||
| No | 76 | 45 |
| Yes† | 93 | 55 |
| Events | ||
| Recurrence / Progression | 57 | 33.7 |
| Death | 67 | 39.6 |
| DICER1 expression†† | 1.87 ± 1.1 * | |
Mean ± standard deviation
Adjuvant therapy included: radiotherapy (n=53), chemotherapy (n=20), radiotherapy + chemotherapy (n=15), hormonal therapy (n=3), radiotherapy + hormonal therapy (n=1) and chemotherapy + hormonal therapy (n=1).
DICER1 arbitrary expression units relative to pooled normal endometria
FIGO: International Federation of Gynecology and Obstetrics
AJCC: American Joint Committee on Cancer
DICER1 mRNA level was not associated with age at diagnosis, race or surgical stage. A clear trend for higher DICER1 transcript levels in lower grade tumors was evident (grade 1: 2.19 ± 1.47 expression units , grade 2: 1.73 ± 1.68 expression units and grade 3: 1.73 ± 1.81 expression units; P = 0.02).
The distribution of tumor DICER1 relative expression according to recurrence status is presented in Table 3. Univariate analysis demonstrated a significant association between lower DICER1 expression and decreased DFS (HR 1.38, 95%CI: 1.07-1.78, P=0.01). Similarly, stage III/IV (AJCC T3,N0,M0; T1-3,N1-2,M0; T4,Nx,M0 and Tx,Nx,M1) disease (HR 2.43, 95%CI: 1.40-4.24, P=0.002) and FIGO grade 3 (HR: 2.05, 95%CI: 1.01-4.18, P=0.04) were all associated with worse DFS. Age, race and use of adjuvant therapy were not associated with DFS. Univariate analysis also demonstrated that older age (HR 1.03, 95%CI: 1.01-1.05, P=0.01), stage III/IV (HR 3.81, 95%CI: 2.21-6.57, P<0.0001) and FIGO grade 3 (HR 2.57, 95%CI: 1.30-5.06, P=0.007) were associated with worse OS. DICER1 expression (HR 1.14, 95%CI: 0.95-1.37, P=0.17), race and use of adjuvant therapy did not appear associated with OS on univariate analysis. Kaplan-Meier survival curves for DFS and OS in relation to DICER1 expression stratified as higher or lower than median expression for the cohort are presented in Figure 1.
Table 3.
Tumor DICER1 Relative Expression According to Recurrence Status
| Relative Expression * | Recurrent (N = 57) | Non Recurrent (N = 112) | ||
|---|---|---|---|---|
| n | (%) | n | (%) | |
| <1 | 22 | 38.6 | 26 | 23.2 |
| 1 - 2 | 24 | 42.1 | 42 | 37.5 |
| >2 - 3 | 8 | 14.0 | 22 | 19.6 |
| >3 - 4 | 2 | 3.5 | 11 | 9.8 |
| >4 - 5 | - | - | 6 | 5.4 |
| >5 – 6 | - | - | 2 | 1.8 |
| >6 | 1 | 1.8 | 3 | 2.7 |
DICER1 arbitrary expression units relative to pooled normal endometria
Figure 1.
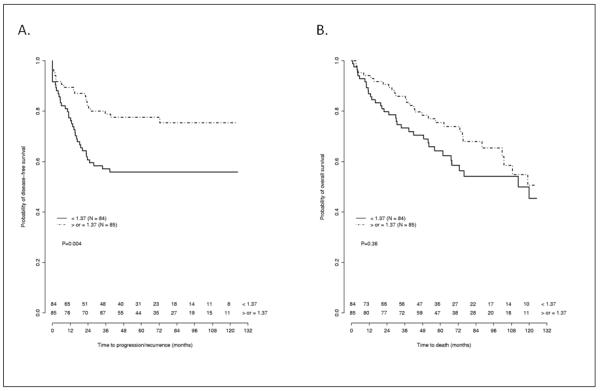
Kaplan-Meier curves for disease free survival (A) and overall survival (B) stratified by median DICER1 expression arbitrary units (<1.37 / ≥ 1.37). Numbers indicate survivors in each group at each censor point.
Multivariate analysis confirmed the association of lower DICER1 expression and worse DFS (HR 1.36, 95%CI: 1.05-1.75, P=0.02). Advanced stage (HR 2.79, 95%CI: 1.59-4.90, P<0.001) also remained associated with worse DFS. Multivariate analysis of DICER1 expression did not reveal a significant association with OS (HR 1.18, 95%CI: 0.98-1.43, P=0.09). Older age (HR 1.04, 95%CI: 1.02-1.06, P<0.0001), advanced stage (HR 6.41, 95%CI: 3.57-11.52, P<0.0001) and high grade (HR 2.96, 95%CI: 1.46-5.99, P=0.003) remained significantly associated with worse OS.
As planned, we studied a sub-set of 87 matched cases (N= 29) and controls (N=58) to further assess the association of DICER1 expression levels and disease recurrence. As expected, tumors that recurred had significantly lower DICER1 transcript levels (median 0.97, range 0.13-7.60 expression units) than tumors that did not recur (median 1.50, range 0.29-12.30 expression units, P=0.01).
LOH and methylation studies
The sub-set of tumors with the highest (N=44) and lowest (N=44) transcript levels, were assessed for DICER1 LOH using two intragenic single nucleotide polymorphisms (rs1057035 and rs1209904). PCR amplification was successfully completed in all cases and 58 tumors were informative for at least one marker (31 “high DICER1 expressors” and 27 “low DICER1 expressors”). Loss of heterozygosity or allelic imbalance was only identified in 3 tumors (DICER1 transcript levels = 0.48, 2.38 and 12.29 expression units respectively). Representative examples of LOH / allelic imbalance demonstrated by RFLP analyses are presented in Figure 2.
Figure 2. Loss of heterozygosity analyses.
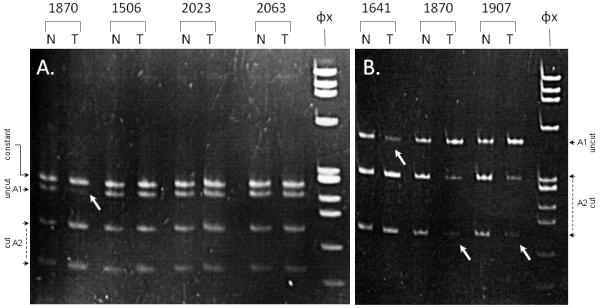
Panel A: PCR amplification products of marker rs1057035 in representative normal (N) / tumor (T) pairs digested with MwoI. Tumor 1870 demonstrates LOH (DICER1 expression = 2.38 arbitrary units).
Panel B: PCR amplification products of marker rs1209904 in representative normal (N) / tumor (T) pairs digested with HpyCH4IV. Tumors 1641 (DICER1 expression = 12.29 arbitrary units), 1870 (DICER1 expression = 2.38 arbitrary units) and 1907 (DICER1 expression = 0.48 arbitrary units) demonstrate allelic imbalance.
White arrows indicate the allelic fragment(s) with reduced intensity.
Three COBRA assays were used to assess the methylation status of 13 CpG pairs in the 5′-UTR region of the DICER1 gene. We evaluated a sub-set of tumors with the highest and lowest DICER1 transcripts. A total of 34 tumors (9 “high DICER1 expressors” and 25 “low DICER1 expressors”) were evaluated with at least two COBRA assays. No methylation was observed in any of the tumors analyzed (data not shown).
Discussion
Considerable effort has gone into developing approaches to better identify endometrial cancer patients at risk for disease progression and recurrence. Despite complete surgical staging, many women recur or progress and ultimately die from their disease. The 1988 FIGO surgical staging that was used for risk stratification for more than two decades as well as complex clinicopathologic algorithms developed for therapeutic triage, lack the desired prognostic precision 4, 5, 7, 19. The need for an accurate risk stratification system for patients with endometrial cancer has motivated multiple efforts aimed at identifying prognostic biomarkers 20-27. Molecular biomarkers may potentially identify women at high risk for recurrence and death, predict response to currently available treatments and direct the development of novel targeted therapies.
Alterations in miRNA expression and processing seem to play important roles in human carcinogenesis 8. In this study, we found that DICER1 expression levels vary widely in primary endometrioid endometrial tumors. Furthermore, lower transcripts are associated with worse clinical outcomes as evidenced by a significantly increased risk of recurrence and an obvious trend towards worse overall survival among patients with tumors with low DICER1 transcripts. Prior studies have suggested that levels of several components of the RNA-induced silencing complex (RISC) are associated with clinical outcomes in various cancer types 9-14. Reduced DICER1 expression has been associated with worse prognosis in lung 9, 10, 28, ovarian 9 and breast cancer 9, 29. On the other hand, higher DICER1 transcript levels seem to represent a poor prognostic factor in prostate cancer 12. Other studies have not found an association between DICER1 expression levels and clinical outcomes in other malignancies such as esophageal cancer 11 and acute myeloid leukemia 14. These discrepancies suggest that specific effects derived from alterations in miRNA processing may be tumor site specific. How DICER1 contributes to the initiation and progression of malignancies is only beginning to be understood. Altered miRNA expression can be expected to occur as a result of variations in pre-miRNA processing by Dicer. Fluctuations in miRNA expression have been shown to regulate expression of key tumor suppressor genes and oncogenes 30. Most recently, Dicer has been shown to play an important role in early apoptotic DNA degradation in C. elegans 31. It is possible that other incompletely understood key cellular processes may also be regulated through similar or novel nuclease activities.
The specific mechanisms responsible for variations in DICER1 transcript levels remain elusive. Kumar et al recently studied the effects of conditional DICER1 mutation on several mouse cancer models and noted that partial loss of DICER1 promotes tumor development 32. The authors also referred to frequent loss of one allele of DICER1 in human tumors (hemizygous deletion) and lack of high-level amplifications or homozygous deletion of the DICER1 locus 32-35. Taken together these findings led the authors to suggest that DICER1 functions as a haploinsufficient tumor suppressor 32. In our evaluation of 58 tumors representing the highest and lowest levels of DICER1 mRNA, we identified LOH and/or allelic imbalance in only 3 cases (5.2%, 2 high-expression and 1 low-expression DICER1 tumors). Interestingly, the tumor with the highest transcript level had evidence of allelic imbalance. Our findings argue against haploinsufficiency as a significant control mechanism of DICER1 expression in endometrioid endometrial tumors. It is important to note that we did not test for mutations (that could lead to nonsense mediated decay) or other unappreciated cis regulatory mechanisms that could explain overall reduction in DICER1 transcript levels.
DNA methylation and concomitant histone modification are frequently associated with decreased transcription in normal and cancer cells. We sought to determine whether methylation of CpG sites in the 5′-UTR region of DICER1 was associated with decreased expression in primary endometrioid endometrial tumors. We found no evidence for methylation. This observation is consistent with a previous report in lung tumors and suggests that different regulatory mechanisms are involved in the transcriptional control of DICER1 10.
In conclusion, we have shown that lower DICER1 transcript levels correlate with worse disease free survival in patients with endometrioid endometrial tumors. The mechanisms responsible for variations in DICER1 expression in endometrial cancer remain unknown. In the future, assessment of DICER1 expression may prove useful for risk assessment. Novel therapies that utilize RNAi are under development 36, 37. Our findings may provide the foundation for utilization of personalized novel RNAi-based therapeutics in patients with endometrioid endometrial cancer at high risk for recurrence.
Supplementary Material
Acknowledgments
Supported by: NCI RO1 CA71754 (P.J.G.), Barnes-Jewish Foundation 00161-0806 (D.G.M. and P.J.G). The Siteman Cancer Center is supported by NCI Cancer Center Support Grant P30 CA91842.
Footnotes
The authors do not have any conflicts of interest to disclose in relation to this manuscript.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Altekruse SFKC, Krapcho M, Neyman N, Aminou R, Waldron W, Ruhl J, Howlader N, Tatalovich Z, Cho H, Mariotto A, Eisner MP, Lewis DR, Cronin K, Chen HS, Feuer EJ, Stinchcomb DG, Edwards BK, editors. SEER Cancer Statistics Review, 1975-2007. National Cancer Institute; Bethesda, MD: 2010. http://seer.cancer.gov/csr/1975_2007/, based on November 2009 SEER data submission, posted to the SEER web site. [Google Scholar]
- 3.Creutzberg CL, van Putten WL, Koper PC, et al. Surgery and postoperative radiotherapy versus surgery alone for patients with stage-1 endometrial carcinoma: multicentre randomised trial. PORTEC Study Group. Post Operative Radiation Therapy in Endometrial Carcinoma. Lancet. 2000;355:1404–1411. doi: 10.1016/s0140-6736(00)02139-5. [DOI] [PubMed] [Google Scholar]
- 4.Creasman W. Revised FIGO staging for carcinoma of the endometrium. Int J Gynaecol Obstet. 2009;105:109. doi: 10.1016/j.ijgo.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 5.Keys HM, Roberts JA, Brunetto VL, et al. A phase III trial of surgery with or without adjunctive external pelvic radiation therapy in intermediate risk endometrial adenocarcinoma: a Gynecologic Oncology Group study. Gynecol Oncol. 2004;92:744–751. doi: 10.1016/j.ygyno.2003.11.048. [DOI] [PubMed] [Google Scholar]
- 6.Morrow CP, Bundy BN, Kruman RJ, et al. Relationship between surgical-pathological risk factors and outcome in clinical stage I and II carcinoma of the endometrium: a gynecologic oncology group study. Gynecologic Oncology. 1991;40:55–65. doi: 10.1016/0090-8258(91)90086-k. [DOI] [PubMed] [Google Scholar]
- 7.Blake P, Swart AM, Orton J, et al. Adjuvant external beam radiotherapy in the treatment of endometrial cancer (MRC ASTEC and NCIC CTG EN.5 randomised trials): pooled trial results, systematic review, and meta-analysis. Lancet. 2009;373:137–146. doi: 10.1016/S0140-6736(08)61767-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merritt WM, Bar-Eli M, Sood AK. The dicey role of Dicer: implications for RNAi therapy. Cancer Res. 70:2571–2574. doi: 10.1158/0008-5472.CAN-09-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merritt WM, Lin YG, Han LY, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karube Y, Tanaka H, Osada H, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugito N, Ishiguro H, Kuwabara Y, et al. RNASEN regulates cell proliferation and affects survival in esophageal cancer patients. Clin Cancer Res. 2006;12:7322–7328. doi: 10.1158/1078-0432.CCR-06-0515. [DOI] [PubMed] [Google Scholar]
- 12.Chiosea S, Jelezcova E, Chandran U, et al. Up-regulation of dicer, a component of the MicroRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006;169:1812–1820. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiosea S, Jelezcova E, Chandran U, et al. Overexpression of Dicer in precursor lesions of lung adenocarcinoma. Cancer Res. 2007;67:2345–2350. doi: 10.1158/0008-5472.CAN-06-3533. [DOI] [PubMed] [Google Scholar]
- 14.Martin MG, Payton JE, Link DC. Dicer and outcomes in patients with acute myeloid leukemia (AML) Leuk Res. 2009;33:e127. doi: 10.1016/j.leukres.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 15.FIGO Annual report on the results of treatment in gynecological cancer. Int J Gynecol Obstet. 1989;28:189. [Google Scholar]
- 16.Giglio S, Monis PT, Saint CP. Demonstration of preferential binding of SYBR Green I to specific DNA fragments in real-time multiplex PCR. Nucleic Acids Res. 2003;31:e136. doi: 10.1093/nar/gng135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zighelboim I, Goodfellow PJ, Gao F, et al. Microsatellite instability and epigenetic inactivation of MLH1 and outcome of patients with endometrial carcinomas of the endometrioid type. J Clin Oncol. 2007;25:2042–2048. doi: 10.1200/JCO.2006.08.2107. [DOI] [PubMed] [Google Scholar]
- 19.Kitchener H, Swart AM, Qian Q, Amos C, Parmar MK. Efficacy of systematic pelvic lymphadenectomy in endometrial cancer (MRC ASTEC trial): a randomised study. Lancet. 2009;373:125–136. doi: 10.1016/S0140-6736(08)61766-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Memarzadeh S, Kozak KR, Chang L, et al. Urokinase plasminogen activator receptor: Prognostic biomarker for endometrial cancer. Proc Natl Acad Sci U S A. 2002;99:10647–10652. doi: 10.1073/pnas.152127499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morrison C, Zanagnolo V, Ramirez N, et al. HER-2 is an independent prognostic factor in endometrial cancer: association with outcome in a large cohort of surgically staged patients. J Clin Oncol. 2006;24:2376–2385. doi: 10.1200/JCO.2005.03.4827. [DOI] [PubMed] [Google Scholar]
- 22.Kodama J, Hasengaowa, Kusumoto T, et al. Prognostic significance of stromal versican expression in human endometrial cancer. Ann Oncol. 2007;18:269–274. doi: 10.1093/annonc/mdl370. [DOI] [PubMed] [Google Scholar]
- 23.Ino K, Yoshida N, Kajiyama H, et al. Indoleamine 2,3-dioxygenase is a novel prognostic indicator for endometrial cancer. Br J Cancer. 2006;95:1555–1561. doi: 10.1038/sj.bjc.6603477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giatromanolaki A, Koukourakis MI, Gatter KC, Harris AL, Sivridis E. BNIP3 expression in endometrial cancer relates to active hypoxia inducible factor 1alpha pathway and prognosis. J Clin Pathol. 2008;61:217–220. doi: 10.1136/jcp.2007.046680. [DOI] [PubMed] [Google Scholar]
- 25.Kamat AA, Merritt WM, Coffey D, et al. Clinical and biological significance of vascular endothelial growth factor in endometrial cancer. Clin Cancer Res. 2007;13:7487–7495. doi: 10.1158/1078-0432.CCR-07-1017. [DOI] [PubMed] [Google Scholar]
- 26.Devetzi M, Scorilas A, Tsiambas E, et al. Cathepsin B protein levels in endometrial cancer: Potential value as a tumour biomarker. Gynecol Oncol. 2009;112:531–536. doi: 10.1016/j.ygyno.2008.10.030. [DOI] [PubMed] [Google Scholar]
- 27.Falcon O, Chirino R, Leon L, et al. Low levels of cathepsin D are associated with a poor prognosis in endometrial cancer. Br J Cancer. 1999;79:570–576. doi: 10.1038/sj.bjc.6690090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 29.Chin K, DeVries S, Fridlyand J, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 30.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakagawa A, Shi Y, Kage-Nakadai E, Mitani S, Xue D. Caspase-dependent conversion of Dicer ribonuclease into a death-promoting deoxyribonuclease. Science. 328:327–334. doi: 10.1126/science.1182374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar MS, Pester RE, Chen CY, et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700–2704. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forbes SA, Bhamra G, Bamford S, et al. The Catalogue of Somatic Mutations in Cancer (COSMIC) Curr Protoc Hum Genet. 2008 doi: 10.1002/0471142905.hg1011s57. Chapter 10:Unit 10 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hill DA, Ivanovich J, Priest JR, et al. DICER1 mutations in familial pleuropulmonary blastoma. Science. 2009;325:965. doi: 10.1126/science.1174334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melo SA, Ropero S, Moutinho C, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. 2009;41:365–370. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Merritt WM, Lin YG, Spannuth WA, et al. Effect of interleukin-8 gene silencing with liposome-encapsulated small interfering RNA on ovarian cancer cell growth. J Natl Cancer Inst. 2008;100:359–372. doi: 10.1093/jnci/djn024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanguino A, Lopez-Berestein G, Sood AK. Strategies for in vivo siRNA delivery in cancer. Mini Rev Med Chem. 2008;8:248–255. doi: 10.2174/138955708783744074. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


