SUMMARY
Decoy Receptor 3 (DcR3), a secreted member of the Tumor Necrosis Factor (TNF) receptor superfamily, neutralizes three different TNF ligands: FasL, LIGHT, and TL1A. Each of these ligands engages unique signaling receptors which direct distinct and critical immune responses. We report the crystal structures of the unliganded DcR3 ectodomain and its complex with TL1A, as well as complementary mutagenesis and biochemical studies. These analyses demonstrate that DcR3 interacts with invariant backbone and side chain atoms in the membrane-proximal half of TL1A which supports recognition of its three distinct TNF ligands. Additional features serve as anti-determinants that preclude interaction with other members of the TNF superfamily. This mode of interaction is unique among characterized TNF:TNFR family members and provides a mechanistic basis for the broadened specificity required to support the decoy function of DcR3, as well as for the rational manipulation of specificity and affinity of DcR3 and its ligands.
INTRODUCTION
The mammalian immune system is a complex network regulated by co-stimulatory signals transmitted by a multitude of secreted and cell surface proteins. The human Tumor Necrosis Factor (TNF) superfamily includes at least 19 members and represents a major class of the co-stimulatory molecules. Most TNF ligands bind unique signaling receptors, and these specific ligand:receptor interactions direct important and diverse biological responses ranging from proliferation to apoptosis (Locksley et al., 2001; Pfeffer, 2003; Ware, 2003). In general, the TNF ligands are homotrimeric type-II membrane proteins with ectodomains that adopt a β-sandwich “jelly-roll” fold. The ectodomains of TNF receptors (TNFR) are elongated type-I membrane proteins containing tandem repeats of one to six cysteine-rich domains (CRDs), which bind the interprotomer grooves formed between adjacent ligand monomers. This mode of interaction results in ligand:receptor assemblies with 3:3 stoichiometries that span two interacting cells. The ligand-mediated clustering of TNFRs directs the recruitment of signaling adapter proteins, such as TNF receptor associated factors (TRAFs) or death domain adapter proteins, and initiates diverse downstream signaling pathways.
Some TNFRs are also produced as soluble forms due to alternate mRNA splicing or selective proteolytic cleavage (Cheng et al., 1994; Locksley et al., 2001; Van Zee et al., 1992). The metalloprotease-mediated cleavage of two TNFα receptors (TNFR1 and TNFR2) has been reported to attenuate proinflammatory TNFα–associated signaling, possibly by reducing the amount of cell surface receptors and by the sequestration/neutralization of free ligands (Galon et al., 2000; Van Zee et al., 1992). By analogy, soluble antagonists that block specific pathways have found wide-spread clinical use. For instance, Etanercept, a soluble fusion protein formed between the ectodomain of TNF receptor 2 (TNFR2) and the Fc region of human immunoglobulin (Ig), is a leading treatment for autoimmune disorders like rheumatoid arthritis (Moreland et al., 1997).
In contrast to most members of the TNFR superfamily, the gene for Decoy Receptor 3 (DcR3, also known as TNFRSF6B) does not encode a cytoplasmic or transmembrane segment, resulting in an obligate secreted protein of 300 amino acids including the signal peptide (Figure 1A). The human DcR3 gene maps to a chromosomal region (20q13.3) associated with both cancer and autoimmune diseases (Bai et al., 2000; Kugathasan et al., 2008; Muleris et al., 1995). DcR3 orthologs have not been identified in the murine genome, suggesting an important difference between the murine and human immune systems (You et al., 2008). In healthy human subjects, low levels of DcR3 mRNA are detected in a broad range of tissues (Bai et al., 2000; Pitti et al., 1998), while the expression of DcR3 is significantly elevated in patients with a range of cancers and autoimmune diseases (Funke et al., 2009; Pitti et al., 1998; Wu et al., 2003). Notably, DcR3 is capable of binding and neutralizing three TNF ligands: FasL, LIGHT and TL1A (Migone et al., 2002; Pitti et al., 1998; Yu et al., 1999). These three ligands belong to the group of conventional TNF ligands, members of which are characterized by a compact trimeric assembly resembling an inverted-bell shape (Compaan and Hymowitz, 2006). However, sequence similarity among FasL, LIGHT and TL1A is modest (~30% identity) and each ligand binds different signaling receptors, which trigger distinct cellular responses (Figure 1B). Recently, the C-terminal region of DcR3, which resides outside the TNF ligand binding domain, was reported to bind heparan sulfate proteoglycans (HSPG) and trigger reverse signaling in antigen presenting cells (APC) (Chang et al., 2006; You et al., 2008).
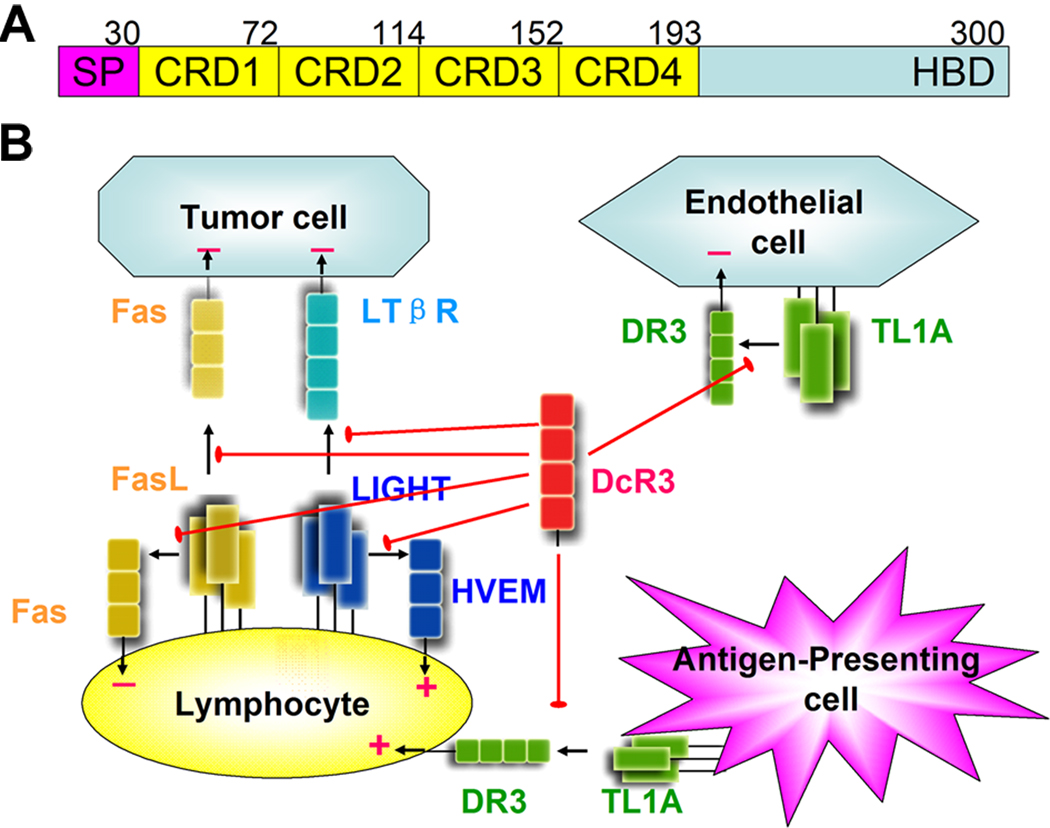
Figure 1. DcR3 is a secreted TNF receptor that neutralizes three TNF ligands and blocks multiple signaling pathways.
(A) DcR3 domain structure. DcR3 is composed of a signal peptide (SP), followed by 4 cysteine-rich domains (CRD) and a heparan-binding domain (HBD) at the C-terminus. The numerals above the schematic refer to the sequence numbers of the last residue in each domain. The DcR3 protein in this report includes residues V30-S195.
(B) TNF ligands (trimers of rectangles), TL1A (green), LIGHT (blue) and FasL (yellow), bind distinct signaling TNF receptors (cubes), DR3 (green), HVEM (blue), LTβR (cyan) and Fas (yellow), directing the proliferation (+) or apoptosis (−) of a variety of cell types. The secreted decoy receptor DcR3 (red) plays a complicated regulatory function by neutralizing the three TNF ligands and blocking multiple signal transduction pathways.
Defining the precise mechanistic contributions of DcR3 to host immunity is complicated by the diverse functions of its three TNF ligands (Figure 1B). LIGHT and its signaling receptor, HVEM, are expressed on the surface of T cells and are essential for the proliferative signaling triggered by T cell-T cell interactions (Wang et al., 2001). The DcR3-mediated blockade of LIGHT was shown to reduce T cell activity in vitro and to ameliorate graft-versus-host responses in vivo (Zhang et al., 2001). In addition, the engagement of TL1A, expressed on antigen presenting cells (Bamias et al., 2006), with its signaling receptor DR3, expressed on activated T cells, promotes T cell proliferation (Migone et al., 2002) and appears essential for the development of several inflammatory diseases including Crohn’s disease and rheumatoid arthritis (Bull et al., 2008; Meylan et al., 2008; Pappu et al., 2008). Inhibition of TL1A-associated pathways, including the administration of DcR3, has been suggested as a promising approach for the treatment of these autoimmune diseases (Young and Tovey, 2006).
The third DcR3 ligand, FasL, and its signaling receptor, Fas, are both highly expressed on the surfaces of activated T cells. In contrast to the immune-stimulatory consequences of the LIGHT:HVEM and TL1A:DR3 interactions, FasL:Fas engagement induces apoptosis of T cells and curbs the activation of immunological responses (Siegel et al., 2000). Disruption of the FasL:Fas interaction is a major contributor to autoimmune lymphoproliferative syndrome (ALPS) in humans (Straus et al., 1999). By competing with Fas and blocking FasL signaling, DcR3 might accelerate overactive immune responses and contribute to the development of autoimmune disorders (Funke et al., 2009; Hayashi et al., 2007).
It is notable that elevated expression of DcR3 is closely correlated with the progression of a variety of cancers (Bai et al., 2000; Pitti et al., 1998; Takahama et al., 2002), and tumor-secreted DcR3 has been proposed to facilitate immune evasion by blocking the apoptotic signals transmitted from FasL and LIGHT expressed on lymphocytes (Ashkenazi, 2002). Furthermore, soluble TL1A induces apoptosis in vascular endothelial cells in an autocrine manner, and neutralization of TL1A by DcR3 was reported to promote neovascularization and tumor growth (Yang et al., 2004) (Figure 1B). These tumor promoting activities suggest that DcR3 antagonists might provide new avenues for cancer therapy (Ashkenazi, 2002).
In order to define and potentially manipulate the functional contributions of DcR3, it is essential to characterize the determinants responsible for its relaxed specificity and decoy strategies. In this work, we focus on the interactions of DcR3 with its three conventional TNF ligands, TL1A, FasL and LIGHT, and report the crystal structures of the human DcR3 cysteine-rich domains and two crystal forms of the human TL1A:DcR3 complex. These studies reveal overall structural similarity to the conventional TNF:TNFR assemblies and highlight the unique features responsible for the broadened specificity of DcR3. In particular, the 3rd CRD (CRD3) of DcR3 forms weak and seemingly non-specific contacts with the upper region of TL1A, which is thought to be a major determinant of specificity in the remainder of the TNF superfamily. Most notably, CRD2 of DcR3 makes major contacts with backbone atoms in the lower region of TL1A and it is the recognition of these invariant features that supports interactions with the three distinct ligands. Additional structural features in CRD2 of DcR3 are responsible for restricting the specificity to these three ligands. These observations are consistent with a series of complementary biochemical and mutagenesis analyses. Our findings provide a basis for understanding and exploiting the roles of DcR3 in a wide range of physiological, pathological and therapeutic settings (Figure 1B).
RESULTS
Overall structure of the TL1A:DcR3 complex
We determined the crystal structures of human DcR3 cysteine-rich domain (CRD) protein and its complex with human TL1A ectodomain in cubic and trigonal crystal forms (Table 1). The overall organization of the TL1A:DcR3 complex is similar to that of the canonical TNF ligand:receptor complex structures (i.e., LTα:TNFR1; PDB code 1TNR and TRAIL:DR5; PDB code 1D0G (Figure S1)) (Banner et al., 1993; Hymowitz et al., 1999; Mongkolsapaya et al., 1999), with each DcR3 molecule contacting the groove between two TL1A subunits, resulting in an assembly with overall 3:3 stoichiometry (Figure 2A,B). Despite distinct lattice packing, the TL1A:DcR3 complexes are highly similar in the two crystal forms (RMSDs of 0.5 Å over 143 TL1A Cα atoms and 1.2 Å over 107 DcR3 Cα atoms). The most significant difference is the ~10° deviation in the angle between CRD2 and CRD3 in DcR3 (Figure 2E).
Table 1.
Statistics for data collection and refinement
| Data collection | |||
|---|---|---|---|
| TL1A:DcR3 Trigonal |
TL1A:DcR3 Cubic |
Unliganded DcR3 |
|
| Space group | P321 | P4332 | H32 |
| Unit-cell lengths (Å) | a = b = 74.89 c = 143.13 | a = b = c = 161.08 | a = b = 130.02 c = 96.03 |
| Wavelength used (Å) | 0.9792 | 0.9791 | 0.9791 |
| Resolution range(Å) | 2.45 – 50.00 | 2.95 – 50.00 | 2.70 – 50.00 |
| Unique reflections (N) | 17791 | 15595 | 8677 |
| Redundancy1 | 7.7 (7.6) | 20.9 (21.2) | 7.3 (5.7) |
| Completeness (%)1 | 99.9 (100.0) | 99.8 (100.0) | 99.6 (99.9) |
| Rmerge1,2 | 0.131 (0.99) | 0.092 (0.898) | 0.073 (0.793) |
| <I/sigmaI>1 | 15.3 (3.2) | 36.6 (3.9) | 18.9 (1.2) |
| Refinement | |||
| Resolution rangse (Å) | 2.45 –36.23 | 2.95 – 25.00 | 2.90(2.80/3.10) – 20.00* |
| Rwork 1,3 | 22.3 (23.5) | 23.2 (32.1) | 28.5 (33.5) |
| Rfree1 | 26.2 (31.4) | 26.1 (37.8) | 31.5 (34.2) |
| Completeness (%) | 99.9 (100.0) | 98.86 (99.73) | 88.17 (18.78) |
| Average B-factors (Å2) | 79.9 | 84.7 | 36.3 |
| R.m.s. bonds (Å) | 0.006 | 0.007 | 0.008 |
| R.m.s. angles (°) | 0.540 | 1.126 | 1.212 |
Values in parenthesis correspond to the highest resolution bin.
Rwork = Σ|Fc − Fo|/ΣFo.
Data after anisotropy correction. (Ellipsoidal truncation with resolution limits of 2.8 Å along the c* directions, 3.1 Å along the a* and b* directions.)
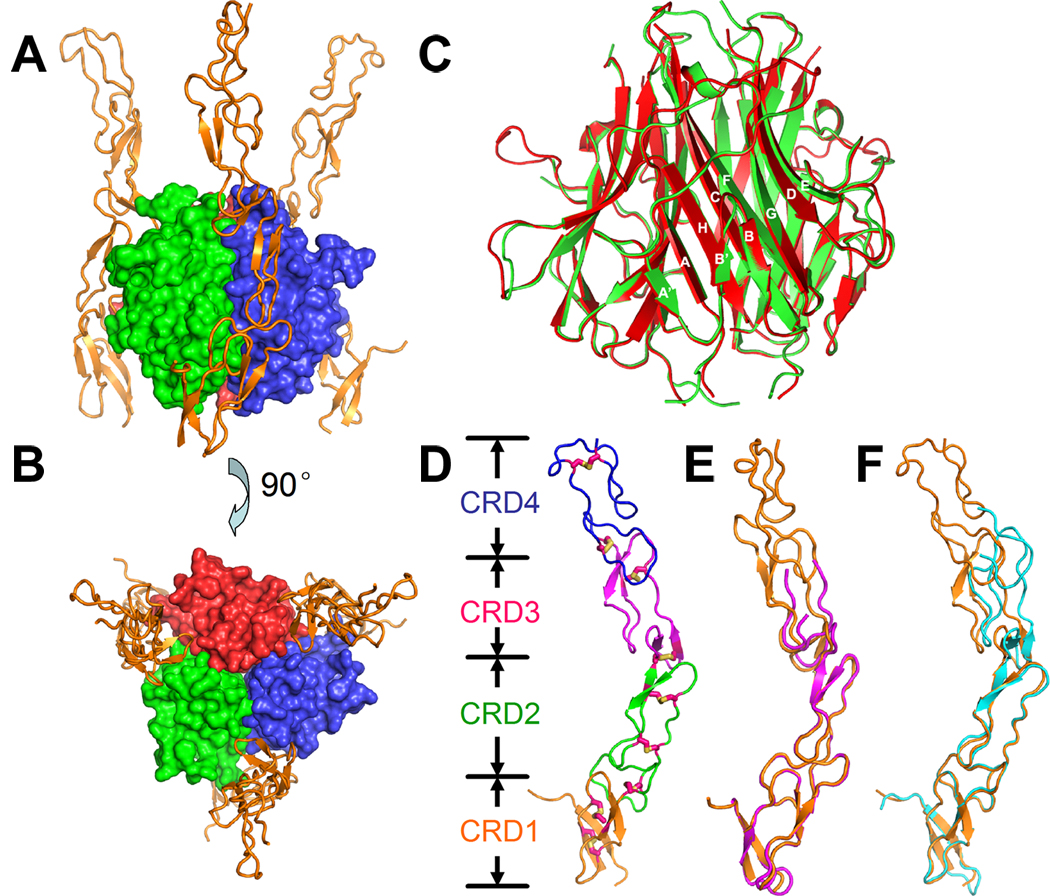
Figure 2. Overall structure of DcR3 and TL1A.
Side-view (A) and top-view (B) of the TL1A:DcR3 complex from the trigonal crystal form. TL1A is a tight homo-trimer, with its three subunits colored in green, blue and red. DcR3 (orange) binds the grooves formed between two TL1A subunits. The structure in (B) is rotated 90° around a horizonal axis relative to (A). (C) Structural superposition of DcR3-bound (red) and unbound (green) TL1A. The TL1A β-strands and connecting loops are labeled according to convention. (D) The structure of DcR3 extracted from the trigonal TL1A:DcR3 complex. DcR3 has 4 cysteine-rich domains (CRD), colored in orange, green, magenta, and blue. Each CRD contains 2 to 3 disulfide bonds, represented as sticks. (E, F) The structure of DcR3 from the trigonal-packing TL1A:DcR3 complex (orange) superimposed with that from the cubic-packing complex (magenta, E) or the unliganded DcR3 (cyan, F).
See also Supplementary Figure 1.
In both the cubic and trigonal crystal forms of the complex, the structure of bound TL1A closely resembles that of the previously reported unbound TL1A, with a typical jelly-roll domain composed of two β-sheets formed by the A′AHCF and B′BGDE strands (Figure 2C) (Jin et al., 2007; Zhan et al., 2009). The bound and unbound structures of the TL1A trimer superpose with an RMSD of 0.584 Å over 402 Cα atoms, indicating that TL1A does not undergo significant tertiary or quaternary reorganization upon binding DcR3 (Figure 2C). DcR3 is an elongated molecule composed of four tandem CRDs, which adopt a slightly kinked organization with a hinge angle between CRD2 and CRD3 of approximately 40°, 30° and 25° in the trigonal complex, cubic complex and unliganded DcR3 crystal forms, respectively (Figure 2D,E,F). This hinge movement is tangential to the TL1A trimer, and reflects modest intrinsic flexibility between CRD2 and CRD3 which is independent of ligand binding. Both CRD3 and CRD4 lack a canonical disulfide bond present in conventional cysteine-rich domains (Figure S2), and exhibit dynamic behavior; electron density for CRD3 and CRD4 in both the cubic form of the complex and the unliganded DcR3 is poor or undetectable and cannot be fully modeled (Figure 2E,F). Given the modest resolution and quality of the electron density, the disulfide bond connectivity of DcR3 modeled in the crystal structures was verified by non-reducing tryptic digestion and FT-ICR-mass spectrometry (Figure S2).
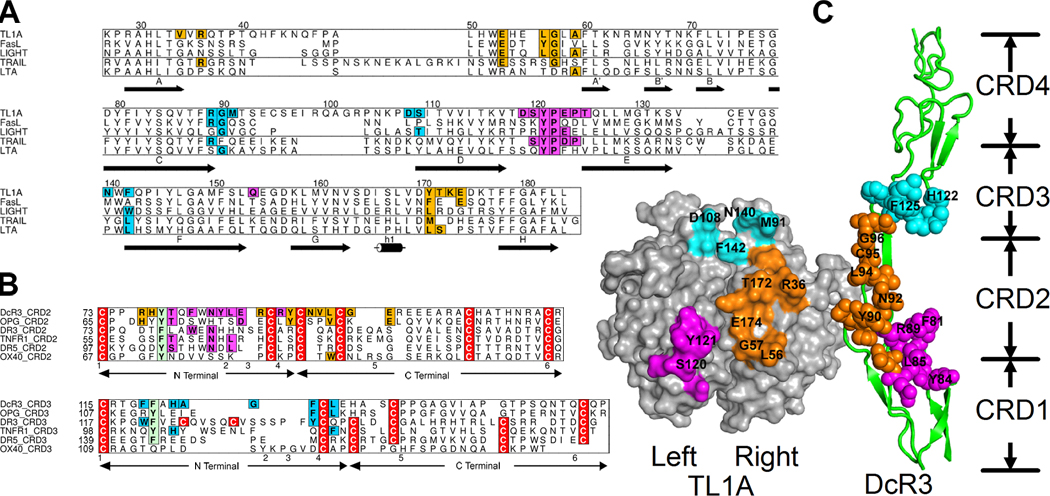
The TL1A:DcR3 interface in the trigonal-packing complex is highlighted in Figure 3 by rotating one DcR3 molecule 180° about a vertical axis. The DcR3-binding residues on TL1A, defined as those burying greater than 20% of their solvent accessible surface area in the interface, can be grouped into three separate regions: the DE loop (the loop connecting D/E β-strands, Magenta in Figure 3A,C), the AA’/GH loops (Orange) and the CD/EF loops (Cyan). This ligand:receptor recognition pattern is unique compared to previously reported structures of TNF:TNFR complexes, and provides a mechanistic basis for the ability of DcR3 to selectively neutralize FasL, LIGHT and TL1A, while precluding interactions with other TNF ligands.
Figure 3. Interaction interface between DcR3 and TL1A.
Structure-based sequence alignment of TNF ligands (A) and receptors (B). The three separate binding patches in the TL1A:DcR3 interface are colored in magenta, orange and cyan; homologous residues in the other ligands/receptors are colored accordingly. In the ligand alignment (A), the numbering at the top is according to TL1A sequence. The secondary structure of TL1A is presented at the bottom, with arrows indicating β–sheets and a cylinder representing the sole α-helical segment. The labels of β–strands are in accordance to those in Figure 2C. In the receptor alignment (B), arrowed lines below the alignment indicate boundaries of N/C-terminal regions of a cysteine-rich domain.
(c) “Open book” view of the TL1A:DcR3 interface. One DcR3 molecule is rotated 180° around a vertical axis to expose the interfaces. The residues are colored as in the alignments of (A) and (B).
Reduced contacts in the specificity-determining membrane-distal region
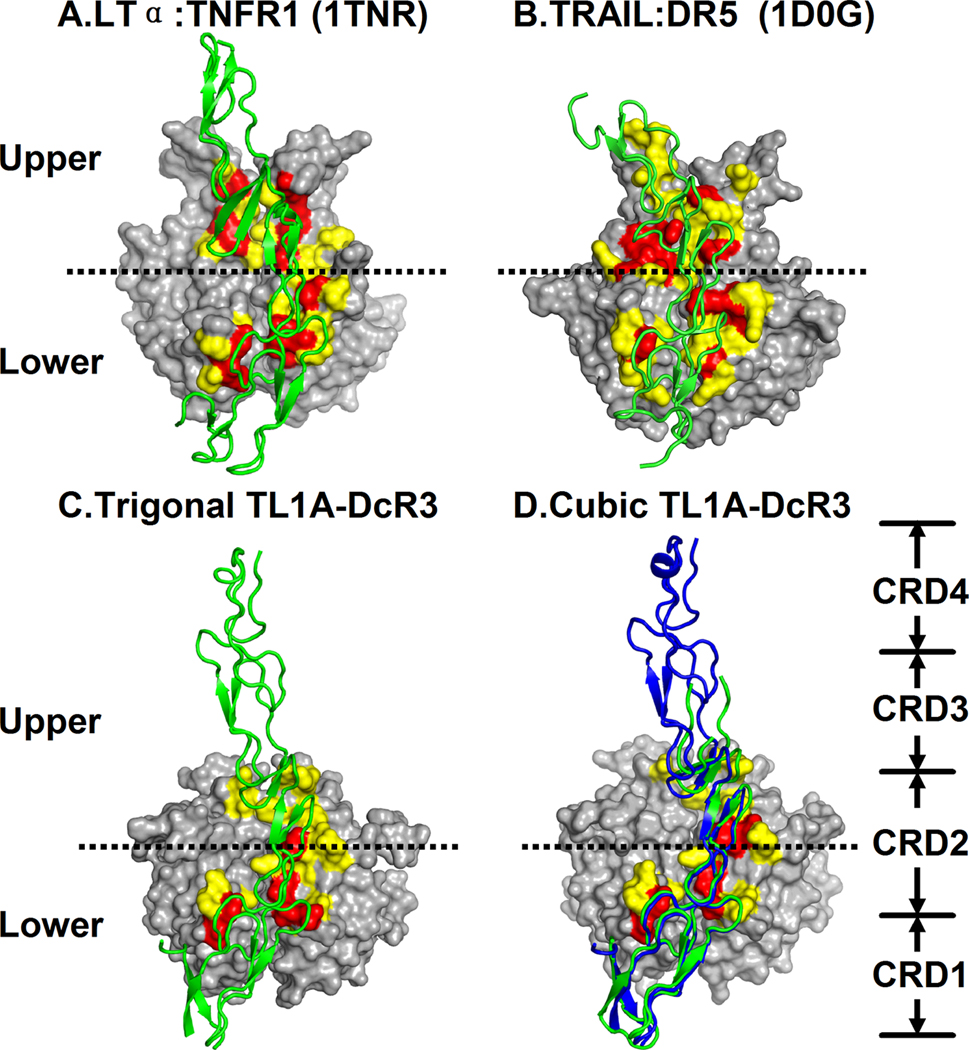
Comparison of the LTα:TNFR1 and TRAIL:DR5 structures suggests that the receptor binding surfaces of the two TNF ligands, LTα and TRAIL, can be divided into a lower region proximal to the ligand membrane (below the dotted lines in Figure 4 panels, hereafter referred as “lower”) and an upper region distal to the membrane (above the dotted lines in Figure 4 panels, hereafter referred as “upper”) (Banner et al., 1993; Hymowitz et al., 1999; Kim et al., 2003). In the membrane-proximal lower region, CRD2 of the two receptors adopt highly similar backbone conformations and interact with the same loop segments (DE and AA’ loops) in their cognate ligands. It has been suggested that these conserved interactions play an important role in defining the overall geometric and architectural features of the ligand:receptor assemblies (Hymowitz et al., 1999). In contrast, in the membrane-distal upper region, CRD3 of the TNFR1 and DR5 receptors adopt different conformations and interact with distinct less conserved areas at the top of the ligands. These interactions in the upper region of the ligand are thought to control the specificities between different members of the TNF and TNFR superfamilies (Banner et al., 1993; Bodmer et al., 2002; Hymowitz et al., 1999).
Figure 4. Comparison of receptor binding interfaces in TNF ligands.
(A) LTα:TNFR1 interface (PDB ID: 1TNR); (B) TRAIL:DR5 interface (PDB IDs: 1D0G,1D4V); (C) Trigonal TL1A:DcR3 interface. In each complex interface, two TNF ligand subunits are displayed by a surface representation and one TNF receptor by a green ribbon. Ligand residues with 20% to 60% of their solvent accessible surface area buried in the interface are colored in orange, and those 60% to 100% buried are colored red. (D) TL1A and DcR3 in the cubic TL1A:DcR3 complex are displayed by surface representation and green ribbon representations as in panels A–C. The blue ribbon represents the DcR3 molecule from the trigonal TL1A:DcR3 complex superimposed onto the cubic TL1A:DcR3 complex.
To compare the TL1A:DcR3 complex with the LTα:TNFR1 and TRAIL:DR5 structures, the ligand residues contributing to the binding interfaces in the three complexes are categorized according to their buried surface area in Figure 4 (20–60% in yellow and 60–100% in red). In both LTα:TNFR1 and TRAIL:DR5, the upper and lower binding regions make significant contributions to the binding interfaces (Figure 4A,B) (Banner et al., 1993; Hymowitz et al., 1999). In contrast, the most heavily buried residues in DcR3-binding interface cluster to the lower half of the TL1A, while the upper part of TL1A makes only modest contact with DcR3 (Figure 4C,D). In the trigonal crystal form of TL1A:DcR3 complex, CRD3 of DcR3 interacts with the bottoms of the CD and EF loops located in the upper region of the groove formed between two TL1A subunits (Cyan in Figure 3). In the cubic crystal form of the complex, CRD3 is more disordered and tilted away from TL1A (Figures 2,4), precluding most of the contacts observed in this region in the trigonal form. The different orientations of CRD3 in the two TL1A:DcR3 crystal forms lead to varied interactions in this upper region, suggesting that these contacts do not make significant energetic contributions to binding. This idea is supported by our previous studies demonstrating that substitutions of multiple residues in the upper region of TL1A do not affect DcR3 binding (Zhan et al., 2009). In the LTα:TNFR1, TRAIL:DR5 and TL1A:DcR3 complexes, the membrane-distal binding upper patches all primarily involve CRD3. However, structure-based sequence comparison shows that the CRD3 in DcR3 is significantly shorter than its counterparts in other receptors (Figure 3B) and this difference may contribute to the reduced contacts at the upper region of TL1A. Together, these observations indicate that DcR3 does not form critical interactions with the “specificity-determining” membrane-distal region of TL1A, consistent with the ability of DcR3 to neutralize three distinct TNF ligands.
Invariant binding determinants in the membrane-proximal region
In the membrane-proximal lower region, both the trigonal and cubic crystal forms of the TL1A:DcR3 complex exhibit virtually identical binding interactions, with CRD2 of DcR3 making two sets of contacts with two adjacent TL1A subunits (Magenta and Orange in Figure 3). As illustrated in Figure 3, one major binding patch (Magenta) is formed between the DE loop of the leftmost TL1A subunit and the N-terminal segment of CRD2 of DcR3. In this patch, the hydroxyl group of Y121 in TL1A forms a hydrogen bond with the backbone of DcR3 and this residue also makes hydrophobic contacts with N83, L85 and R89. Additional polar interactions are formed between the backbone atoms of S120, E123 and P124 in TL1A and Y84 (backbone) and R89 (side chain) in DcR3. These polar interactions are surrounded by additional hydrophobic contacts involving S120, P122 in TL1A and F81, Y84 in DcR3.
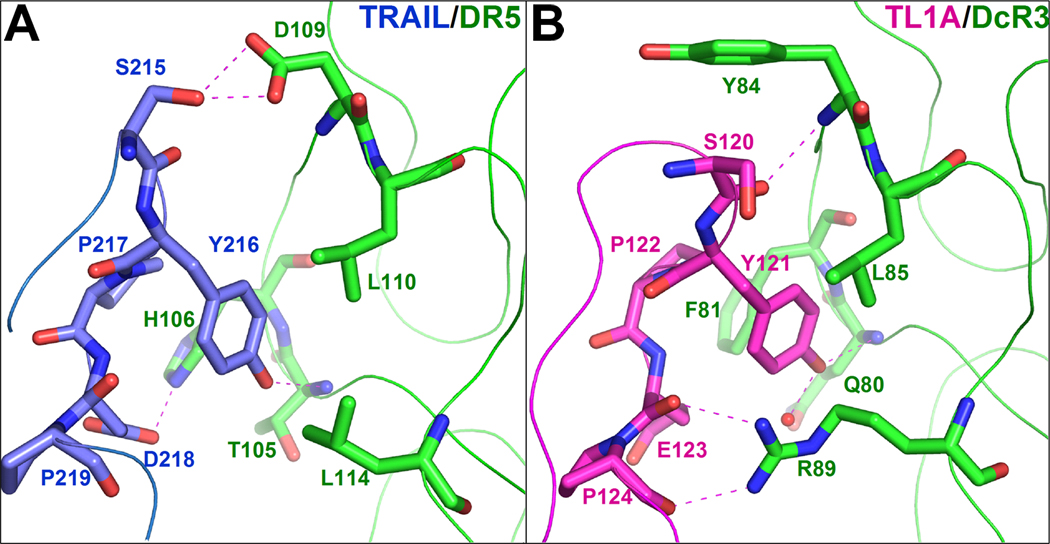
Y121 in the DE loop of TL1A is nearly invariant in the conventional TNF ligands (Zhan et al., 2009). Mutations of this Tyr residue in TL1A and other TNF ligands, including TNFα, LTα, FasL, LIGHT and TRAIL, all severely compromise receptor binding affinities (Goh et al., 1991; Hymowitz et al., 2000; Rooney et al., 2000; Schneider et al., 1997; Yamagishi et al., 1990; Zhan et al., 2009). Following this critical Tyr, Pro122 in TL1A is also conserved in most conventional TNF ligands and provides additional contacts to DcR3. However, the other residues in the DE loop sequences are not conserved among the three ligands of DcR3 (TL1A, FasL and LIGHT). Interestingly, the sequence of TRAIL in this region is almost identical to TL1A (Figure 3A). Thus, it is notable that DcR3 does not bind TRAIL, but instead recognizes TL1A, LIGHT and FasL, which possess more divergent sequences. In TRAIL:DR5, two hydrophobic residues (L110 and L114) and two hydrophilic residues (H106 and D109) from DR5 interact with the side chains of the TRAIL DE loop (S215, Y216 and D218), all of which are conserved in TL1A (S120, Y121 and E123, correspondingly) (Figures 3A,5). However, on the receptor’s side, H106, D109 and L114 of DR5 are substituted with F81, Y84 and R89 in DcR3, respectively (Figures 3B,5). These changes in chemical-physical properties allow DcR3 to form polar interactions with the backbone of the ligand DE loop, and surround this interface with more extensive hydrophobic contacts. The recognition of TL1A DE loop backbone atoms by DcR3 clearly contrasts the recognition of TRAIL sidechain atoms by DR5. Consistent with this model, our previously reported mutagenesis work demonstrated that the S120A/E123A double mutant of TL1A did not significantly decrease the binding affinity with DcR3, as would be expected if the TL1A backbone atoms were involved (Zhan et al., 2009). Thus, in order to function as a generic receptor that neutralizes multiple ligands, we propose that DcR3 possesses broadened specificity as the consequence of recognizing only the invariant side chain (i.e., Y121) and backbone binding determinants in the DE loop. The inability of DcR3 to bind TRAIL may be the consequence of anti-determinants discussed immediately below.
Figure 5. The DE loop interfaces in the TRAIL:DR5 and TL1A:DcR3 complexes.
DR5 (Green in A) makes all of its polar interactions with the side chain atoms of the TRAIL DE loop (Blue). In contrast, DcR3 (Green in B) only forms polar interactions with the invariant backbone atoms and a highly conserved tyrosine residue in the TL1A DE loop (Magenta). The binding residues are represented as sticks, and polar interactions are highlighted by purple dashed lines. The protein backbones are represented by ribbons.
See also Supplementary Figure 3.
Anti-determinants limit more extensive promiscuity
As illustrated in Figure 3, the other major membrane-proximal binding interface (Orange) is formed between the AA’ and GH loops of the rightmost TL1A subunit and CRD2 of DcR3. In the C-terminal end of the TL1A AA’ loop, the physical-chemical properties of the residues corresponding to L56 and G57 are highly conserved only among the DcR3 binding ligands (Figure 3A) (Zhan et al., 2009), and may serve to narrow the specificity of DcR3. These two residues bury ~70% of their solvent accessible area in the interface and form hydrophobic interactions with Y90 in DcR3. The residues corresponding to Y90 in TNFR1 (S72) and DR5 (R115) are both hydrophilic residues which make polar contacts with the C-terminal end of the AA’ loops of their cognate ligands. Mutations of charged residues in the C-terminal end of the AA’ loops of TNFα, LTα and CD40L all significantly reduced receptor binding affinities (Hymowitz et al., 2000), suggesting critical hydrophilic interactions in this region for these receptor-ligand partners. In contrast, our mutagenesis studies demonstrated that the C-terminal end of the AA’ loop of TL1A’s is not critical for DcR3 binding (Zhan et al., 2009). Thus the unique hydrophobic interactions between DcR3 and C-terminal end of the AA’ loop of TL1A may represent yet another specialized mechanistic feature utilized by DcR3 to recognize and discriminate TNF ligands. Specifically, while the hydrophobic residues in this region do not make important contributions to TL1A:DcR3 binding affinity, they instead may serve as anti-determinants to impair the interactions between DcR3 and TNF ligands possessing hydrophilic AA’ loops.
At the center of the groove formed between two TL1A subunits, R36 from the N-terminal segment of the AA’ loop and T172-E174 at the tip of the GH loop are extensively buried in the binding interface and make polar interactions with the C-terminal segment of CRD2 of DcR3 (Top of the orange patch in Figure 3). Consistent with this structural observation, our previous study revealed that the E174A mutation in TL1A significantly decreased binding to DcR3 (Zhan et al., 2009). This hydrophilic region is not conserved between TL1A and the other TNF ligands, suggesting important contacts that are unique to the TL1A:DcR3 interaction. Given this sequence divergence, it is likely that DcR3 makes diverse interactions with TL1A, FasL and LIGHT in this region, which may partially account for the different DcR3 binding affinities exhibited by these three TNF ligands (see below).
Self-assembly of DcR3 molecules
DcR3 cysteine-rich domains pack as parallel dimers in the ligand-free crystal and parallel trimers in the trigonal complex crystal (Figure S1). Consistent with the lack of conserved quaternary organization in the crystalline state, sedimentation equilibrium analyses of DcR3 cysteine-rich domains showed only very weak self-association, with an estimated equilibrium dissociation constant (Kd) greater than 2.4 mM, suggesting that at physiological concentrations (pM concentration in tumor patient serum (Wu et al., 2003)) unliganded DcR3 is predominantly monomeric (Figure S1).
Side chains of S120 and E123 residues in TL1A are critical for DR3 binding
In the interface formed between DE loop of TL1A and CRD2 of DcR3, the backbone atoms of TL1A residues S120 and E123 form polar interactions with DcR3. The S120A/E123A TL1A double mutant only modestly reduced DcR3 binding response by approximately 35% (Figure S3). In contrast, the S120A/E123A TL1A mutant significantly reduced the DR3 binding response by almost 90% (Figure S3). Thus, the side chain atoms of S120 and E123 in TL1A make important contributions to the TL1A:DR3 interaction, while the TL1A:DcR3 interaction is more dependent on recognition of invariant main chain atoms of TL1A at these positions.
Interaction of DcR3 cysteine-rich domains with immobilized TNF ligands
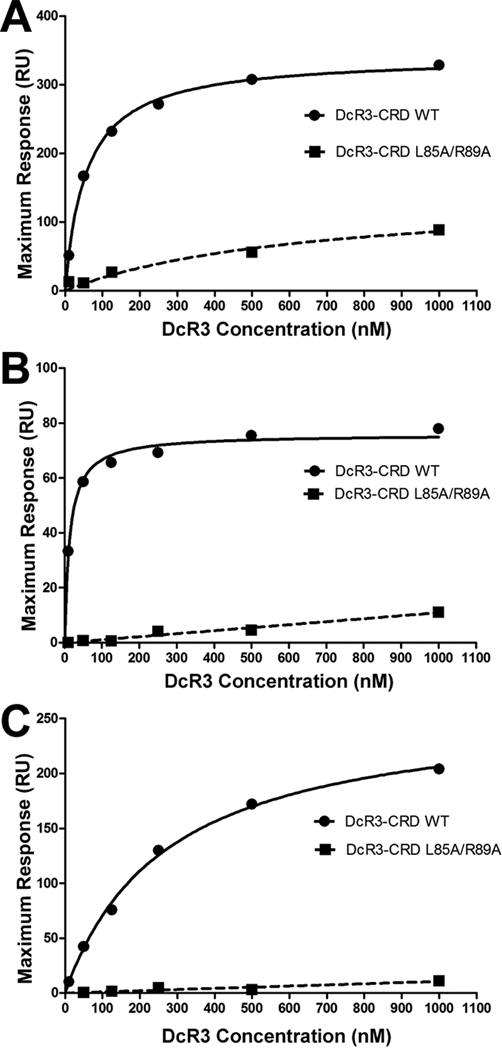
Surface plasmon resonance (SPR) analysis revealed that wild type (WT) human DcR3 CRDs bind immobilized human TL1A (Zhan et al., 2009), LIGHT (R&D Systems) and FasL (R&D Systems) ectodomains with Kds of 56.4±3.7, 14.0±2.0, and 271.4±24.4 nM, respectively (Figure 6). The structure of the TL1A:DcR3 complex shows that side chain atoms of L85 and R89 in CRD2 of DcR3 interact with the TL1A DE loop in the membrane-proximal region of the ligand. To investigate whether these DcR3 residues are important determinants for all three TNF ligands, the L85A/R89A double mutant of DcR3 was expressed and purified, and its affinities for TL1A, LIGHT and FasL were determined as for wild type DcR3. The binding of the L85A/R89A mutant DcR3 to all three ligands is significantly lower than that of WT DcR3 and is too weak to yield confident Kd values; these interactions cannot be saturated at even the highest concentration of DcR3 examined (1000 nM), suggesting affinities in the µM range or weaker. These results suggest that all three ligands utilize a common mode of DcR3 recognition.
Figure 6. TNF ligand binding affinities of DcR3.
(A–C) The affinities of wild type (WT) DcR3 cysteine-rich domains (solid line with circles) and the L85A/R89A DcR3 double mutant (dotted line with squares) to immobilized TL1A (A), LIGHT (B) and FasL (C) were measured by surface plasmon resonance (SPR). Steady state analysis reveals that TL1A, LIGHT and FasL bind WT DcR3 CRDs with Kd values of 56.4±3.7, 14.0±2.0, and 271.4±24.4 nM, respectively. The L85A/R89A DcR3 double mutant exhibited significantly lower binding responses to each ligand.
DISCUSSION
The overall organization of the TL1A:DcR3 interaction is similar to the previously characterized conventional TNF ligand:receptor complexes, LTα:TNFR1 (Banner et al., 1993) and TRAIL:DR5 (Hymowitz et al., 1999; Mongkolsapaya et al., 1999), with an elongated receptor bound at the groove formed by adjacent subunits in the compact homotrimeic ligand, resulting in an assembly with 3:3 ligand:receptor stoichiometry. The extracellular ligand:receptor binding geometry has been suggested to impose similar constrains on the cytoplasmic scaffolding and to facilitate the recruitment of trimeric signaling adapter proteins like TRAFs or death domain proteins (Chattopadhyay et al., 2009). The extracellular C-terminal ends of three TNFRs bound by conventional TNF ligands are separated by 33 Å and 52 Å in the LTα:TNFR1 and TRAIL:DR5 complexes, respectively. The C-termini of each DcR3 CRD construct are separated by approximately 65Å in the TL1A:DcR3 complex, with a modestly flexible hinge between CRD2 and CRD3 that results in a range of subtly different conformations (Figure 2). The greater separation and intrinsic flexibility present in ligand-bound DcR3 may reflect the fact that DcR3, being a decoy receptor, does not directly trigger TNF ligand-mediated signal transduction pathways and thus need not be compatible with the geometric restraints of cytoplasmic signaling molecules.
Full-length DcR3 was reported to bind HSPG via a C-terminal heparan-binding domain. This interaction transmits reverse signaling to dendritic cells by cross-linking cell-surface proteoglycans (Chang et al., 2006; You et al., 2008). However, all studies have utilized dimeric and bivalent Ig-fusion proteins of DcR3. To our knowledge, it is not known whether endogenous monomeric DcR3 is capable of cross-linking proteoglycans and subsequently transmitting reverse signaling in vivo. The present work shows that the self-association of DcR3 CRDs is very weak, with an estimated Kd in the mM range, which contrasts the ligand-independent association of TNFR1 (Naismith et al., 1995) (see supplementary discussion). After binding HSPG through its C-terminal HBD, DcR3 alone may not be able to mediate the cross-linking of the HSPGs; however, binding to the TNF ligands would assemble three DcR3 molecules into a complex that could potentially support crosslinking. Thus, our results suggest that in vivo the HSPG crosslinking and reverse signaling function associated with the C-terminal end of DcR3 may require the simultaneous binding of TNF ligands through its N-terminal cysteine-rich domains.
Despite sharing similar organizational features with the conventional TNF:TNFR assemblies, the binding interface observed in the TL1A:DcR3 complex exhibits important differences relative to other conventional complexes, and provides a mechanistic basis for the broadened specificity and decoy function of DcR3. Our studies demonstrate that DcR3 lacks the specificity-defining determinants for interaction with the membrane-distal region of TL1A, and forms critical interactions with main chain and conserved side chain atoms in the membrane-proximal region of its TNF ligands. This unique strategy allows DcR3 to recognize and neutralize multiple TNF ligands through recognition of invariant determinants. Furthermore, DcR3 makes hydrophobic contacts with the residues conserved only in FasL, LIGHT and TL1A, which may serve as anti-determinants to prevent more wide-spread promiscuity.
The relaxed specificity of DcR3 is the direct consequence of its mode of ligand recogniton. Notably, osteoprotegerin (OPG), the only other secreted decoy TNF receptor, also displays promiscuity by neutralizing both TRAIL and RANKL (Pitti et al., 1998). Interestingly, the sequence of DcR3 is most similar to that of OPG (Figure 3B), and both share a short CRD3, suggesting that reduced contributions by CRD3 might be utilized by both DcR3 and OPG to broaden their ligand binding specificity. In contrast, DcR1, a GPI-linked membrane associated decoy TNFR, specifically binds and neutralizes a single ligand (TRAIL). Based on sequence considerations, ligand binding residues in CRD2 of DcR3 are not conserved in DcR1. Additionally, DcR1 does not share the shortened CRD3 present in DcR3, suggesting that DcR1 behaves more similarly to the conventional signaling TNFRs and makes important interactions with both the membrane-proximal and membrane-distal regions of TRAIL.
DcR3 interaction patterns among TL1A, LIGHT and FasL
SPR analysis demonstrates that wild type DcR3 binds LIGHT approximately 4 and 20 times tighter than TL1A and FasL, respectively (Figure 6). This variation in binding affinities is expected to contribute to the in vivo preferences for TNF ligand binding and neutralization by DcR3; however, these preferences will also be critically dependent on the expression levels and expression patterns of the individual ligands.
Dissection of the interface between DcR3 and TL1A suggests that the most critical binding interface is formed between the DE loop of TL1A and the N-terminal CRD2 segment of DcR3. The L85A/R89A double mutant, which effects two key DcR3 residues in this interface, exhibits significantly lower binding affinity for TL1A, LIGHT and FasL than wild type DcR3 (Figure 6). This behavior suggests that LIGHT and FasL recognize DcR3 in a manner similar to that of TL1A, with residues in the DE loop making crucial contacts with the N-terminal DcR3 CRD2. Differences in affinities between DcR3 and its three ligands are likely due, at least in part, to sequence variation in the ligand AA’ and GH loops which contact the C-terminal segment of CRD2 of DcR3.
Different TL1A-binding mechanisms between DcR3 and DR3
The biological complexity associated with DcR3 is further increased by competition with the cognate signaling receptors for TL1A, LIGHT and FasL. The TL1A-binding affinity of full length DcR3 (1.8nM) is in the same range as that of DR3 (6.5nM), the signaling receptor of TL1A. (Migone et al., 2002). Sequence comparison of DcR3 with DR3 shows that CRD2 from these two receptors have the same disulfide connectivity (Figure 3B). However, the residues in CRD2 of DcR3 that contact the AA’ and DE loops in TL1A are generally not conserved in DR3, switching from predominately hydrophobic in DcR3 to predominately hydrophilic in DR3. These differences suggest that CRD2 of DR3 might form more polar interactions with TL1A compared with DcR3. By substituting conserved hydrophilic residues in TL1A, it might be possible to identify mutations in TL1A that compromise binding to the DR3 signaling receptor, while preserving the interaction with the DcR3 decoy receptor. Consistent with this notion, the S120A/E123A TL1A double mutant exhibited significantly reduced DR3 binding while maintaining a relatively strong DcR3 binding response (Figure S3). Such soluble “non-signaling” TL1A mutants would be predicted to have only a modest effect on normal immune regulation by DR3, but would offer therapeutic efficacy by specifically inhibiting the immune evasion function of DcR3 associated with various cancers and autoimmune diseases (Funke et al., 2009);(Ashkenazi, 2002).
The reduced contacts between DcR3 and the membrane-distal part of TL1A is the consequence of a shortened CRD3 relative to other TNFR family members. The N-terminal CRD3 segment in DR3 features two putative pairs of disulfide bonds and is 10 residues longer than its counterpart in DcR3. Therefore, similar to the LTα:TNFR1 and TRAIL:DR5 complexes (Figure 4A,B), DR3 may utilize this region to form a more extensive interface with the top of the TL1A molecule, distal to the TL1A membrane. As a result, the DR3-binding footprint on TL1A may have a more conventional symmetric distribution between the upper and lower patches compared with the unusual pattern observed with DcR3 (Figure 4C,D).
In conclusion, DcR3 interacts with invariant atoms located in the membrane-proximal half of TL1A. This mode of interaction is distinct from previously characterized TNF ligand:receptor complexes, and provides a mechanistic basis for the decoy strategies utilized by DcR3 to recognize and neutralize multiple TNF family members. These findings also provide insights into the regulation of cross-reactivities in the TNF superfamily and offer a structural basis to engineer TNF ligands and receptors with broadened or tightened specificities for mechanistic and therapeutic applications.
EXPERIMENTAL PROCEDURES
Cloning, expression, mutagenesis and purification of DcR3 and TL1A poteins
DcR3 cDNA was amplified from a human skin cDNA library (BioChain) by PCR. The cysteine-rich domains (CRDs, V30-S195) were cloned into the pMT/BiP/V5-His A vector (Invitrogen) and co-transfected with the pCoBlast (Invitrogen) plasmid at 20:1 ratio into Drosophila S2 cells. Stable blasticidin-resistant cell lines were generated according to the manufacturer’s (Invitrogen) manual. The secreted DcR3 CRD protein was purified to homogeneity by Ni-NTA column (Qiagen) followed by size exclusion chromatography (HiLoad Superdex 75; Amersham). Detailed expression and purification protocol can be found in the supplemented materials.
The cloning, expression and purification of the TL1A protein was previously described (Zhan et al., 2009). In this study, the C95S/C135S TL1A double mutant was used in the crystallization of the TL1A:DcR3 complex, as this mutant is more resistant to aggregation while exhibiting the same structural stability and DcR3 binding affinity as wild type protein (Zhan et al., 2009). DcR3 and TL1A point mutations were generated by QuikChange site-directed mutagenesis (Stratagene), and the mutant proteins were purified as described for the wild type.
Crystallization, structure determination and refinement
Diffraction quality crystals of unliganded DcR3 CRDs and TL1A:DcR3 complex were obtained by sitting drop vapor diffusion at 17 °C. Trigonal and cubic crystal forms of the TL1A:DcR3 complex diffracted to resolutions of 2.45 Å and 2.95 Å, respectively. The unliganded DcR3 diffraction data exhibits strong anisotropy with an effective resolution of 2.9 Å. All structures were determined by molecular replacement and refined by standard methods, resulting in Rwork/Rfree values of 22.3%/26.2%, 23.2%/26.1%, and 28.5%/31.5% for trigonal TL1A:DcR3 complex, cubic TL1A:DcR3 complex, and unliganded DcR3, respectively (Table 1). Atomic coordinates and structure factors of the trigonal and cubic TL1A:DcR3 complex structures and unbound DcR3 structure have been deposited to the PDB and are available under the accession codes 3K51, 3MI8 and 3MHD, respectively.
Surface Plasmon Resonance (SPR) binding assay
SPR binding assays were performed with a BIAcore 3000 optical biosensor (Biacore) at 25 °C, using immobilized recombinant FasL (R&D systemes), LIGHT (R&D systemes) and TL1A C95S/C135S proteins. The wild type and mutant DcR3 CRDs proteins were injected over the chip at a series of concentrations in random order at a flow rate of 20 µl/min. Experimental details are provided in the supplementary materials. After subtracting the response of the blank cell (with no protein immobilized), the averaged maximum response of each experimental cell was plotted against its corresponding concentration. The steady state fits were analyzed with Prism 5 (Graphpad Software), assuming the One site-Total model (Binding model equation: Y=Bmax*X/(Kd+X), where Bmax is the maximum specific binding; Kd is the equilibrium binding constant.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge the staff of the X29 beamline at the National Synchrotron Light Source, and the staff of the 24-ID-E beamline at the Advanced Photon Source. This work was supported by National Institutes of Health Grant AI07289 (to S.G.N. and S.C.A.), the Albert Einstein Cancer Center Grant (National Cancer Institute, P30CA13330) and the Albert Einstein College of Medicine Macromolecular Therapeutics Development Facility.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ashkenazi A. Targeting death and decoy receptors of the tumour-necrosis factor superfamily. Nat Rev Cancer. 2002;2:420–430. doi: 10.1038/nrc821. [DOI] [PubMed] [Google Scholar]
- Bai C, Connolly B, Metzker ML, Hilliard CA, Liu X, Sandig V, Soderman A, Galloway SM, Liu Q, Austin CP, Caskey CT. Overexpression of M68/DcR3 in human gastrointestinal tract tumors independent of gene amplification and its location in a four-gene cluster. Proc Natl Acad Sci U S A. 2000;97:1230–1235. doi: 10.1073/pnas.97.3.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamias G, Mishina M, Nyce M, Ross WG, Kollias G, Rivera-Nieves J, Pizarro TT, Cominelli F. Role of TL1A and its receptor DR3 in two models of chronic murine ileitis. Proc Natl Acad Sci U S A. 2006;103:8441–8446. doi: 10.1073/pnas.0510903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banner DW, D'Arcy A, Janes W, Gentz R, Schoenfeld HJ, Broger C, Loetscher H, Lesslauer W. Crystal structure of the soluble human 55 kd TNF receptor-human TNF beta complex: implications for TNF receptor activation. Cell. 1993;73:431–445. doi: 10.1016/0092-8674(93)90132-a. [DOI] [PubMed] [Google Scholar]
- Bodmer JL, Schneider P, Tschopp J. The molecular architecture of the TNF superfamily. Trends Biochem Sci. 2002;27:19–26. doi: 10.1016/s0968-0004(01)01995-8. [DOI] [PubMed] [Google Scholar]
- Bull MJ, Williams AS, Mecklenburgh Z, Calder CJ, Twohig JP, Elford C, Evans BA, Rowley TF, Slebioda TJ, Taraban VY, et al. The Death Receptor 3-TNF-like protein 1A pathway drives adverse bone pathology in inflammatory arthritis. J Exp Med. 2008;205:2457–2464. doi: 10.1084/jem.20072378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Chan YH, Jackson DG, Hsieh SL. The glycosaminoglycan-binding domain of decoy receptor 3 is essential for induction of monocyte adhesion. J Immunol. 2006;176:173–180. doi: 10.4049/jimmunol.176.1.173. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay K, Lazar-Molnar E, Yan Q, Rubinstein R, Zhan C, Vigdorovich V, Ramagopal UA, Bonanno J, Nathenson SG, Almo SC. Sequence, structure, function, immunity: structural genomics of costimulation. Immunol Rev. 2009;229:356–386. doi: 10.1111/j.1600-065X.2009.00778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Zhou T, Liu C, Shapiro JP, Brauer MJ, Kiefer MC, Barr PJ, Mountz JD. Protection from Fas-mediated apoptosis by a soluble form of the Fas molecule. Science. 1994;263:1759–1762. doi: 10.1126/science.7510905. [DOI] [PubMed] [Google Scholar]
- Compaan DM, Hymowitz SG. The crystal structure of the costimulatory OX40-OX40L complex. Structure. 2006;14:1321–1330. doi: 10.1016/j.str.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Funke B, Autschbach F, Kim S, Lasitschka F, Strauch U, Rogler G, Gdynia G, Li L, Gretz N, Macher-Goeppinger S, et al. Functional characterisation of decoy receptor 3 in Crohn's disease. Gut. 2009;58:483–491. doi: 10.1136/gut.2008.148908. [DOI] [PubMed] [Google Scholar]
- Galon J, Aksentijevich I, McDermott MF, O'Shea JJ, Kastner DL. TNFRSF1A mutations and autoinflammatory syndromes. Curr Opin Immunol. 2000;12:479–486. doi: 10.1016/s0952-7915(00)00124-2. [DOI] [PubMed] [Google Scholar]
- Goh CR, Loh CS, Porter AG. Aspartic acid 50 and tyrosine 108 are essential for receptor binding and cytotoxic activity of tumour necrosis factor beta (lymphotoxin) Protein Eng. 1991;4:785–791. doi: 10.1093/protein/4.7.785. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Miura Y, Nishiyama T, Mitani M, Tateishi K, Sakai Y, Hashiramoto A, Kurosaka M, Shiozawa S, Doita M. Decoy receptor 3 expressed in rheumatoid synovial fibroblasts protects the cells against Fas-induced apoptosis. Arthritis Rheum. 2007;56:1067–1075. doi: 10.1002/art.22494. [DOI] [PubMed] [Google Scholar]
- Hymowitz SG, Christinger HW, Fuh G, Ultsch M, O'Connell M, Kelley RF, Ashkenazi A, de Vos AM. Triggering cell death: the crystal structure of Apo2L/TRAIL in a complex with death receptor 5. Mol Cell. 1999;4:563–571. doi: 10.1016/s1097-2765(00)80207-5. [DOI] [PubMed] [Google Scholar]
- Hymowitz SG, O'Connell MP, Ultsch MH, Hurst A, Totpal K, Ashkenazi A, de Vos AM, Kelley RF. A unique zinc-binding site revealed by a high-resolution X-ray structure of homotrimeric Apo2L/TRAIL. Biochemistry. 2000;39:633–640. doi: 10.1021/bi992242l. [DOI] [PubMed] [Google Scholar]
- Jin T, Guo F, Kim S, Howard A, Zhang YZ. X-ray crystal structure of TNF ligand family member TL1A at 2.1A. Biochem Biophys Res Commun. 2007;364:1–6. doi: 10.1016/j.bbrc.2007.09.097. [DOI] [PubMed] [Google Scholar]
- Kim HM, Yu KS, Lee ME, Shin DR, Kim YS, Paik SG, Yoo OJ, Lee H, Lee JO. Crystal structure of the BAFF-BAFF-R complex and its implications for receptor activation. Nat Struct Biol. 2003;10:342–348. doi: 10.1038/nsb925. [DOI] [PubMed] [Google Scholar]
- Kugathasan S, Baldassano RN, Bradfield JP, Sleiman PM, Imielinski M, Guthery SL, Cucchiara S, Kim CE, Frackelton EC, Annaiah K, et al. Loci on 20q13 and 21q22 are associated with pediatric-onset inflammatory bowel disease. Nat Genet. 2008;40:1211–1215. doi: 10.1038/ng.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locksley RM, Killeen N, Lenardo MJ. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell. 2001;104:487–501. doi: 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- Lupardus PJ, Birnbaum ME, Garcia KC. Molecular basis for shared cytokine recognition revealed in the structure of an unusually high affinity complex between IL-13 and IL-13Ralpha2. Structure. 2010;18:332–342. doi: 10.1016/j.str.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan F, Davidson TS, Kahle E, Kinder M, Acharya K, Jankovic D, Bundoc V, Hodges M, Shevach EM, Keane-Myers A, et al. The TNF-family receptor DR3 is essential for diverse T cell-mediated inflammatory diseases. Immunity. 2008;29:79–89. doi: 10.1016/j.immuni.2008.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migone TS, Zhang J, Luo X, Zhuang L, Chen C, Hu B, Hong JS, Perry JW, Chen SF, Zhou JX, et al. TL1A is a TNF-like ligand for DR3 and TR6/DcR3 and functions as a T cell costimulator. Immunity. 2002;16:479–492. doi: 10.1016/s1074-7613(02)00283-2. [DOI] [PubMed] [Google Scholar]
- Mongkolsapaya J, Grimes JM, Chen N, Xu XN, Stuart DI, Jones EY, Screaton GR. Structure of the TRAIL-DR5 complex reveals mechanisms conferring specificity in apoptotic initiation. Nat Struct Biol. 1999;6:1048–1053. doi: 10.1038/14935. [DOI] [PubMed] [Google Scholar]
- Moreland LW, Baumgartner SW, Schiff MH, Tindall EA, Fleischmann RM, Weaver AL, Ettlinger RE, Cohen S, Koopman WJ, Mohler K, et al. Treatment of rheumatoid arthritis with a recombinant human tumor necrosis factor receptor (p75)-Fc fusion protein. N Engl J Med. 1997;337:141–147. doi: 10.1056/NEJM199707173370301. [DOI] [PubMed] [Google Scholar]
- Muleris M, Almeida A, Gerbault-Seureau M, Malfoy B, Dutrillaux B. Identification of amplified DNA sequences in breast cancer and their organization within homogeneously staining regions. Genes Chromosomes Cancer. 1995;14:155–163. doi: 10.1002/gcc.2870140302. [DOI] [PubMed] [Google Scholar]
- Naismith JH, Devine TQ, Brandhuber BJ, Sprang SR. Crystallographic evidence for dimerization of unliganded tumor necrosis factor receptor. J Biol Chem. 1995;270:13303–13307. doi: 10.1074/jbc.270.22.13303. [DOI] [PubMed] [Google Scholar]
- Pappu BP, Borodovsky A, Zheng TS, Yang X, Wu P, Dong X, Weng S, Browning B, Scott ML, Ma L, et al. TL1A-DR3 interaction regulates Th17 cell function and Th17-mediated autoimmune disease. J Exp Med. 2008;205:1049–1062. doi: 10.1084/jem.20071364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer K. Biological functions of tumor necrosis factor cytokines and their receptors. Cytokine Growth Factor Rev. 2003;14:185–191. doi: 10.1016/s1359-6101(03)00022-4. [DOI] [PubMed] [Google Scholar]
- Pitti RM, Marsters SA, Lawrence DA, Roy M, Kischkel FC, Dowd P, Huang A, Donahue CJ, Sherwood SW, Baldwin DT, et al. Genomic amplification of a decoy receptor for Fas ligand in lung and colon cancer. Nature. 1998;396:699–703. doi: 10.1038/25387. [DOI] [PubMed] [Google Scholar]
- Rooney IA, Butrovich KD, Glass AA, Borboroglu S, Benedict CA, Whitbeck JC, Cohen GH, Eisenberg RJ, Ware CF. The lymphotoxin-beta receptor is necessary and sufficient for LIGHT-mediated apoptosis of tumor cells. J Biol Chem. 2000;275:14307–14315. doi: 10.1074/jbc.275.19.14307. [DOI] [PubMed] [Google Scholar]
- Schneider P, Bodmer JL, Holler N, Mattmann C, Scuderi P, Terskikh A, Peitsch MC, Tschopp J. Characterization of Fas (Apo-1, CD95)-Fas ligand interaction. J Biol Chem. 1997;272:18827–18833. doi: 10.1074/jbc.272.30.18827. [DOI] [PubMed] [Google Scholar]
- Siegel RM, Chan FK, Chun HJ, Lenardo MJ. The multifaceted role of Fas signaling in immune cell homeostasis and autoimmunity. Nat Immunol. 2000;1:469–474. doi: 10.1038/82712. [DOI] [PubMed] [Google Scholar]
- Straus SE, Sneller M, Lenardo MJ, Puck JM, Strober W. An inherited disorder of lymphocyte apoptosis: the autoimmune lymphoproliferative syndrome. Ann Intern Med. 1999;130:591–601. doi: 10.7326/0003-4819-130-7-199904060-00020. [DOI] [PubMed] [Google Scholar]
- Takahama Y, Yamada Y, Emoto K, Fujimoto H, Takayama T, Ueno M, Uchida H, Hirao S, Mizuno T, Nakajima Y. The prognostic significance of overexpression of the decoy receptor for Fas ligand (DcR3) in patients with gastric carcinomas. Gastric Cancer. 2002;5:61–68. doi: 10.1007/s101200200011. [DOI] [PubMed] [Google Scholar]
- Van Zee KJ, Kohno T, Fischer E, Rock CS, Moldawer LL, Lowry SF. Tumor necrosis factor soluble receptors circulate during experimental and clinical inflammation and can protect against excessive tumor necrosis factor alpha in vitro and in vivo. Proc Natl Acad Sci U S A. 1992;89:4845–4849. doi: 10.1073/pnas.89.11.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Lo JC, Foster A, Yu P, Chen HM, Wang Y, Tamada K, Chen L, Fu YX. The regulation of T cell homeostasis and autoimmunity by T cell-derived LIGHT. J Clin Invest. 2001;108:1771–1780. doi: 10.1172/JCI13827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware CF. The TNF superfamily. Cytokine Growth Factor Rev. 2003;14:181–184. doi: 10.1016/s1359-6101(03)00032-7. [DOI] [PubMed] [Google Scholar]
- Wu Y, Han B, Sheng H, Lin M, Moore PA, Zhang J, Wu J. Clinical significance of detecting elevated serum DcR3/TR6/M68 in malignant tumor patients. Int J Cancer. 2003;105:724–732. doi: 10.1002/ijc.11138. [DOI] [PubMed] [Google Scholar]
- Yamagishi J, Kawashima H, Matsuo N, Ohue M, Yamayoshi M, Fukui T, Kotani H, Furuta R, Nakano K, Yamada M. Mutational analysis of structure--activity relationships in human tumor necrosis factor-alpha. Protein Eng. 1990;3:713–719. doi: 10.1093/protein/3.8.713. [DOI] [PubMed] [Google Scholar]
- Yang CR, Hsieh SL, Teng CM, Ho FM, Su WL, Lin WW. Soluble decoy receptor 3 induces angiogenesis by neutralization of TL1A, a cytokine belonging to tumor necrosis factor superfamily and exhibiting angiostatic action. Cancer Res. 2004;64:1122–1129. doi: 10.1158/0008-5472.can-03-0609. [DOI] [PubMed] [Google Scholar]
- You RI, Chang YC, Chen PM, Wang WS, Hsu TL, Yang CY, Lee CT, Hsieh SL. Apoptosis of dendritic cells induced by decoy receptor 3 (DcR3) Blood. 2008;111:1480–1488. doi: 10.1182/blood-2007-09-114850. [DOI] [PubMed] [Google Scholar]
- Young HA, Tovey MG. TL1A: a mediator of gut inflammation. Proc Natl Acad Sci U S A. 2006;103:8303–8304. doi: 10.1073/pnas.0602655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu KY, Kwon B, Ni J, Zhai Y, Ebner R, Kwon BS. A newly identified member of tumor necrosis factor receptor superfamily (TR6) suppresses LIGHT-mediated apoptosis. J Biol Chem. 1999;274:13733–13736. doi: 10.1074/jbc.274.20.13733. [DOI] [PubMed] [Google Scholar]
- Zhan C, Yan Q, Patskovsky Y, Li Z, Toro R, Meyer A, Cheng H, Brenowitz M, Nathenson SG, Almo SC. Biochemical and structural characterization of the human TL1A Ectodomain. Biochemistry. 2009;48:7636–7645. doi: 10.1021/bi900031w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Salcedo TW, Wan X, Ullrich S, Hu B, Gregorio T, Feng P, Qi S, Chen H, Cho YH, et al. Modulation of T-cell responses to alloantigens by TR6/DcR3. J Clin Invest. 2001;107:1459–1468. doi: 10.1172/JCI12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.