Abstract
Recombination is a major force for generating HIV-1 diversity and produces numerous recombinants circulating in the human population. We previously established a cell-based system using green fluorescent protein gene (gfp) as a reporter to study the mechanisms of HIV-1 recombination. We now report an improved system capable of detecting recombination using authentic viral sequences. Frameshift mutations were introduced into the gag gene o that parental viruses do not express full-length Gag; however, recombination can generate a progeny virus that expresses a functional Gag. We demonstrate that this Gag reconstitution assay can be used to detect recombination between two group M HIV-1 variants of same or different subtypes. Using both gfp and gag assays, we found that, similar to group M viruses, group O viruses also recombine frequently. When recombination between a group M virus and a group O virus was examined, we found three distinct barriers for intergroup recombination. First, similar to recombination within group M viruses, intergroup recombination is affected by the identity of the dimerization initiation signal (DIS); variants with the same DIS recombined at a higher rate than those with different DIS. Second, using the gfp recombination assay, we showed that intergroup recombination occurs much less frequently than intragroup recombination, even though the gfp target sequence is identical in all viruses. Finally, Gag reconstitution between variants from different groups is further reduced compared with GFP, indicating that sequence divergence interferes with recombination efficiency in the gag gene. Compared with identical sequences, we estimate that recombination rates are reduced by 3-fold and 10–13-fold when the target regions in gag contain 91% and 72–73% sequence identities, respectively. These results show that there are at least three distinct mechanisms preventing exchange of genetic information between divergent HIV-1 variants from different groups.
Keywords: GFP, Gag, group M, group O, reconstitution
INTRODUCTION
Our current understanding is that independent zoonotic transmission events introduced different simian immunodeficiency viruses (SIVs) into the human population to give rise to four groups of human immunodeficiency virus type 1 (HIV-1), group M, N, O and P.1 Group M viruses are most closely related to SIVcpzptt.2 Group N appears to be a recombinant between an ancient group M virus and another SIVcpzptt strain;2 whereas group O and P are thought to be derived from different strains of SIVgor.3, 4
The four groups of HIV-1 viruses have an uneven distribution across the human population.5 The vast majority of infections in the AIDS pandemic are caused by group M viruses; whereas infections caused by the other three groups of viruses are mainly restricted to regions in Central and West Africa.6 Although thought to be transmitted into the human population via a single zoonotic event, the current group M consists of a large number of genetically different variants.7 Therefore, group M viruses are further divided into subtypes and sub-subtypes, as well as into stable Circulating Recombinant Forms (CRFs) and Unique Recombinant Forms (URFs).8, 9
Multiple mechanisms contribute to the generation of high diversity in the HIV-1 population to allow the divergence of the group M viruses.10 HIV-1 replicates at high rates in many infected people generating high viral loads. Additionally, as a member of the Retroviridae family, one of the essential steps of HIV-1 replication is reverse transcription, which is carried out by the virally encoded reverse transcriptase (RT).11, 12 RT does not have the traditional proof-reading function present in cellular DNA polymerases; as a result, RT-mediated DNA synthesis is error-prone.13 Furthermore, this process has high recombination potential.14 These features of HIV-1 replication provide the basis for generating viral diversity.10
One of the hallmarks of HIV-1 replication is frequent recombination.15 Recombination can occur during DNA synthesis when RT switches between the two copackaged RNAs and uses portions of each RNA as templates for DNA synthesis. If the two copackaged RNAs contain different genetic information, a recombinant distinct from either parent can be generated.16 Many factors are known to affect recombination between two HIV-1 variants.17, 18 RNA copackaging is a prerequisite for retroviral recombination; therefore, the frequency at which RNAs generated from different HIV-1 variants can be copackaged into the same particle influences their recombination potential.19 We have observed that variants with different dimerization initiation signal (DIS) sequences recombine less because their RNAs are copackaged together less frequently.20, 21, 22 Furthermore, the RNA trafficking pathways can affect the recombination potential of two viruses,23 and it has been shown that diversity of nucleotide sequences can affect template switching frequency, and hence, the recombination rate.24, 25, 26, 27
Most of the current studies of HIV-1 recombination focus on group M viruses. Recombination rates of other groups of HIV-1 have not been studied. Dual infection of group O and group M HIV-1 variants have been reported;28 furthermore, intergroup recombinants had been reported.28, 29 We sought to determine the recombination rate of the group O viruses and explore the potential and the barriers for intergroup HIV-1 recombination. To this end, we first establish a recombination assay that can reflect the nature of nucleic acid sequence diversity among variants from different groups. Previously, we employed a system to study HIV-1 recombination using near full-length viruses with marker genes inserted in the nef gene.14 In this system, two near full-length viruses were used, each encoding a surface marker gene and a mutated green fluorescent protein (gfp) gene. The inactivating mutations in the gfp differ in the two HIV-1 vectors; during reverse transcription, recombination can occur to reconstitute a gfp gene encoding a functional protein, which can be detected by flow cytometry. The advantage of the system is the ability to easily score a very large number of infection events using flow cytometry and to directly determine recombination rates. However, the gfp gene does not reflect some of the characteristics of the viral genome undergoing recombination, such as sequence diversity. In this report, we developed an improved dual-marker system that can simultaneously detect recombination in the gfp gene and in the viral genome, using Gag expression as a marker. Using this system, we examined recombination among group M variants, between two group O viruses, and between group M and group O viruses. We found that group O viruses recombine at high rates similar to those of group M viruses. Additionally, group O and group M viruses recombine at low rates even when they carry the same DIS; we have identified distinct mechanisms that contribute to the low intergroup recombination potential.
RESULTS
Strategy to measure recombination using authentic HIV-1 target sequences
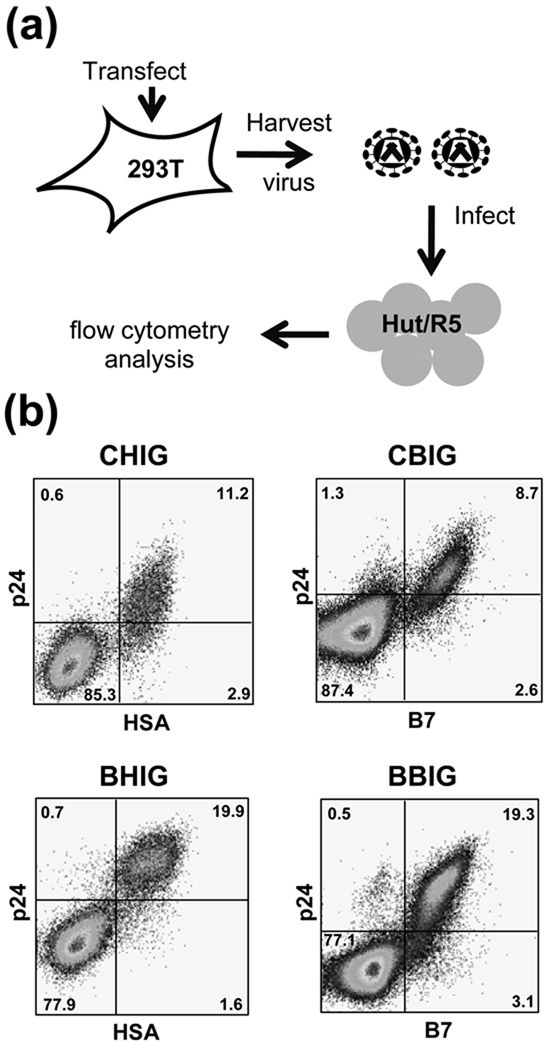
We sought to establish an in vivo system capable of detecting and quantifying recombination in a HIV-1 population using authentic viral sequences. To achieve this, we modified the existing system to allow the detection of Gag only after a recombination event. Frameshift mutations were introduced into the CA region of gag at different positions so that neither parental vector can express a full-length Gag. However, recombination can reconstitute a gag encoding a functional polyprotein, which can be detected by antibody staining and flow cytometry (Fig. 1a). To verify the feasibility of this strategy, first we tested whether Gag expression can be detected reliably by α-p24 antibody staining and flow cytometry. HIV-1 vectors used in the GFP recombination assay contain near full-length genomes, including all cis-acting elements essential for virus replication, and encode gag-pol, tat, and rev. Additionally, a surface marker gene (hsa or B7), followed by IRES and gfp, was inserted in the nef region (Fig. 1b). Control viral vectors (BBIG, BHIG, CBIG, or CHIG) were transfected into 293T cells along with pNLIII(AD8), an HIV-1 Env-expression construct; viruses were harvested and used to infect Hut/CCR5 cells, which were processed and examined by flow cytometry (Fig. 2A). Both subtype B and subtype C Gag can be detected efficiently by this procedure as most of the marker-expressing cells are also positive in Gag expression by antibody staining (Fig. 2b).
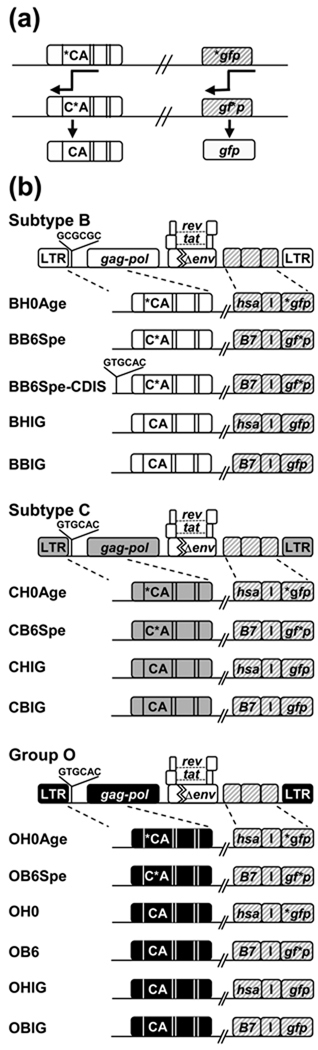
Fig. 1. Experimental strategy and HIV-1 vectors used in the recombination assays.
(a) Schematic representation of recombination in gfp and gag genes. Template switching between inactivating mutations within a gene can generate a recombinant expressing functional GFP and / or Gag containing the CA domain. Slanted double lines indicate parts omitted for simplicity. Direction of DNA synthesis is indicated by arrows. (b) General structures of vectors used in this study. All vectors express functional Tat and Rev and carry a deletion in the env gene. White, grey, and black boxes represent subtype B, C, and group O sequences, respectively. Striped boxes indicate marker genes inserted in the nef reading frame. Asterisk denotes inactivating mutation. DIS sequences, GCGCGC or GTGCAC, are indicated. The name of each construct is descriptive of its structure: the first letter refers to the group or subtype of the viral sequence; the second letter indicates the surface marker gene (H is for hsa and B is for B7); numbers mark the position of the inactivating mutation in GFP (0 is for start of the gene, 6 is for 603 nt downstream); and Age or Spe refers to the frameshift mutation toward the 5’ or 3’ end of CA, which are indicated as *CA or C*A, respectively.
Fig. 2. Detection of Gag expression in infected cells by α-p24-antibody staining and flow cytometry.
(a) Experimental protocol used to generate infected cells for Gag detection. Briefly, constructs were used to transfect 293T cells, viruses were harvested and used to infect target Hut/CCR5 cells, which were processed and analyzed by flow cytometry. (b) Detection of Gag expression in cells infected with subtype C viruses (upper panels) and subtype B viruses (lower panels). Detection of HSA or B7 markers is shown in x-axis, whereas detection of CA (p24) is shown in y-axis.
We then introduced inactivating mutations in the CA region of gag to abolish Gag expression detectable by α-p24 antibody staining and flow cytometry. After testing several mutants (data not shown), two suitable gag mutants were identified. The Age mutant contains a +1 frameshift mutation located near the 5’ end of the CA-encoding region, and the Spe mutant contains a 4-nt insertion in gag near the center of CA (Fig. 1b). The distance between the two mutations is 297 nt and neither mutant exhibits detectable Gag expression (see below).
Comparison of HIV-1 recombination detected by Gag or GFP expression
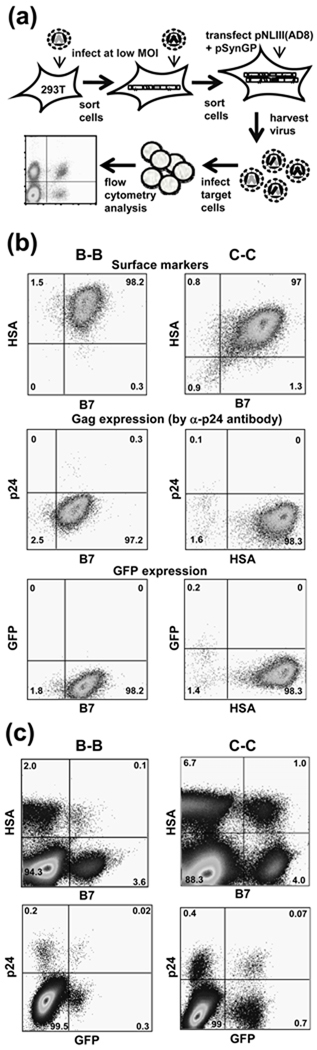
To measure HIV-1 recombination using the Gag reconstitution strategy, we generated cell lines containing two different HIV-1 proviruses, one carrying the hsa marker gene and upstream mutations in both gag and gfp genes, and the other carrying the B7 marker gene and downstream mutations in both gag and gfp genes (Fig. 1b). These cell lines were established by sequentially infecting 293T cells with HIV-1 vectors at low MOIs (Fig. 3a). Infected cells were enriched by several rounds of cell sorting; each final cell line consisted of a pool of more than 100,000 independently infected cells and more than 97% of the cells expressed HSA and B7 markers (Fig. 3b, top row). However, these producer cells do not express Gag or GFP (Fig. 3b, middle and lower panels, respectively), confirming that the inactivating mutations indeed abolished expression of these genes. As these cell lines do not express functional Gag/Gag-Pol or Env, to measure recombination rate, we transfected the producer cells with pSYNGP and pNLIII(AD8), plasmids that express codon-optimized HIV-1 gag-pol and HIV-1 env, respectively. Viruses were harvested and used to infect Hut/CCR5 cells, which were stained with antibodies and analyzed by flow cytometry. An example of the flow cytometry analyses is shown in Fig. 3c. Virus infection was monitored by the expression of HSA or B7 and recombination was detected by GFP and Gag expression. The number of infected cells was converted to MOI; recombination rate was calculated by dividing the GFP or Gag MOI by total infection MOI. To ensure the accuracy of measured recombination rates, we only included data points that have >1000 GFP+ cells or >500 p24+ cells above the background in the negative control (mock-infected cells or uninfected cells).
Fig. 3. Measurements of recombination among group M viruses.
(a) Experimental protocol used to establish dual-infected virus producer cell line and to measure recombination. (b) Expression of surface markers, Gag, and GFP in selected producer cell lines containing subtype B and subtype C proviruses. Gag expression was detected by α-p24 antibody staining. (c) Representative flow cytometry analyses of target Hut/CCR5 cells from recombination experiments.
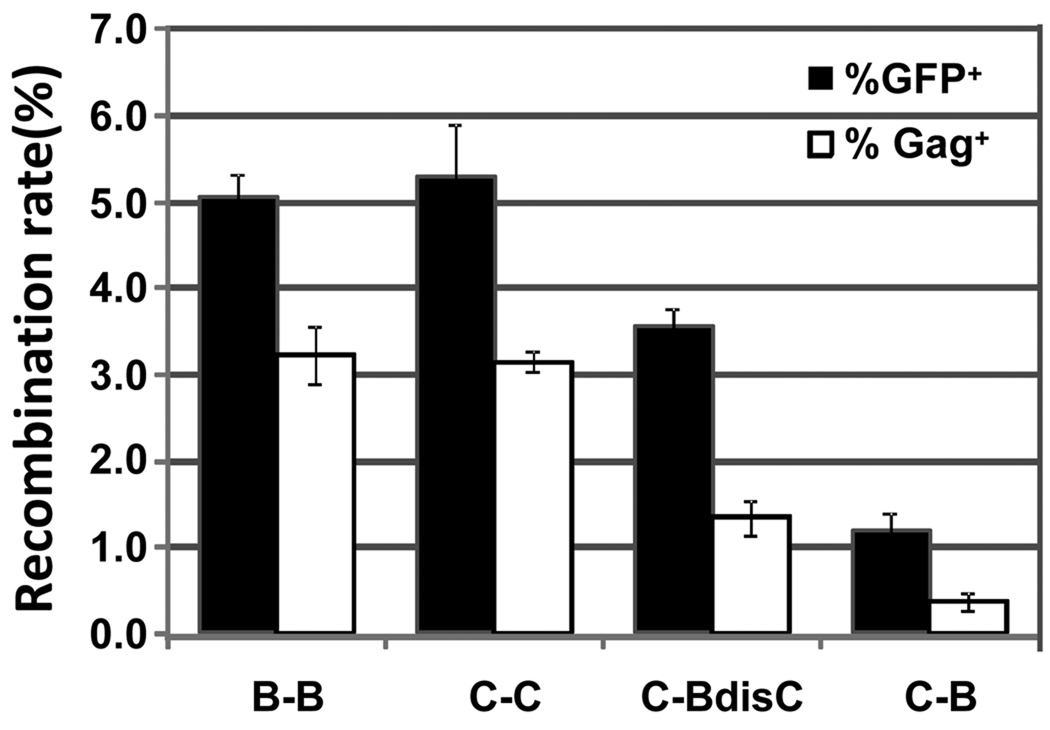
We first measured recombination between two subtype C viruses, CH0Age and CB6Spe (Fig. 1b). Gag expression was detected in 3.2 ± 0.1 % of infection events whereas GFP expression was observed in 5.3 ± 0.6 % of infection events (Fig. 4; marked C-C). The genetic distance between the gag mutations was shorter than that of the gfp mutations (297 nt versus 588 nt), which is likely to account for the lower rate. We also measured the frequencies of GFP and Gag reconstitution between two subtype B viruses, BH0Age and BB6Spe (Fig. 1b), and observed similar rates of recombination, 3.2 ± 0.3% and 5.1 ± 0.3 % for Gag and GFP, respectively (Fig. 4; marked B-B). In these experiments, the gag target sequences between the two vectors are identical; thus to examine intersubtype recombination rates we employed two viruses with different gag sequences.
Fig. 4. Recombination of group M viruses measured by gfp or gag marker gene reconstitution.
Y-axis, the percentage of recombination events measured by GFP+ or Gag+ among infection events. Black bars, percentage of GFP+ events; white bars, percentage of Gag+ events measured by α-p24-antibody staining. Results from three to seven independent experiments were averaged; error bars represent standard deviations.
The recombination rates between a subtype C virus and a subtype B virus, CH0Age and BB6Spe, are much lower in both markers (Fig. 4; C-B); Gag and GFP expression was detected in 0.4 ± 0.1 % and 1.2 ± 0.2 % of infection events, respectively. Subtype B and subtype C HIV-1 contain different DIS sequences, which results in inefficient copackaging of RNAs from these two viruses and a reduced recombination potential. We have previously shown that copackaging of subtype B and C RNAs can be improved by changing the DIS of subtype B virus to that of subtype C20. Therefore, we also examined recombination between a subtype C virus and a subtype B virus containing the subtype C DIS, namely CH0Age and BB6Spe-CDIS (Fig. 4; C-BdisC); in this case 1.3 ± 0.2 % and 3.6 ± 0.2 % of infection events had Gag and GFP expression, respectively. These experiments demonstrated a range of frequencies (3.1% to 0.4%) at which Gag expression was restored in the viral population. Viral elements, such as DIS, that are known to affect the recombination rates detected by GFP also altered the recombination rates measured by Gag expression. The gfp sequences in all of the vectors are identical; thus, GFP reconstitution can be used as an indicator for the recombination potential of these two viruses, taking into accounts issues such as RNA copackaging efficiency. By comparing the Gag and GFP reconstitution frequencies, we can estimate the effect of gag sequence divergence on recombination. For example, GFP reconstituted ~1.6–1.7 fold more efficiently than Gag when the gag target sequences between the two inactivating mutations were identical (Fig. 4, C-C and B-B); GFP reconstituted 2.8 fold more frequently than Gag when the gag sequences were derived from different subtypes of HIV-1 (Fig. 4 C-BdisC). Thus, target sequence divergence probably causes the lower GFP/Gag reconstitution ratios in intersubtype recombination. Taken together, our results established that Gag can be used as a marker to monitor both intrasubtype and intersubtype HIV-1 recombination.
Genetic recombination of group O HIV-1
Unlike group M viruses, group O HIV-1 is thought to be originated from SIVgor.3 To determine the recombination rate of group O HIV-1, we generated group O vectors, pOH0 and pOB6 (Fig. 1B) based on the molecular clone CMO2.5.30 We then generated two producer cell lines using the same procedure described in Fig. 3A. More than 97% of cells in the final cell lines expressed both surface markers and no GFP expression was detected (data not shown). We measured recombination rates using the standard protocol illustrated in Fig. 3a and found that GFP expressed in 4.8 ± 0.2% of the infection events (data not shown in Figures). Therefore, like group M viruses, group O HIV-1 also recombines at high frequency.
Recombination between group M and group O viruses
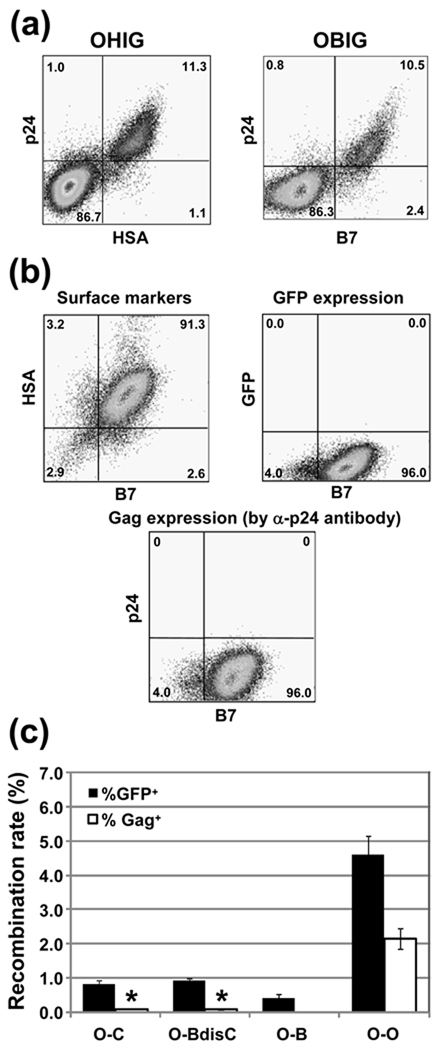
Next, we assessed whether the Gag recombination assay could be used to measure recombination between highly divergent strains, such as group M and O viruses. To ensure that the antibody we used could detect group O Gag, we infected Hut/CCR5 cells with control viruses OHIG or OBIG (Fig. 1b) using the protocol described in Fig. 2a. Infected cells were stained with α-p24 antibody and antibody against HSA or B7, and examined using flow cytometry. As shown in Fig. 5a, Gag expression from group O viruses could be detected efficiently using the α-p24 antibody.
Fig. 5. Measurements of recombination rate between group M and group O viruses.
(a) Detection of group O Gag expression in cells. Target Hut/CCR5 cells were infected with control group O viruses and stained with α-p24 and anti-surface marker antibodies. (b) Expression of surface markers, Gag, and GFP in producer cell line containing two group O proviruses with gag mutations. (c) Inter-group recombination rates. Black and white bars represent % GFP+ and % Gag+ cells in infected population, respectively. Except for those marked by *, results from three to six independent experiments were averaged; error bars represent standard deviations. Each of the two results marked by * is from a single data point although three (O-C) and four (O-BdisC) repetitions were performed each yielding sufficient numbers of GFP+ cells to calculate GFP recombination rate.
To examine recombination between group M and group O viruses, we generated group O viruses with modified gag genes. Group O vectors with either Age or Spe frameshift mutations in the gag gene do not express detectable levels of Gag (data not shown). Using the protocol described in Fig. 3a, we developed a producer cell line that contained OH0Age and OB6Spe proviruses; more than 91% of these cells expressed both HSA and B7 markers but they did not have detectable levels of Gag or GFP (Fig. 5b). Using the procedure described in Fig. 3A, the recombination rates were measured; 4.6 ± 0.5% and 2.2 ± 0.3% of the infection events had GFP and Gag expression, respectively (Fig. 5c).
We then examined recombination between a group O virus and a subtype B virus using two cell lines, one containing OH0Age and BB6Spe and the other cell line containing OB6Spe and BH0Age. The target sequences in CA and gfp in these two cell lines are the same and similar results were obtained; therefore, we combined the results for simplicity (Fig. 5c; O-B). GFP expression was detected in 0.4 ± 0.1% of the infection events, whereas the percentage of events with Gag expression could not be determined accurately due to the limited number of positive cells. Group O viruses, including the molecular clone we used, have the same DIS as subtype C HIV-1, which is different from the DIS sequence of most subtype B variants. To examine whether carrying the same DIS could improve intergroup HIV-1 recombination, we generated a cell line containing a group O virus (OH0Age) and a subtype B virus carrying a subtype C DIS sequence GTGCAC (BB6Spe-CDIS), and detected an increase in GFP+ phenotype to 0.9 ± 0.1 % of the infection events (Fig. 5c; O-BdisC) compared to the 0.4% GFP+ frequency when the DIS sequences were different. Additionally, we generated two cell lines containing one group O virus and one subtype C virus; one cell line containing OH0Age and CB6Spe and one cell line containing CH0Age and OB6Spe (combined results shown as O-C in Fig. 5c). Similarly, GFP expression was detected in 0.8 ± 0.1 % of the infection events.
Results shown in Fig. 5c include 16 sets of data that measure GFP reconstitution between a group M and a group O virus; most of these experiments scored >2000 GFP+ events. If gag reconstitution occurred at similar rates among viruses of different groups as that among different subtypes, we should have been able to score >500 p24+ cells in most of these experiments. Instead, we were only able to measure two data points, one in O-BdisC (0.07%) and the other in O-C (0.08%) despite the fact that three and four sets of experiments were performed, respectively, each yielding sufficient GFP+ cells; these are shown in Fig. 5c with asterisks to indicate that only one datum point was collected. These Gag reconstitution results reveal that, in addition to the barrier revealed by the gfp assay, the sequence divergence between the group O and group M further prevented efficient recombination in the gag gene. Taken together, these results indicated that although matching the DIS sequences did improve the recombination rate between group O and group M viruses, intergroup recombination occurs less frequently than intragroup recombination, indicating that different groups of HIV-1 have a reduced ability to exchange genetic information.
DISCUSSION
In this report we described an improved experimental system to measure recombination rates between different HIV-1 isolates using authentic viral sequences in the gag gene in addition to our previously-established gfp gene. Our findings showed that the gag reconstitution assay can be used to measure recombination between two variants within the same subtype, between viruses from two different subtypes, and even between two viruses from different groups. Using this assay we detected a low level of recombination between a variant from group O and a variant from group M HIV-1, but only obtained limited Gag reconstitution during these intergroup recombination experiments, indicating that intergroup recombination occurs much less frequently than intragroup recombination.
Currently, the Gag reconstitution assay has a smaller dynamic range compared with that of the previous GFP assay, as the genetic distance separating the two gag mutations is shorter than that of gfp. Although the gag gene of HIV-1 is approximately 1.5-kb in length, the assay was constrained to a 297-bp separation of the mutations due to the method of detection and a feature of HIV biology. We used a commercially-available α-p24 antibody that is conjugated with a fluorescent dye and is known to be able to detect full-length Gag. The translation of HIV-1 Gag can be initiated from the start codon of gag in MA or from an AUG in CA using an internal ribosomal entry site.31, 32 As the mutations have to abolish Gag expression from both translation start sites, this severely limited the potential locations at which mutations can be placed. It is foreseeable that in the future the dynamic range of the Gag reconstitution assay can be lengthened significantly if a suitable antibody targeting NC or p6 is available that can reliably detect unprocessed Gag polyprotein.
In this report, we examined for the first time the recombination potential of group O viruses and found that group O viruses, similar to group M viruses, recombine frequently. Although the members of group M viruses can contain different DIS sequences,33, 34 to our knowledge all group O variants contain the DIS sequence, GTGCAC, which is identical to that of most subtype C viruses. Although RNAs containing different DIS sequences can be copackaged into the same particles, it occurred at significantly reduced rate than that of RNAs with the same DIS sequences among group M variants. Our GFP reconstitution assay revealed that the DIS sequences also affects the intergroup recombination rates. However, recombination between a group O and a group M virus is approximately 4-fold lower than that of two group M variants, even when the viruses have the same DIS sequences (Fig. 5 O-C and O-BdisC versus Fig. 4 C-BdisC; p < 0.001, Two way ANOVA). These results suggest that factors other than DIS contribute to reduce recombination rates.
Group O and group M viruses have reduced sequence similarity. Template switching by RT is known to be reduced between two regions with low sequence identity; however, the gfp marker gene used to monitor recombination has the same sequence in the group O and group M vectors. Therefore, sequence diversity should not directly affect template switching in gfp, but was nonetheless capable or reducing the recombination rate in this region. One possible mechanism is that sequence differences may reduce copackaging of group O and group M RNA despite their carrying the same DIS, as additional sequence compatibility is required for efficient copackaging into the same virion. Alternatively, the conformation of the two copackaged RNAs within the core may differ between the viruses, thereby hampering RT switching from one RNA to the other during DNA synthesis. Further examination will be required to distinguish between these two possibilities.
Although the sequence of the gfp gene in all vectors tested was identical, their gag sequences differed. In this case, sequence diversity does play a direct role in reducing Gag reconstitution during recombination. Intragroup recombination with matched DIS sequences (Fig. 4, C-BdisC) was found to be approximately 3-fold more efficient in GFP than in Gag, when the two strains we used have 86% of sequence identity in gag and 91% sequence identity in the 297-nt target region. In intergroup recombination, the molecular clones we used have 69% sequence identity in gag (both O-B and O-C) and 73% (O-B) and 72% (O-C) sequence identity in the 297-nt target region for recombination measurement. Our inability to measure an accurate Gag reconstitution rate for intergroup recombination despite large number of GFP+ events scored in these experiments clearly indicates a much reduced intergroup gag recombination rate compared with the intragroup rate. Our limited data sets (2 measurements) estimated that GFP is reconstituted 10–13-fold more efficiently than Gag even when the two viruses have the same DIS (0.83–0.93% compared with 0.07–0.08%, Fig. 5C, O-C and O-BdisC), indicating that sequence divergence further reduced intergroup recombination in the gag gene. Taken together, these results show that sequence divergence affects intergroup recombination in three distinct ways: a copackaging mechanism mediated by DIS, an indirect mechanism that decreases recombination of homologous sequences (gfp) by affecting copackaging independent of DIS or accessibility of the templates, and a direct mechanisms reducing template switching in divergent regions (gag).
Compared with group M virus infection, group O HIV-1 infection is limited both in the number of infected individuals and in its geographical distribution. Dual infection of group O and group M viruses has been observed 28 and recombinants between them have been isolated from the human population 28, 29. Our findings indicate that group O and group M variants recombine far less frequently than two viruses of the same group. Previously, we have shown that many B/F intersubtype recombinants have difficulty completing the virus life cycle and were eliminated during virus replication 27. Considering that the genetic distance between group O and group M viruses is greater than that of two subtype variants, it is most likely that many of the group O and group M recombinants are not viable and would be eliminated through purifying selection. Therefore, O/M recombinants have to overcome multiple barriers to emerge in the human population. The observations of replication competent O/M recombinants demonstrate the power of HIV-1 recombination and the ability of the HIV-1 genome to evolve.
MATERIAL AND METHODS
Plasmid construction and nomenclature
The names of all plasmids used in this study begin with ‘p’, whereas the names of the viruses derived from these plasmids do not. All of the modified HIV-1-based vectors used in this study contain cis-acting elements essential for virus replication, gag-pol, tat, and rev. Additionally, two marker genes were inserted into the nef gene; the first marker gene is either a mouse CD24 heat-stable antigen gene (hsa) or a human CD80 gene (B7) and the second marker gene is a green fluorescent protein gene (gfp); the translation of GFP is facilitated by the encephalomyocarditis virus internal ribosome entry site (IRES). For simplicity, the first letter of the vector name refers to the group or subtype of the viral sequence; the second letter indicates the surface marker gene (H is for hsa and B is for B7); numbers mark the position of the inactivating mutation in GFP (0 is for start of the gene, 6 is for 603 nt downstream). For example, pBH0 refers to a plasmid containing subtype B HIV-1 modified genome expressing HSA and having a mutated gfp gene with frameshift at the 5’ end of the gene. The gag genes of many vectors contain a frameshift mutation in capsid (CA) region; either a +1 frameshift at the N-terminal of CA (referred to as Age) or a +4 frameshift at the C-terminal of CA (referred to as Spe).
For clarity, the previously described subtype B vectors pON-fHIG and pBIG are referred to as pBHIG and pBBIG;14, 23 these two vectors contain functional gag and gfp and are used as expression and staining controls.
Construct pBH0Age was modified from pON-H0 by adding a +1 frameshift at nucleotide position 1215 (pNL4-3 numbering).35 Plasmid pBB6Spe was modified from pT6-RRE-Spe by adding a +4 frameshift at position 1508 (pNL4-3 numbering) and replacing the thy1.2 marker gene with B7.35 Plasmid pBB6Spe-disC was generated by replacing endogenous DIS (GCGCGC) sequence in pBB6Spe with subtype C DIS (GTGCAC).
Subtype C construct pCH0Age was modified from pCH0 by adding a +1 frame shift at position 1228 (pMJ4 numbering),20 whereas pCB6Spe was modified from pCH0 by adding a +4 frameshift at 1522 position (pMJ4 numbering) and replacing the DNA fragments containing hsa-IRES-*gfp with fragment containing B7-IRES-gfp* from pBB6Spe. Plasmids pCHIG and pCBIG were modified from pCH0, by replacing a DNA fragment containing hsa-IRES-*gfp with DNA fragments containing either hsa-IRES-gfp or B7-IRES-gfp from pBHIG and pBBIG, respectively.
Group O vectors were derived from pCMO2.5 (accession number AY623602),30 a generous gift from Dr. Hans-Georg Krausslich. Plasmid pOH0 was generated by first replacing the LTR-gag-pol region of pON-H0 with that of pCMO2.5 to generate pCMO2.5-X-HO. This was carried out by digesting pCMO2.5 with BsrBI and NdeI restriction enzymes and the fragment containing LTR-gag-pol was cloned into the NdeI and ZraI-digested pON-H0. An XhoI site was introduced into the nef reading frame of pCMO2.5, and the nef-LTR region of the resulting plasmid was used to replace the equivalent region in pCMO2.5-X-HO using XhoI and NgoMIV restriction sites. In plasmid pOH0Age, a +1 frameshift was introduced at nucleotide 1260 (ANT70 numbering) of the pOH0. Plasmid pOB6 was generated by replacing the DNA fragment containing the hsa-IRES-*gfp region of pOH0 with a DNA fragment containing B7-IRES-gfp* from pBB6Spe. Plasmid pOB6Spe was generated by introducing the +4 frameshift at nucleotide 1553 (ANT70 numbering) into pOB6. Control plasmids pOHIG and pOBIG were cloned by replacing the hsa-IRES-*gfp marker cassette in pOH0 with hsa-IRES-gfp or B7-IRES-gfp fragments from pBHIG and pBBIG, respectively.
Standard molecular cloning techniques were used to construct plasmids.36 The general structure of all plasmids was mapped by restriction enzyme digestion. In addition, to ensure the absence of inadvertent mutations introduced by PCR amplification, all plasmid DNA fragments derived from PCR were verified by sequencing.
Cell culture, transfection, infection, and generation of producer cell lines
Human embryonic kidney cell line 293T and human T cell line Hut/CCR5 were maintained in Dulbecco’s modified Eagle’s medium and Roswell Park Memorial Institute-1640 medium, respectively. Both media were supplemented with 5% fetal calf serum, 5% calf serum, penicillin (50 U/ml), and streptomycin (50 U/ml). Additionally, puromycin (1 µg/ml) and G418 (500 µg/ml) were added to the medium used for Hut/CCR5 cells. All cultured cells were maintained at 37°C with 5% CO2.
To generate viral stocks for the production of cell lines, 293T cells were transfected using 10 µM 25kDa branched polyethyleneimine (PEI) (Sigma) with viral constructs, pCMV-G and pSYNGP,37, 38 which express the G protein from vesicular stomatitis virus (VSV-G) and codon-optimized HIV-1 Gag/Gag-Pol, respectively. Viruses were harvested 48 h posttransfection, clarified through a 0.45-µm-pore-size filter, and used immediately or stored at −80°C prior to infection. The producer cell lines were generated by sequential infection of low passage 293T cells by two parental viruses at a low multiplicity of infection (MOI) of less than 0.1. Dually infected cells were enriched by several rounds of cell sorting so that more than 91% of the cells expressed markers encoded by both vectors. Producer cell lines were also stained with anti-p24 antibodies to examine Gag/CA expression.
For recombination experiments, producer cells were transfected using Trans-IT®-LT1 (Mirus) or FugeneHD (Roche) with pSYNGP, which expresses codon-optimized HIV-1 Gag/Gag-Pol and pIIINL(AD8)env,37 which expresses HIV-1 Env. Viruses were harvested 48-h posttransfection, clarified through 0.45-µm-pore-size filter and used to infect target Hut/CCR5 cells by spinoculation in 12-well plates (1 × 106 cells/well) or 6-well plates (2 × 106 cells/well) at 1200 × g for 1.5−2 h at 25°C. Target cells were stained and processed 3 days postinfection and analyzed by flow cytometry.
Flow cytometry analyses and cell sorting
The following fluorescently-labeled antibodies were used in various combinations in this study: phycoerythrin (PE)-conjugated α-mCD24(HSA) antibody (BD Biosciences), Biotin-conjugated α-mCD24(HSA) antibody (BD Biosciences) in combination with PerCP-conjugated streptavidin (BD PharMingen), allophycocyanin (APC)-conjugated α-hCD80(B7) MEM-233 antibody (Novus Biologicals), AlexaFlour®647-conjugated α-hCD80(B7) antibody (Biolegend), and PE-conjugated α-p24 KC57-RD1 antibody (Beckman Coulter).
The following protocol was used to stain cells for flow cytometry analyses: 2 × 106 cells were washed twice in 3 ml of phosphate buffered saline containing 2.5 % fetal calf serum. The resulting cells were incubated with Biotin-conjugated α-HSA antibodies (at 1:500 dilution); washed twice, and incubated with AlexaFlour®647 (or APC)-conjugated α-B7 antibodies (at 1:100 dilution) and streptavidine-PerCP (at 1:500 dilution). Cells were then washed twice, fixed with 4% formaldehyde, washed twice, and incubated with PE-conjugated α-p24 antibodies (at 1:40 dilution), washed twice, and fixed with 2% formaldehyde. All incubations with antibodies and formaldehyde were carried in 200 µl volume for 30 minutes at 25°C in the dark.
Flow cytometry analyses were performed on a FACSCalibur system (BD Biosciences) and the data obtained were analyzed using the FlowJo software (Tree Star). For each recombination experiment, uninfected target cells were used as negative controls. The background levels in negative controls were generally very low and were subtracted from the values in the positive samples during data analysis. Cell sorting was performed on ARIA II system (BD Biosciences).
ACKNOWLEDGEMENT
We thank Hans-Georg Krausslich for Group O molecular clone. This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
ABBREVIATIONS
- DIS
Dimerization Initiation Signal
- RT
Reverse Transcriptase
- SIV
Simian Immunodeficiency Virus
- HSA
Heat-Stable antigen
- MOI
Multiplicity of Infection
- MA
matrix
- CA
capsid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Sharp PM, Hahn BH. The evolution of HIV-1 and the origin of AIDS. Philos Trans R Soc Lond B Biol Sci. 2010;365:2487–2494. doi: 10.1098/rstb.2010.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, Cummins LB, Arthur LO, Peeters M, Shaw GM, Sharp PM, Hahn BH. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 3.Van Heuverswyn F, Li Y, Neel C, Bailes E, Keele BF, Liu W, Loul S, Butel C, Liegeois F, Bienvenue Y, Ngolle EM, Sharp PM, Shaw GM, Delaporte E, Hahn BH, Peeters M. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature. 2006;444:164. doi: 10.1038/444164a. [DOI] [PubMed] [Google Scholar]
- 4.Plantier JC, Leoz M, Dickerson JE, De Oliveira F, Cordonnier F, Lemee V, Damond F, Robertson DL, Simon F. A new human immunodeficiency virus derived from gorillas. Nat Med. 2009;15:871–872. doi: 10.1038/nm.2016. [DOI] [PubMed] [Google Scholar]
- 5.McCutchan FE. Global epidemiology of HIV. J Med Virol. 2006;78 Suppl 1:S7–S12. doi: 10.1002/jmv.20599. [DOI] [PubMed] [Google Scholar]
- 6.Van Heuverswyn F, Peeters M. The Origins of HIV and Implications for the Global Epidemic. Curr Infect Dis Rep. 2007;9:338–346. doi: 10.1007/s11908-007-0052-x. [DOI] [PubMed] [Google Scholar]
- 7.Thomson MM, Perez-Alvarez L, Najera R. Molecular epidemiology of HIV-1 genetic forms and its significance for vaccine development and therapy. Lancet Infect Dis. 2002;2:461–471. doi: 10.1016/s1473-3099(02)00343-2. [DOI] [PubMed] [Google Scholar]
- 8.Leitner T, Korber B, Daniels M, Calef C, Foley B. HIV-1 subtype and circulating recombinant form (CRF) reference sequences. In: Leitner T, Foley B, Hahn BH, Marx PA, McCutchan FE, Mellors JW, Wolinsky S, Korber B, editors. HIV Sequence Compendium. Vol. 2005. Los Alamos, NM: Los Alamos National Laboratory; 2005. pp. 41–48. [Google Scholar]
- 9.Robertson DL, Anderson JP, Bradac JA, Carr JK, Foley B, Funkhouser RK, Gao F, Hahn BH, Kalish ML, Kuiken C, Learn GH, Leitner T, McCutchan F, Osmanov S, Peeters M, Pieniazek D, Salminen M, Sharp PM, Wolinsky S, Korber B. HIV-1 nomenclature proposal. Science. 2000;288:55–56. doi: 10.1126/science.288.5463.55d. [DOI] [PubMed] [Google Scholar]
- 10.Coffin JM. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 11.Temin HM, Mizutani S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature. 1970;226:1211–1213. doi: 10.1038/2261211a0. [DOI] [PubMed] [Google Scholar]
- 12.Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature. 1970;226:1209–1211. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- 13.Mansky LM, Temin HM. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol. 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhodes TD, Nikolaitchik O, Chen J, Powell D, Hu W-S. Genetic recombination of human immunodeficiency virus type 1 in one round of viral replication: effects of genetic distance, target cells, accessory genes, and lack of high negative interference in crossover events. J Virol. 2005;79:1666–1677. doi: 10.1128/JVI.79.3.1666-1677.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robertson DL, Sharp PM, McCutchan FE, Hahn BH. Recombination in HIV-1. Nature. 1995;374:124–126. doi: 10.1038/374124b0. [DOI] [PubMed] [Google Scholar]
- 16.Hu WS, Temin HM. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc Natl Acad Sci U S A. 1990;87:1556–1560. doi: 10.1073/pnas.87.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galetto R, Negroni M. Mechanistic features of recombination in HIV. AIDS Rev. 2005;7:92–102. [PubMed] [Google Scholar]
- 18.Onafuwa-Nuga A, Telesnitsky A. The remarkable frequency of human immunodeficiency virus type 1 genetic recombination. Microbiol Mol Biol Rev. 2009;73:451–480. doi: 10.1128/MMBR.00012-09. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore MD, Hu WS. HIV-1 RNA dimerization: It takes two to tango. AIDS Rev. 2009;11:91–102. [PMC free article] [PubMed] [Google Scholar]
- 20.Chin MPS, Rhodes TD, Chen J, Fu W, Hu W-S. Identification of a major restriction in HIV-1 intersubtype recombination. Proc Natl Acad Sci U S A. 2005;102:9002–9007. doi: 10.1073/pnas.0502522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore MD, Fu W, Nikolaitchik O, Chen J, Ptak RG, Hu W-S. Dimer initiation signal of human immunodeficiency virus type 1: its role in partner selection during RNA copackaging and its effects on recombination. J Virol. 2007;81:4002–4011. doi: 10.1128/JVI.02589-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J, Nikolaitchik O, Singh J, Wright A, Bencsics CE, Coffin JM, Ni N, Lockett S, Pathak VK, Hu W-S. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc Natl Acad Sci U S A. 2009;106:13535–13540. doi: 10.1073/pnas.0906822106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore MD, Nikolaitchik OA, Chen J, Hammarskjöld M-L, Rekosh D, Hu W-S. Probing the HIV-1 Genomic RNA Trafficking Pathway and Dimerization by Genetic Recombination and Single Virion Analyses. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000627. e1000627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.An W, Telesnitsky A. Effects of varying sequence similarity on the frequency of repeat deletion during reverse transcription of a human immunodeficiency virus type 1 vector. J Virol. 2002;76:7897–7902. doi: 10.1128/JVI.76.15.7897-7902.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baird HA, Gao Y, Galetto R, Lalonde M, Anthony RM, Giacomoni V, Abreha M, Destefano JJ, Negroni M, Arts EJ. Influence of sequence identity and unique breakpoints on the frequency of intersubtype HIV-1 recombination. Retrovirology. 2006;3:91. doi: 10.1186/1742-4690-3-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dang Q, Hu WS. Effects of homology length in the repeat region on minus-strand DNA transfer and retroviral replication. J Virol. 2001;75:809–820. doi: 10.1128/JVI.75.2.809-820.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galli A, Kearney M, Nikolaitchik OA, Yu S, Chin MPS, Maldarelli F, Coffin JM, Pathak VK, Hu W-S. Patterns of Human Immunodeficiency Virus type 1 recombination ex vivo provide evidence for coadaptation of distant sites, resulting in purifying selection for intersubtype recombinants during replication. J Virol. 2010;84:7651–7661. doi: 10.1128/JVI.00276-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamaguchi J, Bodelle P, Vallari AS, Coffey R, McArthur CP, Schochetman G, Devare SG, Brennan CA. HIV infections in northwestern Cameroon: identification of HIV type 1 group O and dual HIV type 1 group M and group O infections. AIDS Res Hum Retroviruses. 2004;20:944–957. doi: 10.1089/aid.2004.20.944. [DOI] [PubMed] [Google Scholar]
- 29.Takehisa J, Zekeng L, Ido E, Yamaguchi-Kabata Y, Mboudjeka I, Harada Y, Miura T, Kaptu L, Hayami M. Human immunodeficiency virus type 1 intergroup (M/O) recombination in cameroon. J Virol. 1999;73:6810–6820. doi: 10.1128/jvi.73.8.6810-6820.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tebit DM, Zekeng L, Kaptué L, Gürtler L, Fackler OT, Keppler OT, Herchenröder O, Kräusslich H-G. Construction and characterization of an HIV-1 group O infectious molecular clone and analysis of vpr- and nef-negative derivatives. Virology. 2004;326:329–339. doi: 10.1016/j.virol.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 31.Buck CB, Shen X, Egan MA, Pierson TC, Walker CM, Siliciano RF. The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J Virol. 2001;75:181–191. doi: 10.1128/JVI.75.1.181-191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poon DT, Chertova EN, Ott DE. Human immunodeficiency virus type 1 preferentially encapsidates genomic RNAs that encode Pr55(Gag): functional linkage between translation and RNA packaging. Virology. 2002;293:368–378. doi: 10.1006/viro.2001.1283. [DOI] [PubMed] [Google Scholar]
- 33.St Louis DC, Gotte D, Sanders-Buell E, Ritchey DW, Salminen MO, Carr JK, McCutchan FE. Infectious molecular clones with the nonhomologous dimer initiation sequences found in different subtypes of human immunodeficiency virus type 1 can recombine and initiate a spreading infection in vitro. J Virol. 1998;72:3991–3998. doi: 10.1128/jvi.72.5.3991-3998.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hussein IT, Ni N, Galli A, Chen J, Moore MD, Hu WS. Delineation of the preferences and requirements of the human immunodeficiency virus type 1 dimerization initiation signal by using an in vivo cell-based selection approach. J Virol. 2010;84:6866–6875. doi: 10.1128/JVI.01930-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nikolaitchik O, Rhodes TD, Ott D, Hu WS. Effects of mutations in the human immunodeficiency virus type 1 Gag gene on RNA packaging and recombination. J Virol. 2006;80:4691–4697. doi: 10.1128/JVI.80.10.4691-4697.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Russel DW. Molecular cloning : a laboratory manual. Third Edition. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 37.Yee JK, Miyanohara A, LaPorte P, Bouic K, Burns JC, Friedmann T. A general method for the generation of high-titer, pantropic retroviral vectors: highly efficient infection of primary hepatocytes. Proc Natl Acad Sci U S A. 1994;91:9564–9568. doi: 10.1073/pnas.91.20.9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kotsopoulou E, Kim VN, Kingsman AJ, Kingsman SM, Mitrophanous KA. A Rev-independent human immunodeficiency virus type 1 (HIV-1)-based vector that exploits a codon-optimized HIV-1 gag-pol gene. J Virol. 2000;74:4839–4852. doi: 10.1128/jvi.74.10.4839-4852.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]