Abstract
The pregnane X receptor (PXR) is the molecular target for catatoxic steroids such as pregnenolone 16α-carbonitrile (PCN), which induce cytochrome P450 3A (CYP3A) expression and protect the body from harmful chemicals. In this study, we demonstrate that PXR is activated by the toxic bile acid lithocholic acid (LCA) and its 3-keto metabolite. Furthermore, we show that PXR regulates the expression of genes involved in the biosynthesis, transport, and metabolism of bile acids including cholesterol 7α-hydroxylase (Cyp7a1) and the Na+-independent organic anion transporter 2 (Oatp2). Finally, we demonstrate that activation of PXR protects against severe liver damage induced by LCA. Based on these data, we propose that PXR serves as a physiological sensor of LCA, and coordinately regulates gene expression to reduce the concentrations of this toxic bile acid. These findings suggest that PXR agonists may prove useful in the treatment of human cholestatic liver disease.
For over 40 years it has been understood that certain steroids exhibit a protective effect against various types of intoxication. These “catatoxic” steroids afford protection against harmful chemicals by accelerating their metabolism (1, 2). A comprehensive analysis of steroids in the 1970s identified the synthetic pregnane pregnenolone 16α-carbonitrile (PCN) as a potent catatoxic agent in rodents. Subsequent studies revealed that PCN and other catatoxic compounds stimulate the transcription of the CYP3A subfamily of cytochrome P450 monooxygenases, which are abundant in the liver, where they metabolize a wide variety of xenobiotics and natural compounds including steroids and bile acids (3, 4).
In 1998, we reported (5) the identification of a mouse orphan nuclear receptor, termed the pregnane X receptor (PXR; NR1I2), which is activated by PCN and other catatoxic compounds. We and others (6–10) subsequently cloned and characterized the human, rabbit, and rat orthologs of PXR. The human ortholog of PXR is alternately referred to as the steroid and xenobiotic receptor (SXR; ref. 7) or pregnane-activated receptor (PAR; ref. 6). PXR is activated by the structurally diverse collection of compounds that are known to induce CYP3A expression and binds to xenobiotic response elements present in CYP3A promoters as a heterodimer with the 9-cis retinoic acid receptor (RXR; NR2B1). Recently, it was shown that Cyp3A11 expression is not induced by PCN or dexamethasone in mice lacking functional PXR (11). Together, these data establish PXR as a key regulator of CYP3A expression.
Lithocholic acid (LCA) is a hydrophobic secondary bile acid that is primarily formed in the intestine by the bacterial 7α-dehydroxylation of chenodeoxycholic acid. Administration of LCA and its conjugates to rodents is known to cause intrahepatic cholestasis (12, 13). Cholestasis, functionally defined as a cessation or impairment of bile flow, can cause nutritional imbalance related to malabsorption of lipids and fat-soluble vitamins and, moreover, irreversible liver damage as a result of the accumulation of toxins normally excreted in bile (14). In humans, elevated levels of LCA are found in patients suffering from chronic cholestatic liver disease (15). The potentially harmful effects of LCA and other bile acids are attenuated by two hepatic detoxification pathways, namely hydroxylation and conjugation. These reactions make the bile acid more hydrophilic and facilitate its excretion in the feces or urine. Notably, 6-hydroxylation of LCA is catalyzed by members of the CYP3A subfamily (16, 17).
In 1970, Selye (18) showed that PCN prevented the LCA-induced hepatoxicity and mortality in rodents. Armed with the knowledge that PXR is the PCN receptor, we have now investigated the role of PXR in LCA metabolism. Our data suggest that PXR plays a fundamental role in protecting the body from toxic bile acids.
Materials and Methods
Generation of a PXR-Deficient Mouse Line.
Three genomic clones containing the mouse PXR locus were isolated by screening a 129/Sv genomic BAC library using a cDNA probe derived from mouse PXR cDNA. An ≈15-kb KpnI fragment containing the first coding exon was isolated and sequenced. A replacement-type targeting vector was constructed as shown in Fig. 1A. An ≈8.0-kb SpeI fragment was subcloned into the SpeI site of pBluescript (Stratagene). This 8.0-kb fragment, representing the long arm of the targeting vector, was excised by using the NotI and SalI restriction sites in pBluescript and inserted at the 5′ end of a neomycin (neo) expression cassette that contains the neomycin phosphotransferase gene under the control of the phosphoglycerate kinase I (PGK) promoter. A 3-kb fragment corresponding to the 3′-short arm of the targeting vector was generated by PCR and inserted into the XhoI site downstream of the neo expression cassette. A copy of the herpes simplex virus tk gene, also under the control of the PGK promoter, is located at the 3′ end of the short arm of the vector. This targeting strategy is summarized in Fig. 1A.
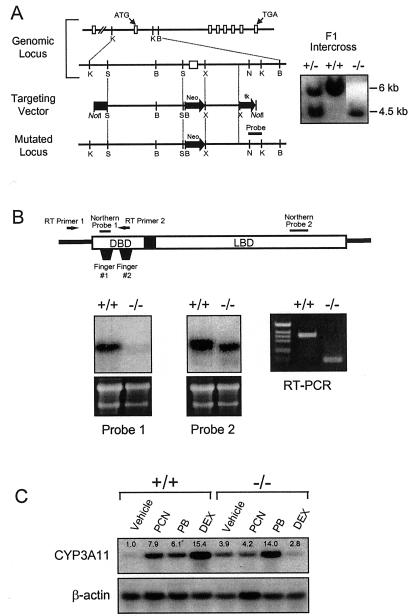
Figure 1.
Disruption of the PXR gene in ES cells and mice. (A) Schematic representation of the PXR locus segment, the targeting vector and the targeted PXR allele. Open boxes indicate exons and labeled boxes indicate the PGK-tk and PGK-neo cassettes. Selected restriction endonuclease sites are indicated: B, BamHI; K, KpnI; N, NheI; S, SpeI; and X, XhoI. A novel BamHI site is introduced into the PXR locus by the homologous recombination event allowing the targeted locus to be distinguished from the wild-type allele by Southern analysis of BamHI digested DNA with the indicated probe (shown at the right). (B) Northern blot analysis of RNA isolated from the livers of wild-type and PXR−/− mice by using the probes indicated in the schematic of the PXR cDNA. Reverse transcription (RT)–PCR was performed using the primers indicated in the schematic. Amplification of the wild-type and disrupted PXR mRNAs yielded products of 111 bp and 363 bp, respectively. (C) Total RNA was prepared from the livers of three wild-type and PXR−/− mice treated with PCN, PB, dexamethasone, or vehicle alone. RNA samples were pooled before Northern blot analysis with probes for Cyp3a11. Bands were quantitated as described in Materials and Methods and represent the mean obtained from three animals in each treatment group. Values are normalized to β-actin and are expressed as fold change relative to wild-type mice receiving vehicle alone.
Embryonic Stem (ES) Cell Culture and Southern Blot Analysis.
Mouse R1 ES cells derived from the 129-Sv strain (19) were cultured and electroporated as described (20). ES cells in which the plasmid integrated by homologous recombination with the endogenous PXR locus were identified by Southern blot analysis of the DNA prepared from individual ES cell colonies using a probe derived from intron 2 (Fig. 1A). The genomic DNA was digested with BamHI, transferred to positively charged nylon membrane (Hybond N+; Amersham Pharmacia), and probed with a ≈400-bp 32P-labeled KpnI-NheI. ES cells carrying the correctly targeted locus were introduced into C57BL/6J blastocysts and the resulting chimeric males bred to C57BL/6 females. Offspring were screened by Southern blot analysis of DNA obtained from tail biopsies to identify those heterozygous for the mutant PXR allele. These heterozygous offspring were then intercrossed to obtain mice homozygous for the PXR mutation and wild-type littermate controls.
Maintenance and Treatment of PXR−/− and Control Mouse Populations.
Adult male wild-type (PXR+/+) and PXR-null (PXR−/−) mice were maintained on standard laboratory chow and were allowed food and water ad libitum. Mice were treated with PCN (0.4 mg/g), dexamethasone (0.1 mg/g), sodium phenobarbital (PB, 0.1 mg/g), or LCA (0.125 mg/g). All inducers were dissolved in corn oil and injected intraperitoneally either once (PCN, dexamethasone, and PB) or twice (LCA) a day for 4 days. Animals were killed 24 h following the final injection. For LCA and PCN cotreatment, PCN was administered by gavage (0.5 mg/g in corn oil) for 3 days. Subsequently, PCN treatment was continued for a further 4 days, during which time animals also received a daily i.p. injection of LCA (0.25 mg/kg in corn oil). Mice were exsanguinated 24 h following the final injection and serum alanine transaminase (ALT) and sorbitol dehydrogenase (SDH) levels determined by using standard techniques. Livers were fixed in neutral-buffered formalin solution Sigma, embedded in paraffin wax, and stained with hematoxylin and eosin. Differences between SDH and ALT levels in vehicle- and LCA-treated and PCN- and LCA-treated animals were determined by using a one-way ANOVA. Significant differences were determined by using Duncan's multiple range post hoc test.
RNA Isolation and Northern Blot Analysis.
Total RNA was isolated from liver by using a commercially available reagent (Trizol; Life Technologies, Grand Island, NY) according to the manufacturer's instructions. Total RNA (10 μg) was resolved on a 1% agarose/2.2 M formaldehyde denaturing gel and transferred to a nylon membrane (Hybond N+; Amersham Pharmacia). Blots were hybridized with 32P-labeled cDNAs corresponding to Cyp3a11 (bases 69 to 1609, GenBank accession no. X60452), rat tyrosine aminotransferase (TAT; bases 95 to 1562, GenBank accession no. X02741), rat CYP7A1 (bases 235 to 460, GenBank accession no. J05460), mouse GR (bases 1982 to 2265, GenBank accession no. M10901), or the 5′-untranslated region of the murine ortholog of rat Oatp2 (C.D.K., unpublished data). Subsequently, blots were stripped and reprobed with a radiolabeled β-actin cDNA (CLONTECH). The intensity of signals was quantitated by using imagequant software (Molecular Dynamics).
Transient Transfection and Scintillation Proximity Binding Assay.
Transient transfection assays and scintillation proximity binding assays were performed as described (10). Bile acids were purchased from Steraloids (Newport, RI) or Sigma.
Quantitation of LCA in Urine.
LCA levels in urine were quantitated by atmospheric pressure ionization–liquid chromatography mass spectrometry (API-LCMS). Briefly, an equal volume of mouse urine and a 5 μg/ml methanolic solution of 2,2,4,4-d4-cholic acid (D4-cholic acid; CDN Isotopes, Quebec, Canada) were combined. Samples were sonicated, centrifuged (3,000 × g for 10 min), and filtered through a 0.45-μm filter unit before injection onto the analytical column of an LCMS instrument (Hewlett–Packard Series 1100 Liquid Chromatograph Mass Selective Detector). LCA and D4-cholic acid were detected as the molecular ions ([M-H−]) 375 and 311 m/z, respectively, in the negative selected ion monitoring mode of the instrumentation. LCA concentrations in the study samples were calculated by comparison to standard solutions of LCA containing D4-cholic acid as internal standard. The significance of differences between mean values was analyzed by using an unpaired Student's t test.
Results
We generated mice lacking functional PXR to study the biological functions of this orphan receptor. A genomic clone encompassing the entire coding region of the murine Pxr gene was isolated and characterized. We replaced the first coding exon, which includes the translation start site and the first zinc finger of the PXR DNA-binding domain (amino acids 1 to 63; ref. 5), with a neo resistance cassette by using homologous recombination (Fig. 1A). As expected, based on the size of this exon, Northern blot and PCR analysis showed that PXR−/− mice expressed a PXR transcript that was ≈200 bp shorter than the wild-type transcript (Fig. 1B). Isolation and sequence analysis of the mutant PXR amplicon revealed that exon 1 was fused to exon 3 in the mutant animal (data not shown). The truncated PXR transcript was present at reduced levels in the PXR−/− mice relative to the wild-type transcript in PXR+/+ animals (Fig. 1B). Because the first zinc finger is essential for DNA binding, we expect that even if this mRNA is translated from a spurious initiation site, the protein produced will not be functional. For simplicity, we will therefore refer to mice homozygous for this mutation as PXR−/−. As described (11), PXR−/− mice were viable, fertile, bred with normal Mendelian distribution, and did not exhibit any overt phenotypic change. Extensive serum analysis did not reveal any significant changes in a number of parameters, including free- and HDL-cholesterol, steroid hormones (progesterone, estradiol, testosterone, and corticosterone), triglycerides, transaminases, albumin, total bilirubin, and total bile acids (data not shown). As anticipated, PXR was required for the induction of Cyp3a11 expression by PCN and dexamethasone (Fig. 1C). Interestingly, basal levels of Cyp3a11 mRNA were increased ≈4-fold in the PXR−/− animals (Fig. 1C), demonstrating an unexpected role for PXR in the repression of Cyp3a11 basal expression. This effect of PXR ablation on basal Cyp3a11 expression was not observed in another PXR−/− mouse line that was derived independently (11). The basis for this difference is not known. Notably, the barbiturate PB induced Cyp3a11 expression in the PXR−/− mice to the same degree as in wild-type mice (Fig. 1C), demonstrating that this xenobiotic can mediate its effects on Cyp3a11 through a PXR-independent signaling pathway. Because PB can activate the orphan nuclear receptor CAR (constitutive androstane receptor; NR1I4; refs. 21 and 22), these data support a role for CAR in the regulation of Cyp3a11 in vivo.
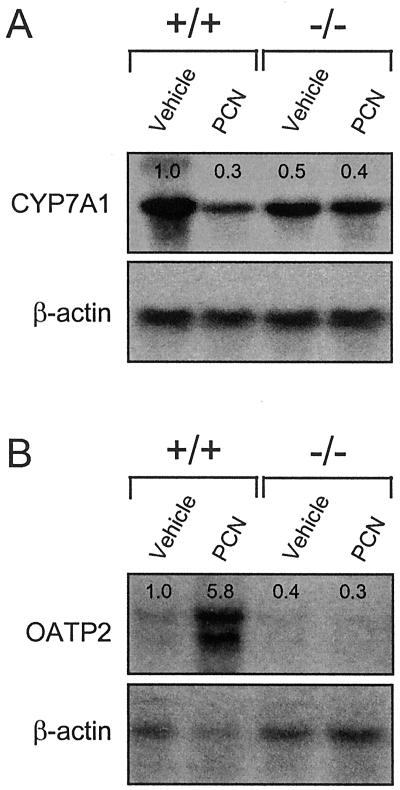
PCN treatment of rats decreases expression of Cyp7a1, which encodes the enzyme responsible for the first and rate-limiting step in the metabolism of cholesterol to bile acids (23). We examined whether PXR has a role in the negative regulation of Cyp7a1 expression in mice. Northern blot analysis of Cyp7a1 mRNA levels in PCN-treated wild-type mice revealed an ≈3-fold decrease in Cyp7a1 expression relative to animals that received vehicle alone (Fig. 2A). Interestingly, Cyp7a1 expression in the PXR−/− mice was dysregulated in two respects: (i) The basal level of Cyp7a1 expression was decreased ≈2-fold in the PXR−/− animals; (ii) Cyp7a1 expression was refractory to PCN treatment (Fig. 2A). These data demonstrate a role for PXR in the regulation of both basal Cyp7a1 expression and its repression by PCN. Expression levels of the nuclear receptors LXRα (NR1H3) and FXR (NR1H4), which stimulate and repress Cyp7a1 in response to oxysterols and bile acids (24), respectively, were unchanged in the PXR−/− mice (data not shown). Moreover, expression of the orphan nuclear receptor small heterodimer partner (SHP, NR0B2), which was recently implicated in bile acid-mediated repression of Cyp7a1 expression (25, 26), was unchanged (data not shown).
Figure 2.
PXR represses Cyp7a1 and induces Oatp2 expression in response to PCN. Total RNA was prepared from the livers of PXR+/+ and PXR−/− mice treated with PCN or vehicle alone. RNA from three animals was pooled and subjected to Northern blot analysis with probes for (A) Cyp7a1, (B) Oatp2, and β-actin. Quantitation of bands was performed as described in Fig. 1.
We also examined the effects of PCN on the expression of other genes involved in the transport and metabolism of bile acids, including those encoding the Na+-dependent taurocholate cotransporting polypeptide (Ntcp), multidrug resistance-associated protein 3 (Mrp3), and the Na+-independent organic anion transporter 2 (Oatp2). PCN treatment had no effect on Ntcp or Mrp3 expression (data not shown). However, PCN treatment strongly induced Oatp2 expression in the PXR+/+ mice, but not in the PXR−/− mice (Fig. 2B). Oatp2 is a basolateral (sinusoidal) transporter that can mediate hepatocellular uptake of a wide range of amphipathic substrates, including bile acids and xenobiotics (27, 28). Interestingly, unlike Cyp3a11, the basal level of Oatp2 expression was not increased in the PXR−/− mice (Fig. 2B).
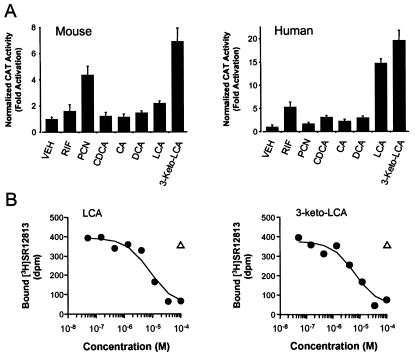
Because Cyp7a1, Oatp2, and members of the CYP3A subfamily are involved in bile acid synthesis, transport, and metabolism (16, 17, 27–30), we examined whether bile acids might directly regulate PXR activity. CV-1 cells were transfected with expression plasmids for either mouse PXR or human PXR (8) and a reporter plasmid containing two copies of the Cyp3a23 PXR response-element upstream of the thymidine kinase (tk) gene promoter and chloramphenicol acetyl transferase (CAT) gene (5). Transfected cells were treated with various bile acids and their taurine or glycine conjugates. Notably, the secondary bile acid LCA was an efficacious activator of human PXR and also activated mouse PXR (Fig. 3A). Neither conjugated LCA nor any of the other conjugated bile acids activated PXR (data not shown). 3-Keto-LCA is the major metabolite of LCA in rats: following LCA feeding, 3-keto-LCA concentrations approach those of LCA (31). 3-Keto-LCA was an efficacious activator of both the human and mouse PXR (Fig. 3A). LCA and 3-keto-LCA bound to human PXR in a scintillation proximity binding assay with half-maximal inhibitory concentrations of 9 μM and 15 μM, respectively (Fig. 3B). Thus, PXR is a low affinity receptor for a subset of bile acids.
Figure 3.
Bile acids bind and activate PXR. (A) CV-1 cells were transfected with expression plasmids for mouse PXR or human PXR and the reporter plasmid (Cyp3a23)2-tk-CAT, and the cells treated with 100 μM concentrations of the indicated bile acids: CDCA, chenodeoxycholic acid; CA, cholic acid; DCA, deoxycholic acid; LCA, lithocholic acid; 3-keto-LCA, 3-keto-lithocholic acid. PCN and rifampicin (RIF) were used as positive controls for mPXR and hPXR, respectively, at 10 μM concentration. Cell extracts were subsequently assayed for CAT activity. Data represent the mean ± SD of assays performed in triplicate. Activation of mPXR by LCA is highly statistically significant as measured by the Student's t test (P < 0.001). (B) Scintillation proximity competition binding assays were performed with human PXR ligand binding domain and 10 nM [3H]SR12813 in the presence of increasing concentrations of LCA and 3-keto-LCA. A single-point negative control (tauro-β-muricholic acid, open triangle) is also shown.
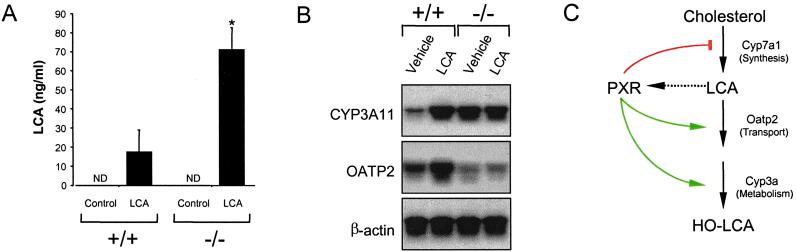
LCA is a highly toxic secondary bile acid that causes cholestasis, a pathogenic state characterized by decreased bile flow and the accumulation of bile constituents in the liver and blood (13, 32). Notably, LCA concentrations of 5–10 μM have been reported in the livers of cholestatic patients and in rat models of biliary cholestasis (33). Because LCA activates PXR at these concentrations, we examined whether PXR−/− mice showed defects in LCA metabolism. LCA feeding resulted in the appearance of a sticky residue in the cages of the PXR−/− mice, but not in the wild-type mice (data not shown). Analysis of the urine revealed that PXR−/− mice had markedly elevated levels of LCA compared with wild-type animals (Fig. 4A). In addition, we observed that shorter term treatment of wild-type mice with LCA resulted in a robust induction of hepatic Cyp3a11 and Oatp2 expression, whereas LCA treatment had no effect on the expression of these genes in PXR−/− mice (Fig. 4B). These data demonstrate that PXR can function as a receptor for LCA and/or LCA metabolites and regulates LCA metabolism in vivo.
Figure 4.
Effects of treating wild-type and PXR−/− mice with LCA. (A) Wild-type and PXR−/− mice were fed a diet supplemented with 0.5% LCA for 8 days. Urine was collected on day 8 of the study and LCA concentrations were determined. Data represent the mean ± SEM of assays performed on urine from eight different mice. ND, not detected; *, Statistically significantly difference between wild-type and PXR−/−, P < 0.05. (B) Total RNA was isolated and pooled from the livers of six wild-type and PXR−/− mice injected for 4 days with LCA or vehicle alone and subject to Northern blot analysis with probes for Cyp3a11, Oatp2, and β-actin. (C) Model for PXR as a bile acid sensor. Activation of PXR by LCA and/or LCA metabolites results in repression of Cyp7a1, which blocks bile acid biosynthesis, and induction of Oatp2 and Cyp3a expression, which promote bile acid uptake and metabolism. The net effect is to prevent the accumulation of bile acids to toxic levels.
We propose that PXR serves as a physiological sensor of LCA and its metabolites in the liver and coordinately regulates gene expression so as to reduce their concentration (Fig. 4C). PXR inhibits Cyp7a expression, which blocks bile acid synthesis. At the same time, PXR induces expression of Oatp2 and Cyp3a11, which may promote the transport and metabolism of LCA. Oatp2 is localized on the basolateral membrane of the hepatocyte and is involved in the cellular uptake of bile acids (27, 28). Its induction by PXR would presumably increase uptake of LCA and other bile acids from sinusoidal blood into the hepatocyte where hydroxylation by Cyp3a11 or other Cyp3a subfamily members could take place. These more hydrophilic bile acid metabolites could then be excreted in the feces or urine. Although members of the CYP3A subfamily are known to hydroxylate LCA (16, 17, 30), it is important to note that bile acid metabolism varies considerably across species with regard to the enzymes involved and the metabolites produced, and it is yet to be determined whether Cyp3a11 is involved in the oxidative metabolism of LCA in mice.
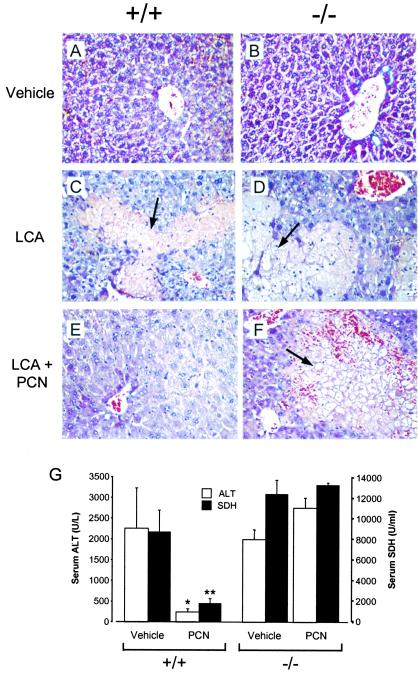
A prediction of this model is that PXR activation will protect the liver against LCA-induced toxicity. To test this prediction, wild-type and PXR−/− mice were treated with LCA in either the presence or absence of the potent PXR ligand PCN. As expected, treatment of either wild-type or PXR−/− mice with LCA resulted in severe liver damage as manifested by the appearance of necrotic foci and high serum levels of the liver ALT and SDH (Fig. 4 A, C, and G). Cotreatment with PCN dramatically reduced the detrimental effects of LCA on the livers of wild-type mice (Fig. 4 E and G). In marked contrast, cotreatment with PCN did not reverse the hepatotoxicity of LCA in the PXR−/− mice (Fig. 4 B, D, F, and G). Thus, PXR activation strongly protects the livers of mice from the harmful effects of LCA.
Summary and Perspectives
It has been almost 30 years since Hans Selye first showed that PCN treatment blocks the hepatotoxicity and mortality caused by LCA treatment in rats (18). In this report, we have used PXR-null mice to demonstrate that the orphan nuclear receptor PXR mediates the hepatoprotective effects of PCN against LCA-induced toxicity. Moreover, we have shown that PXR is activated by LCA and its 3-keto metabolite and coordinately regulates genes involved in the biosynthesis, transport, and metabolism of LCA. Our results indicate that PXR plays a fundamental role in protecting the liver against pathophysiological levels of LCA. PXR thus joins FXR as nuclear receptors that are activated by bile acids (34–36). Although PXR and FXR are activated by distinct sets of bile acids, both receptors are activated by LCA. This raises the interesting possibility that PXR and FXR cooperate to remove LCA from the body when its concentrations reach pathophysiological levels.
Several previous reports suggest that our findings may have implications in the treatment of human cholestatic liver disease. Notably, urine from patients suffering from cholestasis contains elevated levels of 6-hydroxylated bile acids (including the LCA metabolite hyodeoxycholic acid), which are products of CYP3A4 (16, 37). These findings suggest that increased 6-hydroxylation is a relevant mechanism for reducing the levels of toxic bile acids in humans. Elevated levels of 6-hydroxylated bile acids are also observed in the urine of healthy subjects treated with the PXR ligand rifampicin (38). Interestingly, rifampicin has been used successfully in the treatment of pruritus associated with intrahepatic cholestasis and, in some instances, has been reported to induce remission of cholestasis (39–41). The molecular basis for these clinical effects has remained obscure. Based on our data, we suggest that the anticholestatic effects of rifampicin may be mediated through PXR, and that potent PXR ligands may be more efficacious in the treatment of cholestasis, a severe hepatic disease for which there is no known cure.
Figure 5.
PXR activation protects against LCA-induced hepatotoxicity. Wild-type (A, C, and E) and PXR−/− (B, D, and E) mice were treated with vehicle alone (A and B), LCA (C and D), or LCA and PCN (E and F). Liver sections were prepared for histology and stained with hematoxylin and eosin. Areas of severe necrosis in panels C, D, and E are indicated with arrows. (G) Serum ALT (open bars) and SDH (closed bars) levels were measured in wild-type and PXR−/− mice treated with either LCA and vehicle (Vehicle) or LCA and PCN (PCN). ALT and SDH values in untreated animals were 28 ± 10 units/l and 150 ± 20 units/ml, respectively. Data represent the mean ± SEM of assays performed with serum from six different animals. Statistical differences compared with the same genotype treated with LCA and vehicle: *, P < 0.005; **, P < 0.001.
Acknowledgments
We thank Rusty Murray, Barbara Denton, Carolyn Wilson, Mark Justice, Jane Binz, Tula Milliken, and Paul Novak for their patient technical assistance; Jennifer Shenk and Lakshman Ramamurthy for their contributions during the early stages of this work; and Jane Binz for serum analysis. This study was supported in part by National Institutes of Health Grants ES-09649 and ES-07079 (to C.D.K.).
Abbreviations
- PCN
pregnenolone 16α-carbonitrile
- PXR
pregnane X receptor
- LCA
lithocholic acid
- CYP
cytochrome P450
- PB
phenobarbital
- Oatp2
Na+-independent organic anion transporter 2
- SDH
sorbitol dehydrogenase
- PGK
phosphoglycerate kinase I
- ES
embryonic stem
- ALT
alanine transaminase
- neo
neomycin
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Selye H. J Pharm Sci. 1971;60:1–28. doi: 10.1002/jps.2600600102. [DOI] [PubMed] [Google Scholar]
- 2.Kourounakis P, Selye H, Tache Y. Adv Steroid Biochem Pharmacol. 1977;6:35–57. [PubMed] [Google Scholar]
- 3.Guzelian P S. In: Microsomes and Drug Oxidations. Miners J O, Birkett D J, Drew R, McManus M, editors. London: Taylor and Francis; 1988. pp. 148–155. [Google Scholar]
- 4.Maurel P. In: Cytochromes P 450, metabolic and toxicological aspects. Ioannides C, editor. Boca Raton, FL: CRC; 1996. pp. 241–270. [Google Scholar]
- 5.Kliewer S A, Moore J T, Wade L, Staudinger J L, Watson M A, Jones S A, McKee D D, Oliver B B, Willson T M, Zetterstrom R H, et al. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 6.Bertilsson G, Heidrich J, Svensson K, Asman M, Jendeberg L, Sydow-Backman M, Ohlsson R, Postlind H, Blomquist P, Berkenstam A. Proc Natl Acad Sci USA. 1998;95:12208–12213. doi: 10.1073/pnas.95.21.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blumberg B, Sabbagh W, Jr, Juguilon H, Bolado J, Jr, van Meter C M, Ong E S, Evans R M. Genes Dev. 1998;12:3195–3205. doi: 10.1101/gad.12.20.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lehmann J M, McKee D D, Watson M A, Willson T M, Moore J T, Kliewer S A. J Clin Invest. 1998;102:1016–1023. doi: 10.1172/JCI3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H, LeCulyse E, Liu L, Hu M, Matoney L, Zhu W, Yan B. Arch Biochem Biophys. 1999;368:14–22. doi: 10.1006/abbi.1999.1307. [DOI] [PubMed] [Google Scholar]
- 10.Jones S A, Moore L B, Shenk J L, Wisely G B, Hamilton G A, McKee D D, Tomkinson N C O, LeCluyse E L, Lambert M H, Willson T M, et al. Mol Endocrinol. 2000;14:27–39. doi: 10.1210/mend.14.1.0409. [DOI] [PubMed] [Google Scholar]
- 11.Xie W, Barwick J L, Downes M, Blumberg B, Simon C M, Nelson M C, Neuschwander-Tetri B A, Brunt E M, Guzelian P S, Evans R M. Nature (London) 2000;406:435–439. doi: 10.1038/35019116. [DOI] [PubMed] [Google Scholar]
- 12.Fisher M M, Magnusson R, Miyai K. Lab Invest. 1971;25:88–91. [PubMed] [Google Scholar]
- 13.Javitt N B. Nature (London) 1966;210:1262–1263. doi: 10.1038/2101262a0. [DOI] [PubMed] [Google Scholar]
- 14.Trauner M, Meier P, Boyer J L. N Engl J Med. 1998;339:1217–1227. doi: 10.1056/NEJM199810223391707. [DOI] [PubMed] [Google Scholar]
- 15.Fischer S, Beuers U, Spengler U, Zwiebel F M, Koebe H-G. Clin Chem Acta. 1996;251:173–186. doi: 10.1016/0009-8981(96)06305-x. [DOI] [PubMed] [Google Scholar]
- 16.Araya Z, Wikvall K. Biochim Biophys Acta. 1999;1:47–54. doi: 10.1016/s1388-1981(99)00031-1. [DOI] [PubMed] [Google Scholar]
- 17.Teixeira J, Gil G. J Biol Chem. 1991;266:21030–21036. [PubMed] [Google Scholar]
- 18.Selye H. Proc Soc Exp Biol Med. 1972;141:555–558. doi: 10.3181/00379727-141-36821. [DOI] [PubMed] [Google Scholar]
- 19.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder J C. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohn A, Koller B H. In: DNA Cloning. Glover D M, Hames B D, editors. Vol. 4. New York: Oxford Univ. Press; 1995. pp. 143–184. [Google Scholar]
- 21.Honkakoski P, Negishi M. Biochem J. 2000;347:321–337. doi: 10.1042/0264-6021:3470321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei P, Zhang J, Egan-Haffey M, Liang S, Moore D D. Nature (London) 2000;407:920–923. doi: 10.1038/35038112. [DOI] [PubMed] [Google Scholar]
- 23.Li Y C, Wang D P, Chiang J Y. J Biol Chem. 1990;265:12012–12019. [PubMed] [Google Scholar]
- 24.Repa J J, Mangelsdorf D J. Curr Opin Biotechnol. 1999;10:557–563. doi: 10.1016/s0958-1669(99)00031-2. [DOI] [PubMed] [Google Scholar]
- 25.Goodwin B, Jones S A, Price R R, Watson M A, McKee D D, Moore L B, Galardi C, Wilson J G, Lewis M C, Roth M E, et al. Mol Cell. 2000;6:517–526. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 26.Lu T T, Makishama M, Repa J J, Schoonjans K, Kerr T A, Auwerx J, Mangelsdorf D J. Mol Cell. 2000;6:507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- 27.Noe B, Hagenbuch B, Stieger B, Meier P. Proc Natl Acad Sci USA. 1997;94:10346–10350. doi: 10.1073/pnas.94.19.10346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reichel C, Gao B, Van Montfoort J, Cattori V, Rahner C, Hagenbuch B, Stieger B, Kamisako T, Meier P J. Gastroenterology. 1999;117:688–695. doi: 10.1016/s0016-5085(99)70463-4. [DOI] [PubMed] [Google Scholar]
- 29.Chiang J Y L. Front Biosci. 1998;3:D176–D193. doi: 10.2741/a273. [DOI] [PubMed] [Google Scholar]
- 30.Chang T K, Teixeira J, Gil G, Waxman D J. Biochem J. 1993;291:429–433. doi: 10.1042/bj2910429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakai K, Makino T, Kawai Y, Mutai M. Microbiol Immunol. 1980;24:187–196. doi: 10.1111/j.1348-0421.1980.tb00578.x. [DOI] [PubMed] [Google Scholar]
- 32.Miyai K, Mayr W W, Richardson A L. Lab Invest. 1975;32:527–535. [PubMed] [Google Scholar]
- 33.Setchell K D, Rodrigues C M, Clerici C, Solinas A, Morelli A, Gartung C, Boyer J. Gastroenterology. 1997;112:226–235. doi: 10.1016/s0016-5085(97)70239-7. [DOI] [PubMed] [Google Scholar]
- 34.Parks D J, Blanchard S G, Bledsoe R K, Chandra G, Consler T G, Kliewer S A, Stimmel J B, Willson T M, Zavacki A M, Moore D D, et al. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- 35.Makishima M, Okamoto A Y, Repa J J, Tu H, Learned R M, Luk A, Hull M V, Lustig K D, Mangelsdorf D J, Shan B. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- 36.Wang H, Chen J, Hollister K, Sowers L C, Forman B M. Mol Cell. 1999;3:543–553. doi: 10.1016/s1097-2765(00)80348-2. [DOI] [PubMed] [Google Scholar]
- 37.Bremmelgaard A, Sjovall J. Eur J Clin Invest. 1979;9:341–348. doi: 10.1111/j.1365-2362.1979.tb00894.x. [DOI] [PubMed] [Google Scholar]
- 38.Wietholtz H, Marschall H U, Sjovall J, Matern S. J Hepatol. 1996;24:713–718. doi: 10.1016/s0168-8278(96)80268-6. [DOI] [PubMed] [Google Scholar]
- 39.Cancado E L, Leitao R M, Carrilho F J, Laudanna A A. Am J Gastroenterol. 1998;93:1510–1517. doi: 10.1111/j.1572-0241.1998.00472.x. [DOI] [PubMed] [Google Scholar]
- 40.Bachs L, Pares A, Elena M, Piera C, Rodes J. Lancet. 1989;1:574–576. doi: 10.1016/s0140-6736(89)91608-5. [DOI] [PubMed] [Google Scholar]
- 41.Gillespie D A, Vickers C R. J Gastroenterol Hepatol. 1993;8:168–173. doi: 10.1111/j.1440-1746.1993.tb01510.x. [DOI] [PubMed] [Google Scholar]