Abstract
The MutM [formamidopyrimidine DNA glycosylase (Fpg)] protein is a trifunctional DNA base excision repair enzyme that removes a wide range of oxidatively damaged bases (N-glycosylase activity) and cleaves both the 3′- and 5′-phosphodiester bonds of the resulting apurinic/apyrimidinic site (AP lyase activity). The crystal structure of MutM from an extreme thermophile, Thermus thermophilus HB8, was determined at 1.9 Å resolution with multiwavelength anomalous diffraction phasing using the intrinsic Zn2+ ion of the zinc finger. MutM is composed of two distinct and novel domains connected by a flexible hinge. There is a large, electrostatically positive cleft lined by highly conserved residues between the domains. On the basis of the three-dimensional structure and taking account of previous biochemical experiments, we propose a DNA-binding mode and reaction mechanism for MutM. The locations of the putative catalytic residues and the two DNA-binding motifs (the zinc finger and the helix–two-turns–helix motifs) suggest that the oxidized base is flipped out from double-stranded DNA in the binding mode and excised by a catalytic mechanism similar to that of bifunctional base excision repair enzymes.
Keywords: base excision repair/crystal structure/extreme thermophile/MutM protein/Thermus thermophilus HB8
Introduction
In aerobic organisms, cellular DNA is frequently damaged by activated oxygen species from aerobic energy metabolism or oxidative stress. Highly reactive oxygen accelerates the rate of spontaneous mutation and has, therefore, been implicated as a causative agent for ageing and in the pathogenesis of disease, including cancer (Breimer, 1990; Ames et al., 1995). In particular, the 8-oxoguanine (GO) lesion is one of the most stable products of oxidative DNA damage (Dizdaroglu, 1985). GO can form a pair with adenine as well as with cytosine, resulting in a G:C to T:A transversion (Wood et al., 1990; Shibutani et al., 1991). In Escherichia coli, the GO repair system, composed of the MutM protein [EC 3.2.2.23], also called formamidopyrimidine DNA glycosylase (Fpg), accompanied by MutY and MutT, prevents this mutation (Michaels et al., 1992).
The mutM (fpg) gene encoding the MutM protein is highly conserved across a wide range of aerobic bacteria. These enzymes (Mr 30 kDa) possess the invariant N-terminal sequence Pro-Glu-Leu-Pro-Glu-Val-, two strictly conserved lysine residues (Lys52 and Lys147) and a zinc finger motif (-Cys-X2-Cys-X16-Cys-X2-Cys-) at the C-terminus (Figure 1C).
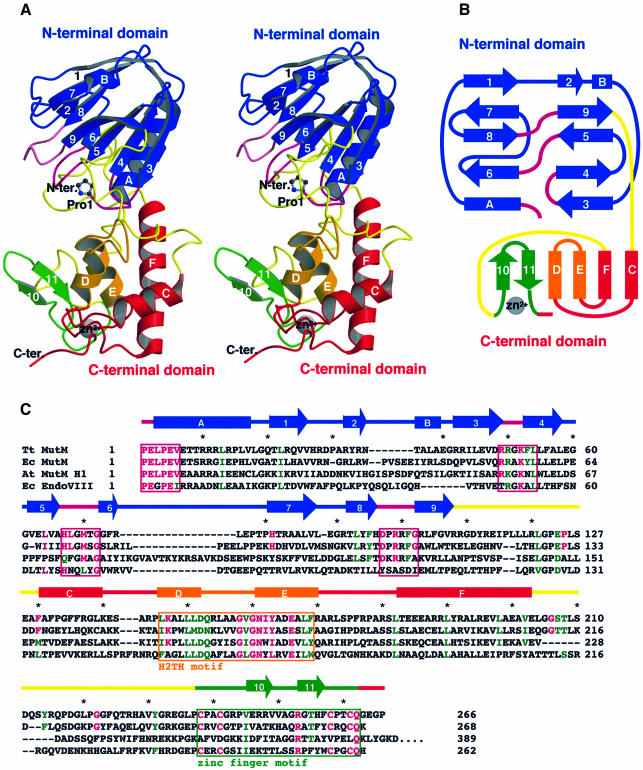
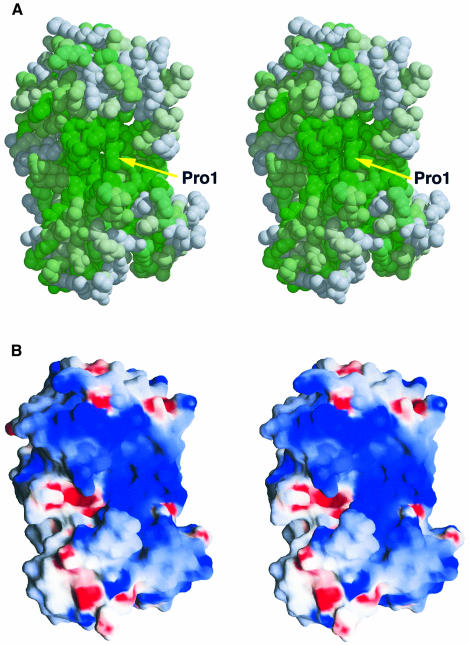
Fig. 1. Overall structure, topology and amino acid sequence of MutM. (A) Stereo ribbon diagram and (B) topology of MutM. The MutM molecule consists of an N-terminal domain (residues 1–109, blue), a C-terminal domain [residues 131–203 (red and orange) and 238–266 (green)] and two long loops [110–130 and 204–237 (yellow)]. The N-terminal domain consists of a two-layered β-sandwich with two α-helices. The C-terminal domain consists of four α-helix bundles (red and orange) and a β-hairpin loop of the zinc finger motif (green). There are two conformers in an asymmetrical unit (see Figure 3B). The conformations of the two long loops (in yellow) in the interdomain cleft differ between the two conformers. These two long loops could work as a hinge in domain movement. Pro1 (shown by a ball-and-stick model), which works as a nucleophile in the glycosylase/AP lyase reaction, is situated at the bottom of the cleft. The highly conserved regions (purple) in the N-terminal domain, the H2TH motif (orange) and the zinc finger motif (green) are located around the cleft (see Figure 2A). (C) Aligned sequences of bacterial T.thermophilus MutM (Tt MutM) (Mikawa et al., 1998), E.coli MutM (Ec MutM) (Boiteux et al., 1987), A.thaliana MutM homologue 1 (At MutM H1) (Murphy and Gao, 1998; Ohtsubo et al., 1998) and E.coli endonuclease VIII (Ec EndoVIII) (Jiang et al., 1997). The α-helices (bars) and β-strands (arrows) are shown above the aligned sequences. The amino acid numbering refers to the T.thermophilus HB8 protein. Coloured letters (homologous residues in magenta, similar residues in green) represent the residues conserved among 16 MutM proteins (see also Figure 2A). The asterisks indicate every tenth amino acid residue of the T.thermophilus sequence. The highly conserved regions in the N-terminal domain (purple), the H2TH motif (orange) and the zinc finger motif (green) are boxed.
MutM was originally isolated from E.coli and characterized as a DNA glycosylase that removes 2,6-diamino-4-hydroxy-5-N-methylformamidopyrimidine (Fapy), the imidazole ring-open form of N7-methylguanine (Boiteux et al., 1987). More recently, MutM was reported to bind to a wide range of oxidatively damaged bases (see Figure 5C), including GO paired to cytosine (Tchou et al., 1991), formamidopyrimidine (FapyG or FapyA) (Boiteux et al., 1992) and 5-hydroxycytosine (5OHC) (Hatahet et al., 1994) and to apurinic/apyrimidinic (AP) sites (Castaing et al., 1992). MutM has at least three enzymatic activities: (i) DNA glycosylase activity, which excises various damaged bases from DNA to produce an aldehydic abasic site; (ii) an AP lyase activity, which cleaves the 3′-phosphodiester bond at AP sites through β-elimination; and (iii) an alternative AP lyase activity, which cleaves the 5′-phosphodiester bond through δ-elimination (O’Connor et al., 1989; Boiteux et al., 1990; Bhagwat and Gerlt, 1996). In terms of the mechanism of MutM glycosylase/AP lyase activity, the initial step has been proposed to be formation of a covalent Schiff base intermediate by nucleophilic attack of the secondary amino group of Pro1 at the N-terminus on the deoxyribose C1′ of the damaged site (see Figure 7) (Tchou and Grollman, 1995; Bhagwat and Gerlt, 1996; Castaing et al., 1999). This is a common mechanism in other N-glycosylase/AP lyase base excision repair enzymes such as E.coli endonuclease III (EndoIII) (Kow and Wallace, 1987; Purmal et al., 1996) and bacteriophage T4 endonuclease V (T4 EndoV) (Schrock and Lloyd, 1991; Purmal et al., 1996). However, the activity of MutM N-glycosylase/AP lyase is quite distinct from those of EndoIII and T4 EndoV, which only cleave the 3′-phosphodiester bond via β-elimination. For EndoIII and T4 EndoV, the nucleophiles are the ε-amino group of Lys120 and the N-terminal α-amino group of Thr2, respectively, and the residues that activate the nucleophile and participate in the β-elimination reaction are Asp138 and Glu23, respectively. In the case of MutM, the Schiff base intermediate was trapped by hydration to identify the α-amino group of Pro1 as a nucleophile (Zharkov et al., 1997). Site-directed mutagenesis of the conserved Lys52 and Lys147 residues suggested their interaction with GO (Rabow and Kow, 1997; Sidorkina and Laval, 1998). It has also been suggested that the zinc finger motif is involved in the catalytic reaction (O’Connor et al., 1993; Tchou et al., 1993). However, the three-dimensional architecture of these residues in the N-glycosylation and β- and δ-elimination reactions remains to be elucidated.
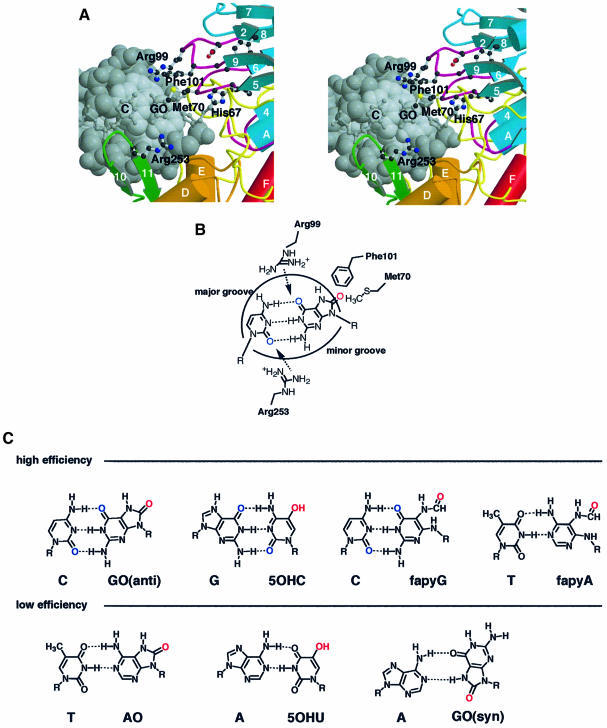
Fig. 5. The structural basis for lesion recognition in MutM. (A) Model of recognition of GO:C-paired DNA (grey) by conserved amino acid residues (ball-and-stick) of MutM. (B) Schematic representation of the proposed lesion-recognition mechanism for the GO:C pair in dsDNA. Arg99 and Arg253 can interact with the keto groups of the GO and C base pair (blue) in both sides of dsDNA to disrupt the pairing hydrogen bonds in the model. The C8 oxo group of the GO base (red) in the DNA major groove could be pulled off by Met70 or Phe101 in the N-terminal domain. Phe101 could be inserted into the vacant site after nucleotide flipping to compensate for base stacking in the dsDNA. (C) Pairing of the oxidized bases in high- and low-efficiency MutM substrates is shown in the upper and lower rows, respectively (Hatahet et al., 1994; Tchou et al., 1994).
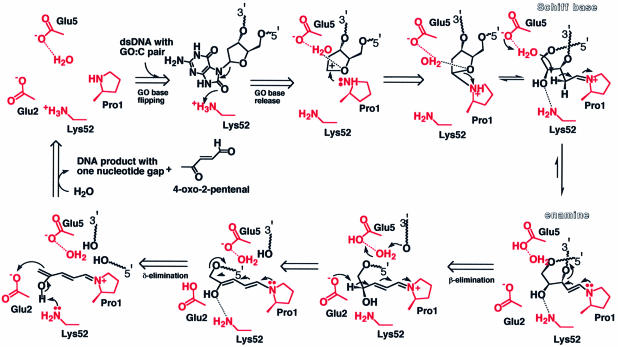
Fig. 7. Schematic representation of the reaction mechanism of MutM N-glycosylase/AP lyase. The reaction scheme is proposed on the basis of the active site architecture according to the mechanism proposed by Castaing et al. (1999). We propose that the invariant amino acid residues (Glu2, Glu5 and Lys52) in the vicinity of the primary catalytic residue Pro1, whose N-terminal amine forms a Schiff base with the C1′ of damaged deoxyribose (Zharkov et al., 1997), act as the additional catalytic residues for the enzyme action and, together with their bound water molecules, form a hydrogen bond network in the active site. In this highly electrostatically positive environment, Lys52 may act as a proton donor for the depurination of the damaged base (Figure 6B). After C2′ of deoxyribose has formed a Schiff base with Pro1, Glu5 could withdraw the proton of C2′ via a bound water, leading to β-elimination. The resulting adduct intermediate (Figure 6C) would deprotonate at C4′ of the opened deoxyribose, leading to δ-elimination. Finally, regain of the proton by Lys52 would release the other product, 4-oxo-2-pentenal, to form the gapped dsDNA product (Bhagwat and Gerlt, 1996). The residues believed to contribute to each reaction step were deduced from the crystal structure and are shown in red.
The primary structures of bacterial (Thermus thermophilus HB8 and E.coli) MutM proteins are similar to those of MutM homologues in Arabidopsis thaliana (At MutM HI), a higher plant (Murphy et al., 1998; Ohtsubo et al., 1998), and E.coli endonuclease VIII (EndoVIII) (Jiang et al., 1997) (Figure 1C; the amino acid numbering refers to the T.thermophilus HB8 protein). These enzymes cleave both 3′- and 5′-phosphodiester bonds of the DNA strand by β- and δ-elimination, respectively, as does MutM protein (Melamede et al., 1994; Ohtsubo et al., 1998). However, AtMMH has no zinc finger motif and EndoVIII’s substrate specificity is different from that of MutM. Recently, 8-oxoguanine DNA glycosylase 1 (OGG1) proteins from several eukaryotes, including humans, and ribosomal protein S3 from Drosophila melanogaster (Krokan et al., 1997) have been characterized as GO:C-specific N-glycosylase/AP lyases with activities identical to those of MutM.
The crystal structures of several DNA glycosylases and their complexes with double-stranded DNA (dsDNA) have revealed common structural features, including a helix–hairpin–helix (HhH) motif and nucleotide flipping. The HhH motif, first reported in E.coli endonuclease III (EndoIII) (Thayer et al., 1995), has also been found in E.coli AlkA (Labahn et al., 1996; Yamagata et al., 1996; Hollis et al., 2000) and MutY (Guan et al., 1998) and in human OGG1 proteins (Bruner et al., 2000). Comparison of the amino acid sequence of MutM with those of these DNA glycosylases has not yet revealed common motifs or catalytic residues in MutM, despite their similar enzymatic properties.
Here we report the X-ray crystallographic structure of the MutM enzyme from an extremely thermophilic bacterium, T.thermophilus HB8, determined at 1.9 Å resolution by multiwavelength anomalous diffraction (MAD) phasing of the Zn2+ ion. The crystal structure shows a zinc finger motif, a helix–two-turns–helix (H2TH) motif instead of an HhH motif and putative catalytic residues analogous to those of the other DNA glycosylases. We propose a model for the structure of the dsDNA–MutM complex. This model readily leads to a reaction mechanism for the enzyme and to an understanding of the catalytic reaction mechanism of related endonucleases.
Results and discussion
Overall structure
MutM measures ∼50 × 30 × 30 Å3 and consists of two compact domains connected by a hinge region (Figure 1A). The N-terminal domain (residues 1–109, coloured blue) is a two-layered β-sandwich structure composed of nine antiparallel β-strands (β1–β9), except for strand β2, with an α-helix on each side of the β-sandwich (αA and αB) (Figure 1A and B). The C-terminal domain (residues 131–266, coloured red, orange or green) has four helices (αC–αF) and two β-strands (β10 and β11) that form a zinc finger motif (Figure 1A and B). There are two molecules in an asymmetrical unit in the crystal. The different overall conformations of these two molecules show that there is significant hinge movement, whereas the corresponding domains of the two molecules in an asymmetrical unit are substantially identical (Figure 3B).
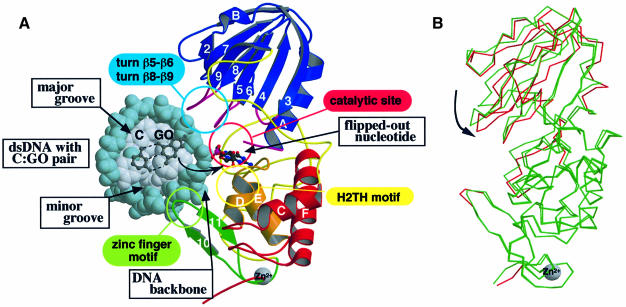
Fig. 3. The model of the MutM–DNA complex and its hinge movement. (A) The model of the complex between the flipped out DNA and MutM in the closed form was obtained by molecular dynamics calculation. The viewing direction is similar to that in Figure 1A. The kinked DNA is drawn as a CPK model with backbones coloured in blue and with bases in grey. C and GO bases and their sugar residues before and after flipping out are shown by ball-and-stick models. All four conserved regions [(i) turns β5–β6 and β8–β9 in the sky-blue circle, (ii) the catalytic site in the pink circle, (iii) the H2TH motif in the yellow circle and (iv) the zinc finger motif in the light-green circle] are in the large cleft of the MutM molecule. The N-terminal domain can access the major groove of DNA and the zinc finger motif of the C-terminal domain can access the minor groove. The H2TH motif of the C-terminal domain is situated near the active site and may interact with the DNA backbone with the damaged base. (B) The superimposed structures of the two molecules in an asymmetrical unit. The superimposition was carried out to minimize the r.m.s.d. of the main chain in the C-terminal domain only. The main chain that deviates by >1 Å from the other molecule is only coloured red in the closed form monomer.
MutM differs structurally from other base excision repair enzymes. A search of the protein structural database for the whole protein and for each domain showed no similar folding (the DALI server; Holm and Sander, 1993).
Most of the conserved amino acid residues in this family of proteins are clustered in the large cleft between the two domains (Figures 1A and 2A). The putative essential residues for the enzymatic activities and the two DNA-binding motifs, the H2TH and zinc finger motifs, are located in this region. The N-terminal proline, whose α-amino group has been identified as a nucleophile in the DNA glycosylase/AP lyase reaction (Zharkov et al., 1997), occupies the bottom of the cleft (Figure 2A). In addition, the molecular surface inside the cleft is highly positive electrostatically (Figure 2B). Thus, the invariant residues in the cleft are candidates for the residues essential for DNA binding and catalysis.
Fig. 2. Conserved residues and electrostatic potential surface of MutM. (A) Stereo CPK model of MutM looking towards the interdomain large cleft. The surface is shown by a colour gradient according to residue conservation, with absolutely conserved residues in dark green. Homology was based on multiple alignment of the MutMs from T.thermophilus HB8 (Mikawa et al., 1998), E.coli (Boiteux et al., 1987), Neisseria meningitidis (Swartley and Stephens, 1995), Bacillus firmus (Ivey, 1990), Synechococcus elongatus naegeli (Floss et al., 1997), Bacillus subtilis (Lapidus et al., 1997), Salmonella typhimurium (Suzuki et al., 1997), Lactococcus lactis subsp. cremoris (Duwat et al., 1995), Haemophilus influenzae (sp|P44948), Synechocystis PCC6803 (sp|P74290), Mycobacterium leprae (emb|CAA19197.1), Mycobacterium tuberculosis (sp|Q10959), Streptococcus mutans (sp|P55045), Streptomyces coelicolor A3(2) (emb|CAB63194.1), Mycoplasma pneumoniae (sp|P75402) and Mycoplasma genitalium (sp|P55825). Homologous sequences were identified with PSI-BLAST (Altschul et al., 1997) and sequences were aligned with CLUSTAL W (Thompson et al., 1994). (B) The solvent-accessible surface is coloured according to electrostatic potential, with positive in blue and negative in red. The deep cleft between the two domains is electrostatically positive. Damaged DNA may bind to this large cleft.
From sequence comparison among the 16 MutM proteins, we propose the following four highly conserved regions involved in DNA binding and catalysis: (i) the invariant N-terminal six residues and the loop between β3 and β4 (coloured magenta in Figure 1A and B and the boxes coloured magenta in Figure 1C); (ii) the two loops between β5 and β6 and between β8 and β9 (coloured magenta); (iii) the H2TH motif (αD and αE, coloured orange), which resembles the HhH motif that interacts with the DNA phosphodiester backbone (Thayer et al., 1995; Mullen et al., 1997; Burner et al., 2000; Hollis et al., 2000); and (iv) the zinc finger motif (β10 and β11, coloured green) forming a β-hairpin loop directed to the domain interface.
MutM–DNA interactions
A MutM–DNA footprinting experiment showed that MutM interacts with six nucleotides of the damaged strand, two nucleotides on the 3′ side of the lesion and three on the 5′ side (Castaing et al., 1999). In the enzymatic reaction, the α-imino group of Pro1 forms a Schiff base with C1′ of the deoxyribose in GO as a reaction intermediate (Zharkov et al., 1997). Lys52 has been suggested to interact with the C8 oxygen of the flipped GO base (Sidorkina et al., 1998). As mentioned above, there are two DNA-binding motifs in the MutM molecule, the H2TH and zinc finger motifs in the cleft. These regions are presumed to interact with dsDNA.
Taking these results into account, we tried to construct the structure of the complex of MutM with dsDNA, using the molecule with the narrower cleft in an asymmetrical unit (Figure 3B), on the basis of several crystal structures of complexes of glycosylases and dsDNA. The structure selected as the initial model was calculated by molecular dynamics using GO-flipped dsDNA bent by 45°, as in the uracil-DNA-glycosylase–dsDNA complex (Parikh et al., 1998).
The calculated model shows that the GO-flipped DNA fits nicely into the cleft of MutM (Figure 3). In the model complex, the DNA can interact with all four conserved regions mentioned above (Figure 1): the H2TH motif (yellow circle in Figure 3A), the zinc finger motif (light green circle), the two turns of β5–β6 and β8–β9 (sky-blue circle) and the putative catalytic site (pink circle). We will discuss the role of these regions further.
H2TH motif
The αD–αE region (residues 146–169) in the C-terminal domain has a characteristic H2TH motif structure with two turns between the two helices. The amino acid residues in the region are highly conserved among MutM homologues, including EndoVIII (Figure 1C). Similar consensus sequences are found in E.coli EndoIII (Thayer et al., 1995) and Pyrococcus furiosus Flap endonuclease-1 (FEN-1) (Hosfield et al., 1998) (Figure 4A).
Fig. 4. Two DNA-binding motifs in the MutM structure. (A) Superimposition of the MutM H2TH motif (purple) upon the HhH motif of E.coli EndoIII (green; PDB code 2ABK) (Thayer et al., 1995) and the H3TH motif of Pyrococcus furiosus Flap endonuclease-1 (FEN-1) (cyan; PDB code 1B43) (Hosfield et al., 1998). In the H2TH motif of MutM, there are two turns between α-helix D (αD) and α-helix E (αE). The amino acid sequences are aligned based on structural superimposition. The amino acid residues conserved among MutM, EndoIII and FEN-1 are coloured red. The HhH motif in EndoIII and the H3TH motif in FEN-1 are thought to interact with the dsDNA backbone (Doherty et al., 1996; Artymiuk et al., 1997). Therefore, the H2TH motif of MutM is presumed to play a similar role to the HhH and H3TH motifs and may contribute to non-sequence-specific recognition. (B) The ε-amino group of Lys147 forms hydrogen bonds (dashed lines) to the side chain of conserved residues and the carbonyl oxygen atoms of the main chain (ball-and-stick) at the interface of the H2TH motif (orange) and in the zinc finger motif (green). The decrease in catalytic efficiency caused by replacement of Lys147 with alanine (Rabow and Kow, 1997) can be attributed to the disruption of these hydrogen bonds and loss of folding integrity in this region of DNA interaction.
The HhH motif is found in E.coli EndoIII (Figure 4A, green) (Thayer et al., 1995) as a common non-specific DNA-binding motif and is implicated in binding to the DNA backbone phosphate. It is also observed in E.coli AlkA (Labahn et al., 1996; Yamagata et al., 1996; Hollis et al., 2000) and MutY (Guan et al., 1998) and in human OGG1 proteins (Bruner et al., 2000). These common HhH motifs in monofunctional glycosylases and bifunctional glycosylase/AP lyases suggest similar roles in DNA recognition, although the catalytic mechanisms of these enzymes are somewhat different (Labahn et al., 1996; Hollis et al., 2000). The helix–three-turns–helix (H3TH) motif, three turns between two helices, is found in P.furiosus FEN-1 (Figure 4A, cyan) (Hosfield et al., 1998). Both the HhH and H3TH motifs are found in a wide range of non-specific DNA-binding proteins, including nucleases, N-glycosylases, ligases, helicases, topoisomerases and polymerases, which are responsible for DNA synthesis or DNA repair. By analogy with the DNA polymerase β structure, it is suggested that each of these motifs binds to DNA in a non-specific manner by means of the formation of hydrogen bonds between amide nitrogen atoms of the protein backbone and phosphate oxygen atoms of the DNA backbone (Doherty et al., 1996; Artymiuk et al., 1997).
Our model of the complex between MutM and damaged dsDNA (Figure 3A) shows that the H2TH motif can access the DNA backbone at the bottom of the cleft. Therefore, we conclude that the H2TH motif plays a similar role to the HhH and H3TH motifs, which participate in non-sequence-specific DNA recognition.
Zinc finger motif
MutM also has another DNA-binding motif, a zinc finger motif at the C-terminus (Figures 1, 3 and 4B). The zinc finger motif is a well known DNA-binding motif found in various sequence-specific DNA-binding proteins (O’Connor et al., 1993). The zinc finger motif of MutM (-Cys-X2-Cys-X16-Cys-X2-Cys-) is a simple β-hairpin loop that extends into the inter-domain cleft of MutM. Such a β-hairpin loop has also been reported in human 3-methyladenine DNA glycosylase (AAG) complexed with DNA (Lau et al., 1998), in which the tyrosine residue at the top of the hairpin loop interacts directly with the DNA lesion in the minor groove and may be essential for base-flipping. The [4Fe–4S] cluster loop motif of EndoIII also extends into the inter-domain cleft and is thought to intercalate into the minor groove of DNA (Thayer et al., 1995). The [4Fe–4S] cluster loop motif has a conserved basic residue at the edge of the loop. Thus, the positively charged Arg253 on the edge of the β-hairpin loop may interact with the minor groove of DNA in a specific manner similar to that of EndoIII, since the arginine residue is invariant among MutM family proteins (Figure 5A).
Lys147 of MutM is a key structural residue in hydrogen bond networks between the H2TH motif and the zinc finger motif, maintaining the proper arrangement of the H2TH motif relative to the zinc finger motif (Figure 4B). Replacement of Lys147 by alanine causes a 50-fold decrease in the cleavage of GO:C substrates but does not affect AP site substrates (Rabow and Kow, 1997). The zinc finger motif, which specifically recognizes the GO:C pair, is firmly fixed with respect to the C-terminal domain by the hydrogen bond network, so the Lys147→Ala substitution would be expected to disrupt the interaction, resulting in a drastic decrease of activity against GO:C substrates. In contrast, the conformation of the H2TH motif at the core of the C-terminal domain, which is proposed to interact with the phosphate of the DNA backbone, would not be disturbed structurally by the mutation, and excision of the sugar moiety from the flipped out AP site would not be affected.
Recognition of a wide range of oxidatively damaged bases
MutM has broad specificity and high efficiency for oxidatively damaged lesions, including (i) GO paired to cytosine (Tchou et al., 1991); (ii) Fapy (Boiteux et al., 1992); and (iii) 5-hydroxycytosine (5OHC) paired to guanine (Hatahet et al., 1994) and (iv) AP sites (Castaing et al., 1992) (Figure 5C).
The proposed model of the MutM complex (Figures 3 and 5A and B) supplies many clues for interpreting the specificity determinants of these substrates. The C8 oxygen of GO is located in the major groove of the model B-form dsDNA (Lipscomb et al., 1995) (Figure 5A and B). In the model, the damaged base interacts with turns β5–β6 and β8–β9 in the N-terminal domain, in which residues His67, Met70, Arg99 and Phe101 are conserved. These residues probably contribute to lesion recognition; in particular, Arg99 may form a hydrogen bond with the C6 oxo group at the damaged position.
The minor groove of DNA interacts with the β-hairpin loop of the zinc finger motif in the model of the DNA complex. Arg253 is the only invariant residue in the β-hairpin loop; it can interact specifically with the C2 oxo group of cytosine in the GO:C pair (Figure 5B). The two arginine residues, Arg99 and Arg253, interact with both sides of the docked DNA to cleave the pairing hydrogen bonds between the GO:C bases. Substrates with low cleavage efficiency lack these oxo groups (Figure 5C) (Hatahet et al., 1994; Tchou et al., 1994). The C8 oxo group in the major groove may be pushed out by a large hydrophobic residue, either Met70 or Phe101, during binding of the dsDNA, using the hinge movement suggested by the existence of two molecules with different conformations in an asymmetrical unit (Figure 3B). The hinge movement of the domains may also contribute to the cleavage of GO:C hydrogen bonds. When nucleotide flipping of the GO residue occurs, Phe101 can be inserted into the abasic site to compensate for the absence of the base.
On the basis of amino acid sequence, this model may be applicable to another GO:C base-excision repair enzyme with different substrate specificity, EndoVIII (Figure 1C). Met70, Arg99 and Phe101, which may contribute to substrate specificity, are not conserved in EndoVIII, although the putative catalytic groups Pro1, Glu2 and Glu5 are conserved between these enzymes.
Architecture of catalytic groups in the active site
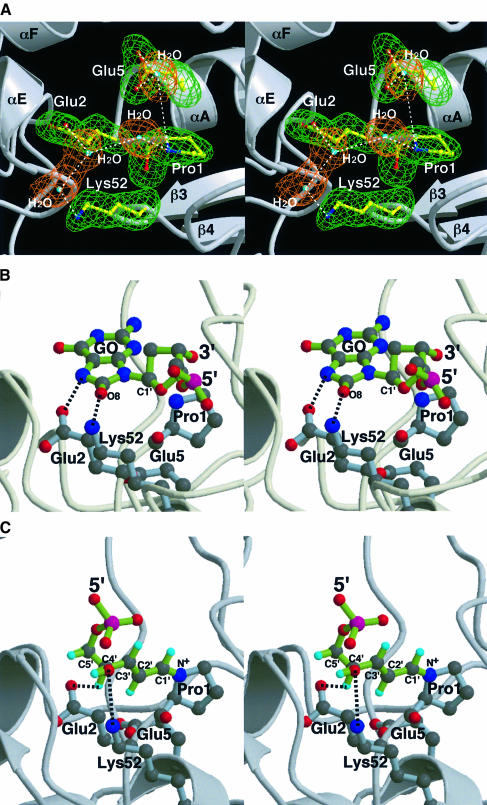
The crystal structure of MutM reveals that the nucleophile Pro1, the primary residue for enzymatic catalysis, is positioned at the bottom of the cleft and surrounded by the invariant charged residues Glu2, Glu5 and Lys52 with a hydrogen bonding network including several bound water molecules (Figure 6A). The charged residues Glu2, Glu5 and Lys52 are likely to be involved in the enzyme action. The hydrophilic environment around the N-terminal proline, which forms a Schiff base intermediate with C1′ of the deoxyribose of GO (Zharkov et al., 1997), is electrostatically highly positive (Figure 2B). Such a microenvironment in the active cleft would make Lys52 a proton donor at neutral pH. On the other hand, the side chain carboxylic acids of Glu2 and Glu5 are likely to be deprotonated in these conditions and so could be proton acceptors.
Fig. 6. Architecture of the MutM active site. (A) The nucleophile Pro1 of MutM is surrounded by the invariant charged residues Glu2, Glu5 and Lys52, accompanied by several bound water molecules. The hydrophilic active site is very positive electrostatically (Figure 2B), so Lys52 is expected to be a proton donor for glycosidase activity in MutM. (B) Model of docking of the flipped out GO nucleotide to MutM active site in Figure 3A. The 8-oxo group of the GO base is located near the ε-amino group of Lys52, which as a proton donor may help the protonation of the oxo group and lead to depurination. (C) The putative reaction intermediate adduct with Pro1 after β-elimination, which is configurationally definitive due to its conjugated double bonding (Bhagwat and Gerlt, 1996), can fit into the active site and the hydroxyl group at C4′ of the opened deoxyribose reaches the carboxylic acid of Glu2, which is a good candidate for a proton acceptor.
Docking studies of the model complex with dsDNA show that the flipped out GO base could accommodate the hydrophilic active site during ‘gripping’ of the DNA; the 8-oxo group of the GO base is placed at the ε-amino group of Lys52 (Figure 6B). In the bound mode of the flipped GO nucleotide, the bound water molecule between the side chain carboxylic group of Glu5 and the secondary amino group of the primary catalytic residue, Pro1, would form hydrogen bonds with the oxygen atom of the pentose ring in the GO nucleotide, and thus may help the depurination reaction by neutralizing the resulting carbonium ion, followed by Schiff base formation with the N-terminal amine of Pro1. The hydrogen bond network of the water and δ-carboxylic acid of Glu5 may act as the proton acceptor for β-elimination. However, the highly restrained structure of the DNA complex and so many possible degrees of torsion in the flipped nucleotide make the model rather arbitrary (Figure 6B). Another putative intermediate after the β-elimination reaction has a rather constrained configuration because of its conjugated double bonding with the planar pyrrolidinium ring of Pro1. The putative intermediate can be readily placed into the active site, and the C4′ of the opened deoxyribose reaches to the δ-carboxylate of Glu2, which may act as a proton acceptor for the following δ-elimination reaction (Figure 6C) (Bhagwat and Gerlt, 1996). From these insights into the architecture of the active site, including the docking models, we propose that Glu2, Glu5 with its bound water, and Lys52 are catalytic residues additional to the primary residue Pro1 in the trifunctional DNA glycosylase/β-lyase/δ-lyase action.
Catalytic mechanism of MutM
On the basis of the three-dimensional structures, including the docking models of dsDNA and its reaction intermediate, we propose the following catalytic mechanism for the MutM N-glycosylase/AP lyase reaction. This mechanism also takes into account biochemical studies on the reaction mechanism (Bhagwat and Gerlt, 1996; Castaing et al., 1999) (Figure 7). (i) Initial binding of MutM to GO-flipped DNA with a ‘gripping’ motion at the hinge region. The ammonium cation of Lys52 acts as proton donor for scission of the glycosidic bond in the damaged nucleotide to release the GO base. (ii) The resulting carbonium ion at C1′ of deoxyribose is attacked by the N-terminal amino group of Pro1 on C1′ as the nucleophile, and the cation is stabilized by a hydrogen-bonding network, including the bound water and the carboxylate of Glu5. (iii) Protonation of Glu5 and concerted electron rearrangement breaks the pentose ring of the deoxyribose and forms a Schiff base between the C1′ atom of the opened deoxyribose and the ring nitrogen atom of Pro1. (iv) The enamine forms via mesomeric equilibrium, followed by (v) 3′-phosphoester breakage with β-elimination. (vi) Proton withdrawal by Glu2 causes transfer of the conjugated diene. (vii) Subsequent δ-elimination results in esterification of the 5′-phosphoester bond. (viii) Protonation of Lys52 and release of the deoxyribose product 4-oxo-2-pentenal leave the dsDNA with a one-nucleotide gap (Figure 7) (Bhagwat and Gerlt, 1996).
The almost concerted reaction to form the Schiff base intermediate would result in depurination occurring at the same time as ‘gripping’ of the DNA in the hydrophilic active pocket, as in T4 EndoV (Figure 3B) (Vassylyev et al., 1995), and the released base of the damaged nucleotide could be accommodated in the highly hydrophobic cavity lined by the aromatic residues Phe53 and His67 in the vicinity of the active site. A potent inhibitor based on a reduced AP site analogue (Castaing et al., 1999) would fit well to MutM, as this analogue of the Schiff base complex intermediate would form a hydrogen bond between the 4′-hydroxyl group of the opened deoxyribose analogue and the ε-amino group of Lys52 without configurational restraint in the hydrophilic environment, like step (iii) in the reaction scheme (Figure 7). The possibility cannot be excluded that the concerted mechanism of Schiff base formation initiates the reaction with base ring opening (Rabow and Kow, 1997), although the mechanism proposed above seems more likely because MutM acts strongly on AP site substrates (Tchou et al., 1994). In order to confirm the detailed mechanism of the MutM N-glycosylase/AP lyase reactions, mutational analyses of the active-site residues are under way.
Conclusions
The crystal structure and sequence alignment of MutM reveal DNA-binding motifs and some important invariant and conserved residues that may be involved in catalysis. On the basis of these docking studies with GO-flipped dsDNA, together with biochemical results, we propose mechanisms for recognition of oxidatively damaged DNA and for N-glycosylase/AP lyase activity.
Materials and methods
Crystallization and X-ray diffraction data collection
Thermus thermophilus HB8 MutM crystals (Mikawa et al., 1998) were prepared as described previously (Sugahara et al., 2000). MAD data were collected at two wavelengths near the zinc absorption edge and at 1.04 Å as remote data (Table I) using one crystal at beamline BL45XU at SPring-8 (Yamamoto et al., 1998). Before data collection, the crystal was soaked for several seconds in a cryoprotectant solution prepared by adding glycerol to the mother liquor to a final concentration of 20% (v/v) and flash cooled in a nitrogen stream at 100 K. Data were processed and scaled using the programs DENZO and SCALEPACK (Otwinowski and Minor, 1997).
Table I. Data collection and refinement statistics.
| Parameter | λ1 (native) | λ2 (peak) | λ3 (edge) |
|---|---|---|---|
| Data collection | |||
| resolution (Å)a | 50–1.9 (1.93–1.90) | 50–2.2 (2.24–2.20) | 50–2.2 (2.24–2.20) |
| wavelength (Å) | 1.04 | 1.2823 | 1.2829 |
| observations | 402 397 | 260 102 | 260 684 |
| independents | 37 893 (1729) | 25 078 (992) | 25 134 (994) |
| completeness (%) | 90.0 (82.1) | 92.7 (73.5) | 92.7 (73.1) |
| Rmerge (%)b | 7.0 (62.0) | 6.2 (17.9) | 6.3 (20.0) |
| <I>/<σ(I)> | 29.1 (3.2) | 32.1 (10.6) | 31.6 (8.4) |
| Refinement statistics | |||
| resolution (Å) | 50–1.9 | ||
| reflections | 36 536 | ||
| Rcrystc | 0.214 | ||
| Rfreed | 0.258 | ||
| average B factor | 30.82 | ||
| number of non-hydrogen atoms | 4335 | ||
| R.m.s. deviations from ideality | |||
| bond length (Å) | 0.006 | ||
| angles (°) | 1.35 | ||
| dihedral angles (°) | 23.37 | ||
| improper angles (°) | 0.84 |
aValues in parentheses are for the outermost shell of data.
bRmerge = Σ|Iobs – <I>|/ΣIobs
cRcryst = Σ||Fobs| – |Fcalc||/Σ|Fobs|
dRfree is monitored with 10% of the reflection data excluded from refinement.
MAD phasing, model building and refinement
The structure of MutM was solved with MAD phasing. The two zinc atoms in an asymmetrical unit were located in the Bijvoet difference and dispersive anomalous difference Patterson maps. SOLVE (Terwilliger and Berendzen, 1999) was used to refine the parameters of zinc atoms and to calculate phases to 2.2 Å resolution (an overall figure of merit of 0.40). The MAD phases were further improved by solvent flattening (47% solvent content) by program DM in PERT mode (an overall figure of merit of 0.69) (CCP4, 1994). At this stage, the map showed the folding of the polypeptide backbone. A polyalanine model with secondary structural elements was built using the program O (Jones et al., 1991). This model indicated that the two MutM molecules in an asymmetrical unit (Sugahara et al., 2000) had different conformations, so the phases were improved by combination of density modification and non-crystallographic symmetry (NCS) multidomain averaging [program DM (CCP4, 1994)] using two separate masks and matrices (CCP4, 1994). The NCS matrix was determined with program LSQKAB (CCP4, 1994) and the molecular mask for NCS averaging was generated using programs O (Jones et al., 1991) and RAVE (Kleywegt and Jones, 1994). A 2.2 Å map (an overall figure of merit of 0.80) of excellent quality was obtained by using the improved phases and enabled us to build >90% of amino acid residues. This initial model was refined using diffraction data from 50 to 1.9 Å resolution by Powell conjugate gradient minimization and torsion angle-restrained molecular dynamics without NCS restraint using the Crystallography & NMR System package (CNS version 0.9; Brünger et al., 1998). The remaining residues were fitted to the electron density maps calculated from experimental and model phase information. The progress of the structural refinement was evaluated at each stage by the free R factor (Brünger, 1992) and by inspection of stereochemical parameters with the program PROCHECK (Laskowski et al., 1993). The Ramachandran plot showed no residues in the disallowed region and indicated that 86.6% of residues lay in favourable regions, 12.7% in allowed regions and 0.7% in generously allowed regions. The current model consists of the entire 266 amino acid residues coded by the T.thermophilus HB8 mutM gene with a zinc atom in each monomer and 292 water molecules. All of the main-chain atoms and most side-chain atoms are clearly visible in the 2(Fo – Fc) electron density map contoured at the 1σ level. The final R values and root-mean-square deviations from ideality are given in Table I.
Molecular dynamic simulation
The model of MutM complexed with GO-flipped DNA was obtained from molecular dynamic (MD) simulation using the CNS version 0.9 program (Brünger et al., 1998). The coordinates of dsDNA with a flipped nucleotide were extracted from the crystal structure of the human uracil-DNA glycosylase–DNA complex (Parikh et al., 1998). The three-dimensional intermediate fragment was generated by ChemOffice (CambridgeSoft Corporation). On the basis of some reported assumptions for MutM–DNA interaction (see Results and discussion), the initial coordinates of the MutM and dsDNA were manually docked with program O (Jones et al., 1991). The MutM–DNA model was then surrounded by a 5 Å layer of water and subjected first to 200 steps using conjugate gradient minimization with all atoms except water molecules fixed. Subsequently, MD simulation was run by fixing the conformation of the protein backbone (except for two loops consisting of residues 110–128 and 204–237) and the dsDNA (except for the flipped GO nucleotide) for 30 ps at 298 K with a time step of 1 fs.
All molecular images were produced using Raster3D (Merritt and Murphy, 1994) and Molscript (Kraulis, 1991), except for Figure 2B, which was produced with GRASP (Nicholls et al., 1993).
Accession numbers
Coordinates reported in this paper have been deposited in the Protein Data Bank under accession No. 1ee8.
Acknowledgments
Acknowledgements
We are greatly indebted to Dr Tetsuya Ishikawa, RIKEN Harima Institute, SPring-8, and to Dr Tatzuo Ueki, Japan Synchrotron Radiation Research Institute (JASRI), for providing us with the opportunity to use the X-ray facilities; to Professor Noriyoshi Sakabe of the University of Tsukuba, Dr Ryoji Masui, Dr Jun Ishijima and Noriko Nakagawa, our colleagues in Osaka University, for critical discussions; and to Dr Masashi Miyano, RIKEN Harima Institute, SPring-8, for his critical reading of this manuscript and for many illuminating discussions regarding the mechanism of action of MutM.
References
- Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames B.N., Gold,L.S. and Willett,W.C. (1995) The causes and prevention of cancer. Proc. Natl Acad. Sci. USA, 92, 5258–5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artymiuk P.J., Ceska,T.A., Suck,D. and Sayers,J.R. (1997) Prokaryotic 5′–3′ exonucleases share a common core structure with γδ-resolvase. Nucleic Acids Res., 25, 4224–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagwat M. and Gerlt,J.A. (1996) 3′- and 5′-strand cleavage reactions catalyzed by the Fpg protein from Escherichia coli occur via successive β- and δ-elimination mechanisms, respectively. Biochemistry, 35, 659–665. [DOI] [PubMed] [Google Scholar]
- Boiteux S., O’Connor,T.R. and Laval,J. (1987) Formamidopyrimidine-DNA glycosylase of Escherichia coli: cloning and sequencing of the fpg structural gene and overproduction of the protein. EMBO J., 6, 3177–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux S., O’Connor,T.R., Lederer,F., Gouyette,A. and Laval,J. (1990) Homogeneous Escherichia coli FPG protein. A DNA glycosylase which excises imidazole ring-opened purines and nicks DNA at apurinic/apyrimidinic sites. J. Biol. Chem., 265, 3916–3922. [PubMed] [Google Scholar]
- Boiteux S., Gajewski,E., Laval,J. and Dizdaroglu,M. (1992) Substrate specificity of the Escherichia coli Fpg protein (formamidopyrimidine-DNA glycosylase): excision of purine lesions in DNA produced by ionizing radiation or photosensitization. Biochemistry, 31, 106–110. [DOI] [PubMed] [Google Scholar]
- Breimer L.H. (1990) Molecular mechanisms of oxygen radical carcinogenesis and mutagenesis: the role of DNA base damage. Mol. Carcinogen., 3, 188–197. [DOI] [PubMed] [Google Scholar]
- Bruner S.D., Norman,D.P.G. and Verdine,G.L. (2000) Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature, 403, 859–866. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. (1992) Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature, 355, 472–475. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Castaing B., Boiteux,S. and Zelwer,C. (1992) DNA containing a chemically reduced apurinic site is a high affinity ligand for the E. coli formamidopyrimidine-DNA glycosylase. Nucleic Acids Res., 20, 389–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaing B., Fourrey,J.L., Hervouet,N., Thomas,M., Boiteux,S. and Zelwer,C. (1999) AP site structural determinants for Fpg specific recognition. Nucleic Acids Res., 27, 608–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project No. 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Dizdaroglu M. (1985) Formation of an 8-hydroxyguanine moiety in deoxyribonucleic acid on γ-irradiation in aqueous solution. Biochemistry, 24, 4476–4481. [DOI] [PubMed] [Google Scholar]
- Doherty A.J., Serpell,L.C. and Ponting,C.P. (1996) The helix–hairpin–helix DNA-binding motif: a structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res., 24, 2488–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duwat P., de Oliveira,R., Ehrlich,S.D. and Boiteux,S. (1995) Repair of oxidative DNA damage in gram-positive bacteria: the Lactococcus lactis Fpg protein. Microbiology, 141, 411–417. [DOI] [PubMed] [Google Scholar]
- Floss B., Igloi,G., Cassier-Chauva,T.C. and Muehlenhoff,U. (1997) Molecular cloning and overexpression of the petF gene from the thermophilic cyanobacterium Synechococcus elongatus. Photosyn. Res., 54, 63–71. [Google Scholar]
- Guan Y., Manuel,R.C., Arvai,A.S., Parikh,S.S., Mol,C.D., Miller,J.H., Lloyd,R.S. and Tainer,J.A. (1998) MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nature Struct. Biol., 5, 1058–1064. [DOI] [PubMed] [Google Scholar]
- Hatahet Z., Kow,Y.W., Purmal,A.A., Cunningham,R.P. and Wallace,S.S. (1994) New substrates for old enzymes. 5-Hydroxy-2′-deoxycytidine and 5-hydroxy-2′-deoxyuridine are substrates for Escherichia coli endonuclease III and formamidopyrimidine DNA N-glycosylase, while 5-hydroxy-2′-deoxyuridine is a substrate for uracil DNA N-glycosylase. J. Biol. Chem., 269, 18814–18820. [PubMed] [Google Scholar]
- Hollis T., Ichikawa,Y. and Ellenberger,T. (2000) DNA binding and a flip-out mechanism for base excision by the helix–hairpin–helix DNA glycosylase, Escherichia coli AlkA. EMBO J., 19, 758–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Hosfield D.V., Mol,C.D., Shen,B. and Tainer,J.A. (1998) Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity. Cell, 95, 135–146. [DOI] [PubMed] [Google Scholar]
- Ivey D.M. (1990) Nucleotide sequence of a gene from alkaliphilic Bacillus firmus RAB that is homologous to the fpg gene of Escherichia coli. Nucleic Acids Res., 18, 5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D., Hatahet,Z., Melamede,R.J., Kow,Y.W. and Wallace,S.S. (1997) Characterization of Escherichia coli endonuclease VIII. J. Biol. Chem., 272, 32230–32239. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.-Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kleywegt G.J. and Jones,T.A. (1994) Halloween … masks and bones. In Bailey,S., Hubbard,R. and Waller,D. (eds), From First Map to Final Model. SERC Daresbury Laboratory, Warrington, UK, pp. 59–66. [Google Scholar]
- Kow Y.W. and Wallace,S.S. (1987) Mechanism of action of Escherichia coli endonuclease III. Biochemistry, 26, 8200–8206. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Krokan H.E., Standal,R. and Slupphaug,G. (1997) DNA glycosylases in the base excision repair of DNA. Biochem. J., 325, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labahn J., Schärer,O.D., Long,A., Ezaz-Nikpay,K., Verdine,G.L. and Ellenberger,T.E. (1996) Structural basis for the excision repair of alkylation-damaged DNA. Cell, 86, 321–329. [DOI] [PubMed] [Google Scholar]
- Lapidus A., Galleron,N., Sorokin,A. and Ehrlich,S.D. (1997) Sequencing and functional annotation of the Bacillus subtilis genes in the 200 kb rrnB–dnaB region. Microbiology, 143, 3431–3441. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., McArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK—A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Lau A.Y., Scharer,O.D., Samson,L., Verdine,G.L. and Ellenberger,T. (1998) Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: mechanisms for nucleotide flipping and base excision. Cell, 95, 249–258. [DOI] [PubMed] [Google Scholar]
- Lipscomb L.A., Peek,M.E., Morningstar,M.L., Verghis,S.M., Miller,E.M., Rich,A., Essigmann,J.M. and Williams,L.D. (1995) X-ray structure of a DNA decamer containing 7,8-dihydro-8-oxoguanine. Proc. Natl Acad. Sci. USA, 92, 719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamede R.J., Hatahet,Z., Kow,Y.W., Ide,H. and Wallace,S.S. (1994) Isolation and characterization of endonuclease VIII from Escherichia coli. Biochemistry, 33, 1255–1264. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Murphy,M.E.P. (1994) Raster3D version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D, 50, 869–873. [DOI] [PubMed] [Google Scholar]
- Michaels M.L., Tchou,J., Grollman,A.P. and Miller,J.H. (1992) A repair system for 8-oxo-7,8-dihydrodeoxyguanine. Biochemistry, 31, 10964–10968. [DOI] [PubMed] [Google Scholar]
- Mikawa T., Kato,R., Sugahara,M. and Kuramitsu,S. (1998) Thermostable repair enzyme for oxidative DNA damage from extremely thermophilic bacterium, Thermus thermophilus HB8. Nucleic Acids Res., 26, 903–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen G.P. and Wilson,S.H. (1997) DNA polymerase β in abasic site repair: a structurally conserved helix–hairpin–helix motif in lesion detection by base excision repair enzymes. Biochemistry, 36, 4713–4717. [DOI] [PubMed] [Google Scholar]
- Murphy T.M. and Gao,M.J. (1998) Two cDNAs (accession nos. AF099970 and AF099971) encoding Arabidopsis homologs of bacterial formamidopyrimidine–DNA glycosylase genes are produced by alternative processing (PGR98-204). Plant Physiol., 118, 1535. [Google Scholar]
- Nicholls A., Bharadwaj,R. and Honig,B. (1993) GRASP—graphical representation and analysis of surface properties. Biophys. J., 64, A116. [Google Scholar]
- O’Connor T.R. and Laval,J. (1989) Physical association of the 2,6-diamino-4-hydroxy-5N-formamidopyrimidine-DNA glycosylase of Escherichia coli and an activity nicking DNA at apurinic/apyrimidinic sites. Proc. Natl Acad. Sci. USA, 86, 5222–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor T.R., Graves,R.J., de Murcia,G., Castaing,B. and Laval,J. (1993) Fpg protein of Escherichia coli is a zinc finger protein whose cysteine residues have a structural and/or functional role. J. Biol. Chem., 268, 9063–9070. [PubMed] [Google Scholar]
- Ohtsubo T., Matsuda,O., Iba,K., Terashita,I., Sekiguchi,M. and Nakabeppu,Y. (1998) Molecular cloning of AtMMH, an Arabidopsis thaliana ortholog of the Escherichia coli mutM gene, and analysis of functional domains of its product. Mol. Gen. Genet., 259, 577–590. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Parikh S.S., Mol,C.D., Slupphaug,G., Bharati,S., Krokan,H.E. and Tainer,J.A. (1998) Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J., 17, 5214–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purmal A.A., Rabow,L.E., Lampman,G.W., Cunningham,R.P. and Kow,Y.W. (1996) A common mechanism of action for the N-glycosylase activity of DNA N-glycosylase/AP lyases from E. coli and T4. Mutat. Res., 364, 193–207. [DOI] [PubMed] [Google Scholar]
- Rabow L.E. and Kow,Y.W. (1997) Mechanism of action of base release by Escherichia coli Fpg protein: role of lysine 155 in catalysis. Biochemistry, 36, 5084–5096. [DOI] [PubMed] [Google Scholar]
- Schrock R.D. III and Lloyd,R.S. (1991) Reductive methylation of the amino terminus of endonuclease V eradicates catalytic activities. Evidence for an essential role of the amino terminus in the chemical mechanisms of catalysis. J. Biol. Chem., 266, 17631–17639. [PubMed] [Google Scholar]
- Shibutani S., Takeshita,M. and Grollman,A.P. (1991) Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature, 349, 431–434. [DOI] [PubMed] [Google Scholar]
- Sidorkina O.M. and Laval,J. (1998) Role of lysine-57 in the catalytic activities of Escherichia coli formamidopyrimidine-DNA glycosylase (Fpg protein). Nucleic Acids Res., 26, 5351–5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugahara M., Mikawa,T., Kato,R., Kumasaka,T., Yamamoto,M., Fukuyama,K., Inoue,Y. and Kuramitsu,S. (2000) Crystallization and preliminary X-ray crystallographic studies of Thermus thermophilus HB8 MutM protein involved in repairs of oxidative DNA damage. J. Biochem. (Tokyo), 127, 9–11. [DOI] [PubMed] [Google Scholar]
- Suzuki M., Matsui,K., Yamada,M., Kasai,H., Sofuni,T. and Nohmi,T. (1997) Construction of mutants of Salmonella typhimurium deficient in 8-hydroxyguanine DNA glycosylase and their sensitivities to oxidative mutagens and nitro compounds. Mutat. Res., 393, 233–246. [DOI] [PubMed] [Google Scholar]
- Swartley J.S. and Stephens,D.S. (1995) Co-transcription of a homologue of the formamidopyrimidine-DNA glycosylase (fpg) and lysophosphatidic acid acyltransferase (nlaA) in Neisseria meningitidis. FEMS Microbiol. Lett., 134, 171–176. [DOI] [PubMed] [Google Scholar]
- Tchou J. and Grollman,A.P. (1995) The catalytic mechanism of Fpg protein. Evidence for a Schiff base intermediate and amino terminus localization of the catalytic site. J. Biol. Chem., 270, 11671–11677. [DOI] [PubMed] [Google Scholar]
- Tchou J., Kasai,H., Shibutani,S., Chung,M.H., Laval,J., Grollman,A.P. and Nishimura,S. (1991) 8-Oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc. Natl Acad. Sci. USA, 88, 4690–4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchou J., Michaels,M.L., Miller,J.H. and Grollman,A.P. (1993) Function of zinc finger in Escherichia coli Fpg protein. J. Biol. Chem., 268, 26738–26744. [PubMed] [Google Scholar]
- Tchou J., Bodepudi,V., Shibutani,S., Antoshechkin,I., Miller,J., Grollman,A.P. and Johnson,F. (1994) Substrate specificity of Fpg protein. Recognition and cleavage of oxidatively damaged DNA. J. Biol. Chem., 269, 15318–15324. [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen, J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer M.M., Ahern,H., Xing,D., Cunningham,R.P. and Tainer,J.A. (1995) Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J., 14, 4108–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassylyev D.G., Kashiwagi,T., Mikami,Y., Ariyoshi,M., Iwai,S., Ohtsuka,E. and Morikawa,K. (1995) Atomic model of a pyrimidine dimer excision repair enzyme complexed with a DNA substrate: structural basis for damaged DNA recognition. Cell, 83, 773–782. [DOI] [PubMed] [Google Scholar]
- Wood M.L., Dizdaroglu,M., Gajewski,E. and Essigmann,J.M. (1990) Mechanistic studies of ionizing radiation and oxidative mutagenesis: genetic effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry, 29, 7024–7032. [DOI] [PubMed] [Google Scholar]
- Yamagata Y. et al. (1996) Three-dimensional structure of a DNA repair enzyme, 3-methyladenine DNA glycosylase II, from Escherichia coli. Cell, 86, 311–319. [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Kumasaka,T., Fujisawa,T. and Ueki,T. (1998) Trichromatic concept at SPring-8 RIKEN beamline I. J. Synchrotron Rad., 5, 222–225. [DOI] [PubMed] [Google Scholar]
- Zharkov D.O., Rieger,R.A., Iden,C.R. and Grollman,A.P. (1997) NH2-terminal proline acts as a nucleophile in the glycosylase/AP-lyase reaction catalyzed by Escherichia coli formamidopyrimidine-DNA glycosylase (Fpg) protein. J. Biol. Chem., 272, 5335–5341. [DOI] [PubMed] [Google Scholar]