Abstract
The Tec family tyrosine kinase, Itk, is a key component of the T cell receptor signaling pathway. Biochemical studies have shown that Itk activation requires recruitment of Itk to the membrane via its pleckstrin homology domain, phosphorylation of Itk by the Src kinase, Lck, and binding of Itk to the SLP-76/LAT adapter complex. However, the regulation of Itk enzymatic activity by Itk domain interactions is not yet well understood. Here we show that full length Itk self-associates in an intermolecular fashion. Using this information, we have designed an Itk variant that exhibits reduced self-association but maintains normal binding to exogenous ligands via each of its regulatory domains. When expressed in insect cells, the Itk substrate PLCγ1 is phosphorylated more efficiently by the Itk variant than by wild-type Itk. Furthermore, expression of the Itk variant in primary murine T cells induced higher ERK activation and increased calcium flux following TCR stimulation compared to wild-type Itk. Our results indicate that the Tec kinase Itk is negatively regulated by intermolecular clustering, and that disruption of this clustering leads to increased Itk kinase activity following TCR stimulation.
Introduction
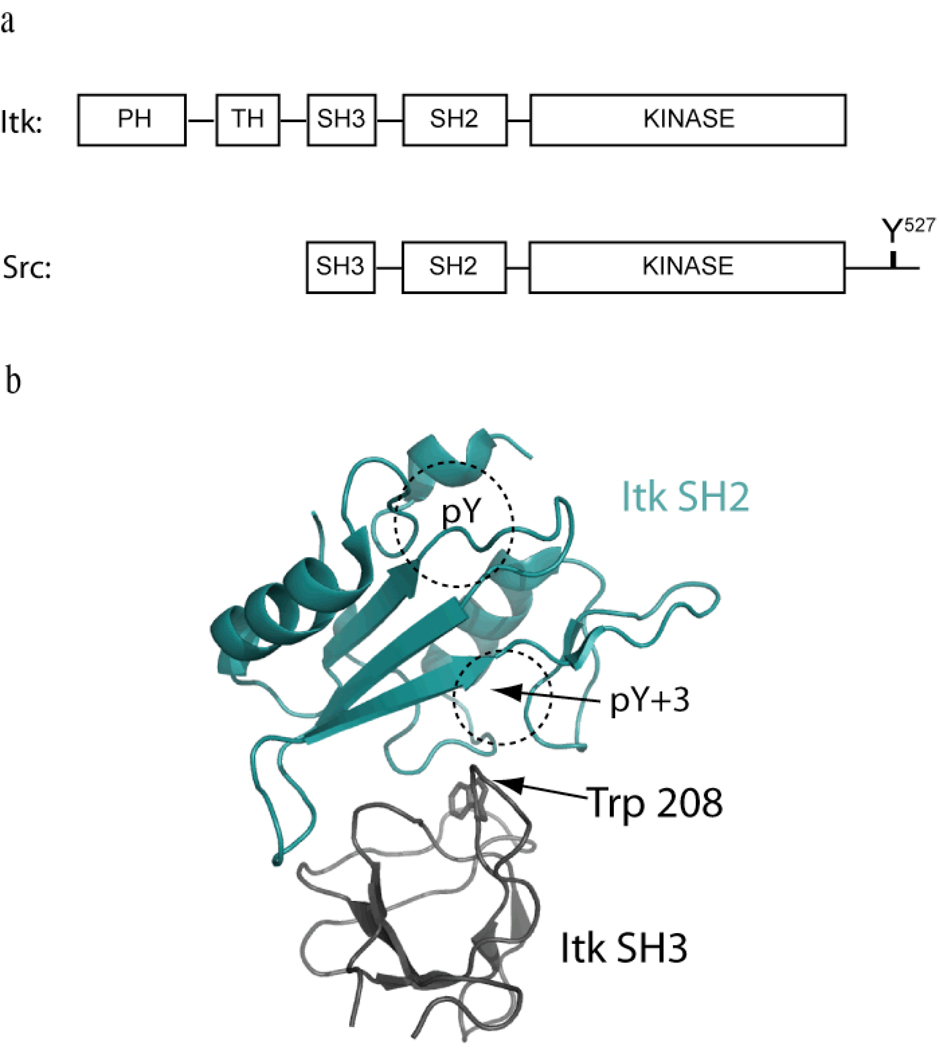
Interleukin-2 tyrosine kinase (Itk) is a non-receptor protein tyrosine kinase of the Tec family that is expressed in T cells, mast cells and NK cells (1–5). Itk participates in signaling processes following T cell receptor engagement by phosphorylating and activating phospholipase C-γ1 (PLC-γ1), leading to production of two second messengers, DAG and IP3 (6–9). In addition to Itk, the Tec family includes Btk, Tec, Rlk and Bmx, each of which shares a similar domain structure with the Src kinase family members (10). Both families contain a Src homology 3 (SH3) domain, a Src homology 2 (SH2) domain and the catalytic domain. With the exception of Rlk, the Tec kinases also contain a Pleckstrin Homology (PH) domain and a Tec Homology (TH) domain at the amino terminus (Fig. 1a).
Figure 1. Negative regulation of Itk; domain structure differences with Src suggest an alternative mode of regulation.
(a) Domain structures of Itk and Src. Both kinases contain the SH3-SH2-Kinase domain cassette but differ at both the amino- and carboxy termini. Itk contains the Pleckstrin homology/Tec homology (PH-TH) region that is absent in Src. Src contains the negative regulatory tail region lacking in Itk. Y527 in the Src tail gets phosphorylated and binds in an intramolecular fashion to the Src SH2 domain to negatively regulate Src kinase activity (15). Itk is not regulated in the same manner but the SH3 and SH2 domains associate with each other in an intermolecular sense (18). (b) The three-dimensional structure of the Itk SH3/SH2 complex (pdb: 2K79) has been solved (16). The Itk SH2 domain (shown in cyan) binds to the ligand-binding pocket on the Itk SH3 domain (shown in black). Trp 208 is located in the SH3 binding pocket and mediates binding of the SH3 domain to both the Itk SH2 domain and classical proline-rich ligands. The classical phospholigand binding pockets (pY and pY+3) are labeled on the SH2 domain. The SH3 binding site on the SH2 domain partially overlaps with phospholigand binding on SH2.
The mechanisms promoting Itk activation following TCR stimulation have been well described. To date, three upstream signals are required for Itk activation, including Itk recruitment to PIP3 in the membrane via its PH domain, Itk binding to the SLP-76/LAT adapter complex via its SH2 and SH3 domains, and finally, Itk phosphorylation by Lck at the activation loop tyrosine in its kinase domain (11), However, the structural changes in Itk that accompany this activation process, as well as the mechanism(s) by which Itk activity is turned off when TCR signaling is terminated, have not been established. One reason for this lack of information is that, despite similarities to the Src kinases in primary structure, there are also significant differences between Itk and Src (Fig. 1a). Most notably, the Tec kinases all lack the carboxy-terminal autoinhibitory sequence that serves to negatively regulate the Src kinases (12–15). Thus, the well characterized inhibitory interaction in Src between the Src SH2 domain and phosphorylated Y527 in the carboxy-terminal tail cannot occur for the Tec kinase family. The absence of the Src regulatory tail sequence in Itk and the related Tec family members raises questions about the domain interactions and conformational changes that regulate Itk activity during the course of T cell signaling.
We and others have previously reported detailed structural studies for regulatory domain fragments of Itk, Btk, Tec and Rlk (16–25). An emerging theme for each of these kinases is that the non-catalytic domains form dimeric and higher order oligomeric structures in solution. For Itk, self-association of the regulatory domains occurs via intermolecular interactions between the SH3 domain and the SH2 domain (18). The structure of the intermolecular Itk SH3/SH2 complex has been solved (16) providing a molecular basis for probing the functional significance of intermolecular association. An intermolecular interaction has also been described for the Itk PH domain (26). The isolated PH domain interacts both with itself and with the PH domain within full length Itk in co-immunoprecipitation experiments suggesting that multiple contacts across the regulatory domains stabilize a self-associated form of Itk. Finally, a split YFP system has previously allowed visualization of intermolecular interactions between full-length Itk molecules in cells (27). Thus, abundant data point to intermolecular clustering of Itk, most likely mediated by multiple Itk regulatory domains; yet to date, the functional significance of this self-association has not been explained.
Here, we extend the earlier studies of Itk domain fragments and demonstrate that full-length Itk self-associates in an intermolecular fashion in vitro. To evaluate the functional significance of the observed self-association, we then designed a mutant Itk molecule that retains all of the structural features of the wild-type enzyme, yet exhibits diminished self-association. We next examined substrate phosphorylation levels by the wild type and mutant Itk molecules, as well as the signaling properties of wild type and mutant Itk following expression in primary CD4+ T cells. The results of these experiments indicate that the Itk mutant exhibiting diminished self-association has increased activity and signaling capacity both in vitro and upon T cell receptor engagement in primary T cells. Based on these findings, we discuss a mechanistic explanation for this observation and propose a model for the control of Itk activity during T cell receptor signaling.
Materials and Methods
Constructs and baculovirus production
V5 or myc tagged proteins were cloned into the pcDNA3.1D/V5-His-TOPO vector (Invitrogen). Itk(BtkSH3) was generated by replacing Itk SH3 domain sequence from Pro171 to Asn232 with the human Btk sequence spanning Ser214 to Ser275 by PCR. Flag tagged full-length wild-type (mouse Itk) or Itk(BtkSH3) were cloned into the pENTR/D-TOPO vector (Invitrogen) by TOPO cloning. Point mutations (Y180F) were introduced using the QuickChange Site-Directed Mutagenesis Kit (Stratagene). The pENTR vectors with various inserts were recombined in vitro with BaculoDirect C-Term Linear DNA (Invitrogen) according to the manufacturers instructions (Invitrogen) for virus production. The PLCγ1 baculovirus has been described previously (9).
Immunoprecipitation and western blot
NIH 3T3 cells were transfected with myc or V5 tagged DNA using Effectene™ transfection reagent from Qiagen. Twenty-four hours post transfection, cells were lysed in buffer containing 0.5% NP 40, 50mM Tris (pH 7.4), 150mM NaCl, 10mM MgCl2, and protease inhibitors (Roche Applied Science). Immunoprecipitation, western transfer and western blotting were performed using standard techniques. Antibodies used throughout are as follows: anti-Erk (Cell Signaling Technology), anti-phosphoErk (Cell Signaling Technology), anti-Itk (Upstate), anti-PLCgamma1 (Upstate), anti-pY783 (Upstate), anti-Myc (Invitrogen), anti-V5 (Invitrogen), anti-Btk pY223 (used to probe the analogous site in Itk, pY180) and anti-Btk pY551 (used to probe the analogous site in Itk, pY511) are from Dr. Owen Witte.
Protein expression and purification
The purification method for bacterially expressed protein has been described previously (18). For production of Itk enzyme for in vitro kinase assays, baculovirus encoding either full-length wild-type Itk or the Itk(BtkSH3) along with Lck virus (if indicated) were used to infect Sf9 cells in a 1:1 ratio. Proteins were purified using methods reported previously (28–30). A kinase inactive mutant of Itk is used to ensure that activity measurements reflect Itk activity alone and not co-purifying Lck. Co-expression of Itk (or Itk(BtkSH3)) with PLCγ1 in Sf9 cells did not include Lck expression to avoid spurious phosphorylation of the substrate by Lck. It has been demonstrated previously that Itk produced in this manner is active (31).
NMR spectroscopy
NMR spectra were recorded at 298 K on a Bruker DRX500 spectrometer operating at a 1H frequency of 499.867 MHz. Protein concentrations were adjusted to 1 mM. Global least squares parameter fitting of the titration data was performed using the Matlab (version 5.3.1, The Mathworks Inc.) suite of programs.
Phosphorylation status
Purified wild-type full-length Itk, Itk(BtkSH3) and Y180F full-length Itk were subjected to autophosphorylation assays by incubation at room temperature in a buffer containing 50mM Hepes (pH 7.0), 10mM MgCl2, 1mM DTT, 1mg/ml BSA, 0.2mM ATP at RT for 30min at 0.8 µM final enzyme concentration. The reaction was stopped by the addition of SDS-loading dye, run on an SDS-PAGE gel and western blotted.
Initial velocity measurements
The in vitro kinase assay (described in (28)) is adapted from previous in vitro kinase assays developed for Itk (29, 30). Biotin labeled peptide-B (AnaSpec Inc.), was used as a substrate. For initial velocity measurements at varying enzyme concentrations, the peptide B concentration was maintained at 400 µM.
Purification of primary CD4+ T cells
Spleen and lymph node cells were isolated from wild-type or Itk−/− (6) C57BL/10 (Jackson Labs) mice, and CD4+ T cells were purified by positive selection using anti-CD4 antibody-coated magnetic microbeads (Miltenyi Biotec).
Retrovirus production
Itk and Itk(BtkSH3) were cloned into the retroviral mouse stem cell virus vector MSCV2.2-IRES-GFP (32). Phoenix-E retroviral packaging cells were transfected with each retrovirus construct plus the pCL-Eco retrovirus Packaging vector (Imgenex) as described previously (9), and virus particles were harvested and stored at −80°C. For each infection, 2 × 106 primary CD4+ T cells were stimulated with 5ng/ml of PMA (Sigma) and 375ng/ml of Ionomycin (Calbiochem) for 24 hrs and then incubated with 2ml of viral supernatant plus IL-2 (30ng/ml) and lipofectamin (Invitrogen). After 3–4 days, CD4+GFP+ infected cells were sorted by flow cytometry and cultured a further two weeks in IL-2. For analysis, 3 × 106 cells were restimulated by incubation with biotinylated-anti-CD3 Ab (25 µg/ml; eBioscience) for 10 min, followed by streptavidin (50 µg/ml; Pierce Protein Research Products) crosslinking for 5 min. Cell lysates were analyzed for phospho-ERK by immunoblot; alternatively cells were permeabilized and stained with anti-phosphoERK antibody followed by flow cytometry.
Ca++ flux assay
Wild-type Itk and Itk(BtkSH3) were cloned in to the retroviral vector pMX-IRES-hCD8 (33). Four days after infection, cells were sensitized with 10 µg/ml biotin-αCD3 for 10 minutes on ice in OPTI-MEM, washed, and incubated with 16 µM Fura-Red (Invitrogen) and 16 µM Fluo-3 (Invitrogen) in RPMI-3% FBS in 37°C for 45 min. Cells were washed twice with RPMI-3% FBS, incubated at room temperature in Tyrode’s buffer-BSA for 30 minutes. Cells were analyzed by flow cytometry with the addition of streptavidin at 30 seconds. Tyrodes’ buffer-BSA for Ca++ influx assay: 137 mM NaCl, 1.2 mM HEPES, 2.7 mM KCl, 0.04mM NaH2PO4·H2O, 5.6 mM Glucose, 0.5 mM CaCl2·H2O, 1mM MgCl2·6H2O, 2% FBS.
Statistics
Statistical analysis was performed using the student’s t test.
Results
The Itk SH3 domain interacts specifically with the Itk SH2 domain in a non-classical manner
SH2 and SH3 domains are well-studied signal transduction modules that mediate protein-protein interactions to alter protein localization and to promote proximity for enzyme-substrate interactions. SH2 and SH3 binding domains typically mediate interactions with target proteins that contain phosphotyrosine motifs or proline-rich sequences, respectively. For instance, the Itk SH2 domain binds to phosphorylated tyrosine residues on SLP-76, and the Itk SH3 domain binds to proline-rich regions in SLP-76 and Vav1 (11). However, the regulatory SH2 and SH3 domains of Itk interact with each other via an intermolecular association that does not fit these established paradigms for SH2 and SH3 recognition (18). The Itk SH3/SH2 interaction is neither phosphotyrosine-dependent nor mediated by a polyproline motif (16). Thus, the Itk amino acid residues mediating this self-association are distinct from those involved in binding to other signaling proteins, such as SLP-76. Given this distinction, Itk provides us with an ideal opportunity to specifically disrupt self-association by altering the SH3 and/or SH2 domains in a manner that does not affect binding to canonical proline-rich or phosphotyrosine ligands. These alterations would then permit functional assays allowing us to compare full-length, wild-type Itk to a full-length Itk variant in which self-association is disfavored.
Molecular determinants of the intermolecular SH3/SH2 interaction
The three dimensional structure of the binary Itk SH3/SH2 complex is shown in Figure 1b. Using this structure, we aimed to identify sequence changes that would abolish the interaction between the Itk SH3 and SH2 domains but maintain the classical ligand binding functions of the SH3 and SH2 domain, proline-rich peptide recognition and phosphotyrosine binding, respectively. A simplistic approach to this goal would be mutation of the central residue in the SH3 binding cleft, Trp 208, which has been previously shown to disrupt the interaction between the SH2 domain and SH3 domain of Itk (18). However, this conserved tryptophan also makes extensive contacts to cognate proline-rich ligands (34, 35) and is therefore not an appropriate target for mutation in our current study. In fact, the SH2/SH3 interface in this complex either fully or partially overlaps with the binding sites on these domains that contact the classical phospholigand and proline-rich ligand sequences. Since our goal is to selectively disrupt just the SH3/SH2 interaction without affecting the other binding functions of the Itk SH3 and SH2 domains, we turned our attention to an approach that would disrupt the SH3/SH2 interaction but maintain the canonical ligand binding characteristics of each of these regulatory domains.
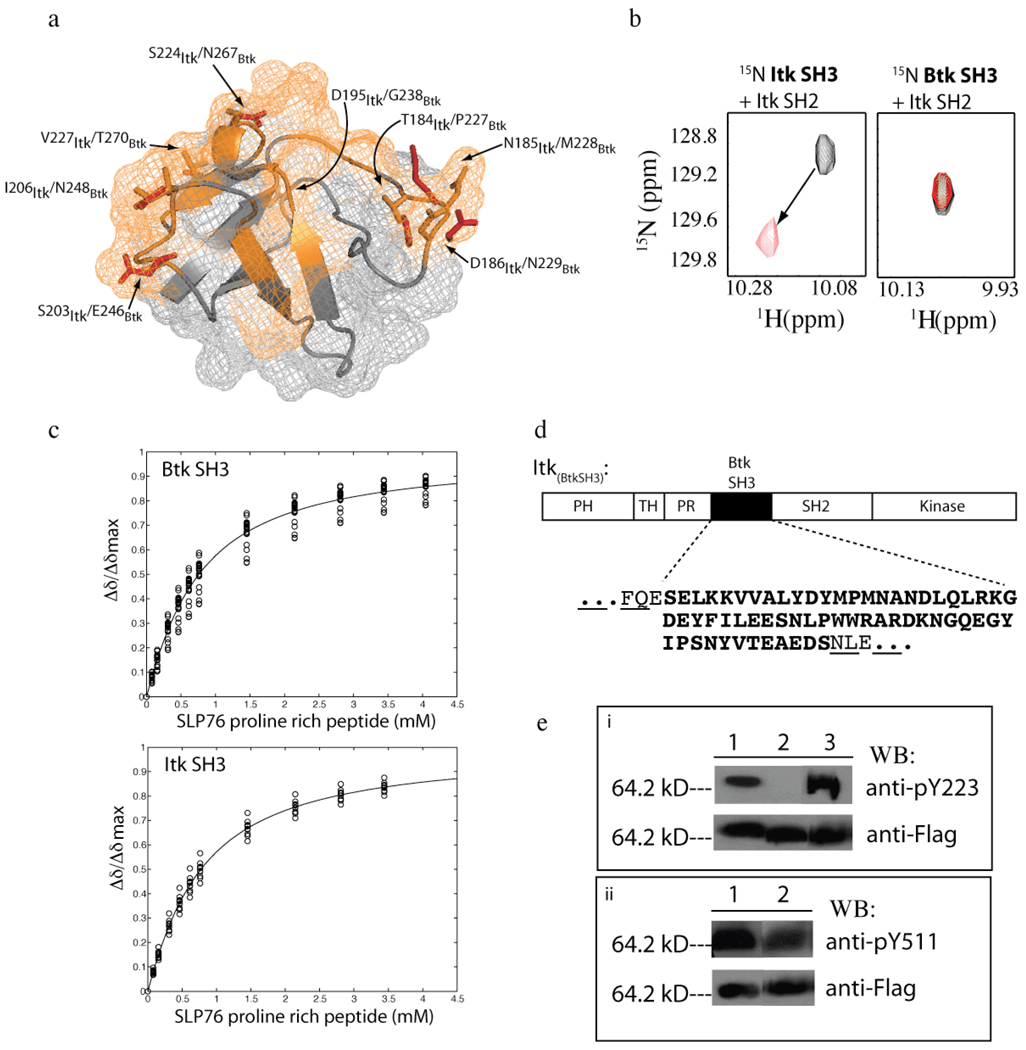
The Btk SH3 domain is 51% identical to that of Itk. Of the Itk SH3 domain surface residues that directly contact the Itk SH2 domain in the SH3/SH2 complex, there are eight residues that differ between Itk and Btk SH3 domains (Fig 2a). These sequence differences are enough to eliminate the intermolecular SH3/SH2 interaction. Using NMR spectroscopy we measured binding between the Itk SH2 domain and the Btk SH3 domain. Changes in NMR peak position (chemical shift perturbations) indicate specific binding between proteins. Unlike the Itk SH3/Itk SH2 interaction that is readily detected by NMR, we find no chemical shift perturbations are observed in NMR spectra upon mixing the Btk SH3 domain with the Itk SH2 domain (Fig. 2b). Thus, the Itk SH2 domain and the Btk SH3 domain do not show detectable binding to each other.
Figure 2. Specific disruption of the intermolecular SH3/SH2 interaction.
(a) The Itk SH3 and Btk SH3 domain structures are superimposed (only labeled side chains from the Btk SH3 domain are shown for clarity). The Itk SH3/SH2 interaction surface is indicated in orange on the SH3 structure (18). Within this binding surface, only eight residues are not conserved between Itk and Btk. Each of the non-conserved pairs of residues are labeled and indicated on the structure (orange are Itk side chains and red are Btk side chains). (b) Select region of heteronuclear single quantum coherence (HSQC) spectra of 15N-labeled SH3 domains (from either Itk (left) or Btk (right)) indicating the extent of chemical shift perturbations associated with binding to unlabeled Itk SH2 domain. For both, the black spectrum is 15N labeled SH3 protein alone and the red spectrum has been acquired following addition of equimolar unlabeled Itk SH2 domain in each case. The arrow indicates the change in resonance frequency for a representative peak upon addition of Itk SH2 to 15N labeled Itk SH3 domain. Addition of unlabeled Itk SH2 domain to 15N labeled Btk SH3 domain results in no change in chemical shift values. This result confirms the absence of an interaction between Itk SH2 and Btk SH3. (c) Concentration dependence of the normalized 1H chemical shift (Δδ/Δδmax) for representative residues of 15N labeled Btk SH3 domain (top) and 15N labeled Itk SH3 domain (bottom) upon titration of the Slp76 derived proline-rich peptide. The titration data were acquired and analyzed as previously described (21). (d) Design of the Itk(BtkSH3) protein. The Itk SH3 sequence within full-length Itk is replaced with the SH3 sequence of Btk. The sequence of the Btk SH3 domain is shown in bold below the domain map of Itk and the surrounding Itk residues within the resulting Itk(BtkSH3) are non-bold and underlined. (e) Western blot analysis using phosphotyrosine specific antibodies to the autophosphorylation site in the SH3 domain (Y180 for Itk/Y223 for Btk) and the activation loop tyrosine (Y511). (i) Lane 1: wild-type full-length Itk, lane 2: full-length, mutant Itk(Y180F) and lane 3: Itk(BtkSH3) were expressed as fusions with the flag peptide and purified from insect cells, resolved by SDS-PAGE, transferred to PDVF membrane and blotted with the Btk antibody to pY223 (Btk numbering; this tyrosine corresponds to Y180 in Itk) to detect the autophosphorylation status in the SH3 domain (top panel). Cross reactivity of the Btk pY223 antibody for Itk pY180 has been previously demonstrated (9). (ii) Lane 1: wild type full length Itk, lane 2: Itk(BtkSH3) were prepared as described above and blotted with the anti-pY551 antibody to detect the activation loop phosphorylation status (top panel). Itk protein levels are indicated in the bottom panel for both (i) and (ii).
To ensure that the Btk SH3 sequence binds the canonical proline-rich ligand in a manner similar to the Itk SH3 domain, we next compared the affinities of the Itk and Btk SH3 domains for a proline-rich peptide derived from Slp-76 (Q184QPPVPPQRPMA195), previously shown to bind the SH3 domain of Itk (36). Separate titrations of the QQPPVPPQRPMA peptide into 15N labeled Itk or Btk SH3 domains induced chemical shift perturbations that were analyzed to determine the respective dissociation constants. The binding curves generated upon addition of increasing concentration of the Slp-76 peptide into the Btk and Itk SH3 domains are indistinguishable (Fig. 2c). Thus, the Btk SH3 domain does not interact with the Itk SH2 domain, yet maintains normal proline binding characteristics. These data provide a strategy for altering the full-length Itk sequence to disfavor the observed self-association of wild-type Itk, while maintaining classical proline ligand binding characteristics. A full-length Itk protein (designated Itk(BtkSH3)) that contains the Btk sequence within the region that normally spans the Itk SH3 domain (Fig. 2d) was constructed. All other regions of Itk(BtkSH3) are identical to wild-type full-length Itk.
Using the newly constructed Itk(BtkSH3) protein, we examined the phosphorylation status of both Tyr 511 in the Itk activation loop and Tyr 180 within the Itk SH3 domain. Tyr 180 is strictly conserved among SH3 domains and the corresponding residue in the Btk SH3 domain is Y223. Phosphorylation at both tyrosine sites (Y511 and Y180) accompanies activation of Itk in T cells and if disrupted would interfere with Itk function in T cells (9). Full length wild type Itk and Itk(BtkSH3) were separately expressed as flag tagged fusion proteins and were co-expressed with Lck in insect cells. The proteins were purified, separated by SDS-PAGE and then probed by western blotting with a Btk pY223 specific antibody (previously shown to recognize specific phosphorylation on both Itk Y180 and Btk Y223 (9)) or an Itk pY511 specific antibody. Both wild type Itk and Itk(BtkSH3) are phosphorylated to the same degree at the expected sites; Tyr 180 and Tyr 511 (Fig. 2e). Thus, sequence changes in the full length Itk(BtkSH3) diminish intermolecular self-association but do not alter the normal phosphorylation patterns associated with wild type Itk activation.
To examine the extent that the intermolecular Itk SH3/SH2 interaction mediates self-association of the full-length Itk kinase, we generated differentially tagged versions of full-length, wild-type Itk and Itk(BtkSH3) for co-immunoprecipitation experiments (Fig. 3a). First, the myc and V5 epitope tagged wild type Itk were transiently co-transfected into NIH 3T3 cells and subjected to immunoprecipitation using anti-myc antibody. The immunoprecipitated fraction was then resolved by SDS-PAGE and probed with an antibody to the V5 tag. As a control, NIH 3T3 cells were also transiently transfected with V5 tagged Itk and/or the empty vector alone. The V5 tagged full-length Itk is readily detected in anti-myc immunoprecipitates from co-transfected NIH 3T3 cells indicating that full-length wild-type Itk self-associates in an intermolecular fashion (Fig. 3a). These data are consistent with previous in vivo data that also indicates intermolecular association of full length Itk molecules (27).
Figure 3. Wild-type Itk co-immunoprecipitates with itself, Itk(BtkSH3) does not.
(a) Myc and V5 tagged full-length wild type Itk were transiently transfected into NIH 3T3 cells (lane 1). As controls, V5 tagged protein or pcDNA3 vector alone (lanes 2 & 3, respectively) were transiently transfected into NIH 3T3 cells. In all three lanes, anti-myc antibody was used to precipitate myc tagged protein from cell lysates; V5 antibody was used to detect V5 tagged Itk that co-immunoprecipitates with myc tagged Itk (top panel). The presence of V5 and myc-tagged proteins is confirmed in the center and bottom panels. (b) Same experiment as in (a) using V5-and myc-tagged full-length Itk(BtkSH3). The absence of the coIP band in lane 1 (top panel) indicates a loss of the intermolecular interactions within Itk(BtkSH3) that stabilize the self-associated form.
Btk SH3 sequence limits Itk self-association
In a manner identical to the wild-type Itk sequence, myc and V5 epitope tagged Itk(BtkSH3) were transiently co-transfected into NIH 3T3 cells and subjected to co-immunoprecipitation. Consistent with the absence of an intermolecular interaction between the isolated Itk SH2 domain and the Btk SH3 domains, co-immunoprecipitation data indicate that full-length Itk(BtkSH3) does not self-associate to a level that is detected in the co-IP experiments for wild-type Itk (Fig. 3b). The observed differences in the co-immunoprecipitation data for Itk and Itk(BtkSH3) suggest that disruption of the non-canonical Itk SH3/SH2 interaction reduces intermolecular self-association of full-length Itk compared to wild type protein. The functions of these Itk proteins were next examined both in vitro and in primary T cells.
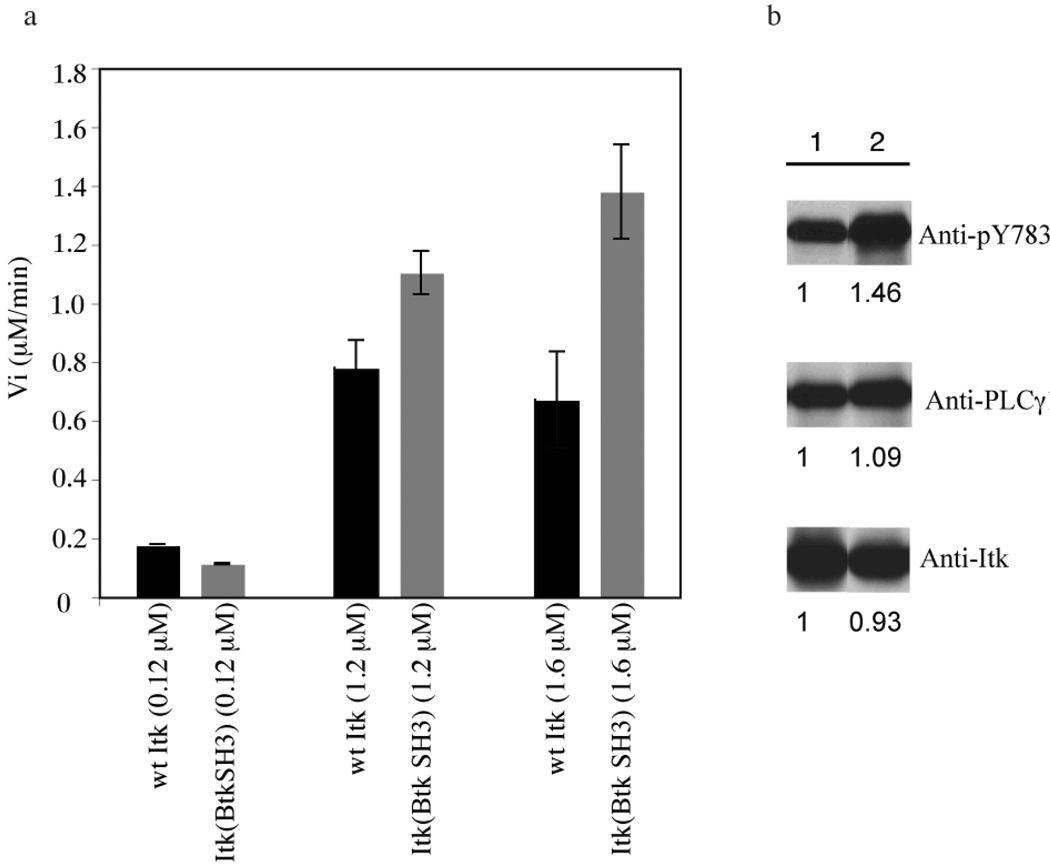
Recombinant flag-tagged, wild-type Itk and Itk(BtkSH3) were separately purified from baculovirus-infected insect cells and used for in vitro kinase assays. Using the in vitro kinase assay that we have described previously for wild-type Itk (28), a biotinylated peptide substrate (30) was used to measure the initial velocity (Vi) of wild type Itk and Itk(BtkSH3) at different concentrations of enzyme (Fig. 4a). The concentration of peptide substrate in these assays is 400 µM (~five times Km), to ensure that substrate is not limiting (28). At higher enzyme concentrations, the Itk(BtkSH3) mutant is more active than wild type Itk at the same concentration (Fig. 4a). At the lowest enzyme concentration, the initial velocities of wild type Itk and Itk(BtkSH3) are similar (Fig. 4a) suggesting that (1) the differences observed at the higher enzyme concentration are not due to intrinsic activity differences between the wild type Itk and Itk(BtkSH3) constructs and (2) the increase in activity is a concentration dependent phenomenon, consistent with a role for the SH3/SH2 intermolecular interaction in regulating Itk kinase activity. We next compared the activity of both wild type Itk and Itk(BtkSH3) by co-infecting Sf9 insect cells with the Itk substrate, PLC-γ1, and either wild-type Itk or Itk(BtkSH3). Tyrosine phosphorylation of PLCγ1 on Tyr783 was more robustly induced by Itk(BtkSH3) than by wild-type Itk, suggesting enhanced kinase activity for Itk(BtkSH3) compared to wild-type Itk on its native substrate (Fig 4b).
Figure 4. Increased activity of Itk(BtkSH3) versus wild-type Itk in vitro.
(a) Initial velocity (Vi in µM/min) is measured for the wild-type Itk enzyme and the Itk(BtkSH3) enzyme at three different enzyme concentrations, 0.12 µM, 1.2 µM and 1.6 µM, using Biotin labeled peptide-B as a substrate. At each concentration the wild type Itk data are shown in black and the Itk(BtkSH3) data are in grey. Experiment was carried out in duplicate and the data shown are representative of three independent experiments. (b) Full-length wild-type Itk (lane 1) or Itk(BtkSH3) (lane 2) were co-expressed with PLCγ1 in Sf9 insect cells. After three days, cell lysates were prepared, and immunoblotted for phospho-PLCγ1 (top panel), total PLCγ1 (middle panel) or Itk (bottom panel). The bands on the western blot are quantified using Quantity One software and setting wild-type Itk to one.
The in vitro data shown in Figure 4 prompted us to introduce wild-type Itk and Itk(BtkSH3) into Itk−/− primary CD4+ T cells by retrovirus transduction to compare the signaling properties of the two Itk constructs. With this retrovirus vector, GFP is translated from the same mRNA transcript as Itk; thus Itk and Itk(BtkSH3) were expressed at comparable levels in Itk−/− deficient cells, as indicated by GFP fluorescence (Fig. 5a). Previous studies have shown that optimal activation and phosphorylation of the p42/44 ERK Map-kinases following T cell activation are dependent on Itk (8, 37). To compare wild type Itk and Itk(BtkSH3) function in primary T cells, transduced T cells were stimulated by T cell receptor (TCR) engagement, and ERK phosphorylation was assessed. Itk−/− T cells expressing wild-type Itk were ~20% positive for ERK phosphorylation, in contrast to Itk−/− T cells transduced with the retrovirus vector alone (~10% phospho-ERK+). Expression of Itk(BtkSH3) further enhanced ERK activation, leading to ~30% phospho-ERK+ cells. Average data from three experiments are shown in Figure 5b. Of note, the levels of wild type Itk and Itk(BtkSH3) in retrovirus-infected Itk−/− T cells are substantially lower than the normal level of endogenous Itk in wild-type T cells (Fig. 5c). Nonetheless, even this low level of Itk(BtkSH3) induces ERK phosphorylation that is similar if not greater than that seen in wild-type T cells following TCR stimulation (Fig. 5c). These data indicate that Itk(BtkSH3) is able to function in the TCR signaling pathway of primary T cells and compared to wild type Itk expressed at the same level, the non-clustering Itk mutant exhibits enhanced TCR signaling.
Figure 5. Itk(BtkSH3) exhibits enhanced signaling in T cells.
(a–c) Itk−/− primary CD4+ T cells were infected with the retrovirus vector alone (MSCV, solid line), a retrovirus expressing wild-type Itk (Itk, long dashed line) or a retrovirus expressing Itk(BtkSH3) (short dashed line); GFP+ cells were sorted on day 4 post-infection. (a) Histograms of GFP fluorescence at the time of analysis are shown compared to non-infected control cells (filled histogram). Cells were stimulated with anti-CD3 antibody for 5 minutes, fixed, permeabilized, and stained with anti-phospho-ERK antibody. The percentages of phospho-ERK+ cells are indicated on each histogram. The non-stimulated control shown (NS) is MSCV infected cells. Non-stimulated controls for all cell lines are provided in Supplementary Materials. (b) The graph shows data from three experiments described in (a). Bars indicate mean±S.D. of the percentage of phospho-ERK+ T cells. *, p<0.0005; **, p<0.0006; ***, p<0.004 (using Paired t-test). (c) Itk−/− T cells infected with vector alone (lane 3), retrovirus expressing wild-type Itk (lane 4) or retrovirus expressing Itk(BtkSH3) (lane 5) were stimulated with crosslinking anti-CD3 antibody for 5 minutes, lysed, and immunoblotted for Itk (top panel), phospho-ERK (middle panel) and total ERK (bottom panel). Lanes 1 and 2 represent primary Itk+/+ CD4+ T cells stimulated with anti-CD3 antibody for 0 and 5 minutes, respectively. (d and e) Itk+/+ primary CD4+ T cells were infected with the retrovirus vector alone (MSCV, solid line), a retrovirus expressing wild-type Itk (Itk, long dashed line) or a retrovirus expressing Itk(BtkSH3) (short dashed line) and treated in a manner identical to the data experiment shown in (a) and (b). (f) Wild-type or Itk−/− CD4+ T cells were infected with pMX-IRES-hCD8 vector alone or vector containing Itk constructs. Four days after infection, hCD8+ T cells were gated on and analyzed for calcium mobilization in response to anti-CD3 stimulation. Levels of wild-type Itk and Itk(BtkSH3) protein in the transduced Itk−/− T cells were comparable, as assessed by hCD8 staining (See Supplementary Materials). Data are representative of three independent experiments with similar results.
We also expressed wild-type Itk or Itk(BtkSH3) in Itk+/+ primary CD4+ T cells. According to our model, the active pool of Itk is the monomeric fraction and further addition of wild-type Itk to the endogenous Itk supply should not significantly increase the pool of free monomer due to its self-association. In contrast, we expect that addition of Itk(BtkSH3) should increase the population of monomer supply since the Itk(BtkSH3) molecules are less likely to interact with themselves or with wild-type Itk. Indeed, expression of Itk(BtkSH3) in Itk+/+ T cells enhanced ERK phosphorylation following TCR stimulation compared to T cells transduced with the control retrovirus, whereas transduction of Itk+/+ T cells with wild-type Itk had no effect (Fig. 5d, e). These results indicate that Itk(BtkSH3) enhances Itk functional activity in the presence of wild-type Itk, suggesting that co-expression of the two forms of Itk leads to an increase in Itk monomers in T cells.
Given the increased activity of Itk(BtkSH3) measured in vitro (Fig. 4a), the higher levels of PLCγ1 phosphorylation induced by Itk(BtkSH3) compared to wild type Itk (Fig. 4b) and the increase in phosphorylated Erk in primary T cells expressing Itk(BtkSH3) (Fig. 5a–e), we next monitored calcium flux in stimulated Itk−/− T cells expressing either wild-type Itk, Itk(BtkSH3) or vector alone (Fig. 5f). As can be seen, Itk−/− cells transduced with vector alone have a markedly impaired calcium response compared to Itk-sufficient wild-type CD4+ T cells; expression of wild-type Itk in Itk−/− cells partially restores this response. Importantly, Itk−/− T cells expressing Itk(BtkSH3) showed enhanced calcium flux compared to cells expressing wild type Itk, consistent with the earlier observations of increased activity for the non-clustering Itk mutant. Efforts to assess IL-2 responses by the retrovirus-transduced Itk−/− T cells indicated a trend towards higher activity in cells expressing Itk(BtkSH3) versus wild-type Itk, but the differences seen were not statistically significant. Nonetheless, these data overall provide further support for a model in which disruption of intermolecular Itk clustering leads to an increase in Itk mediated signaling following TCR stimulation.
Discussion
Strength and duration of TCR signaling are important in regulating T cell development and lineage commitment. As a key component of the TCR signaling pathway, Itk has been found to modulate several aspects of thymocyte maturation, including positive selection, negative selection, and conventional versus innate CD8+ T cell development (11, 38). These observations suggest that Itk activity is involved in fine-tuning TCR signaling, and that this, in turn, regulates T cell fate decisions. Further investigation of this process would be greatly aided by a mouse model in which Itk kinase activity is increased relative to wild-type Itk. Previous efforts to increase Itk activity by overexpression have uniformly been unsuccessful (36). The data presented in this report provide a biochemical explanation for these failures. Quite unlike the Src family kinases, there are to date no known mutations that activate Itk signaling. To our knowledge, Itk(BtkSH3) is the first Itk variant to show activity greater than that of the wild type enzyme in T cells. Thus, in the future, a knock-in mouse expressing Itk(BtkSH3) in place of wild-type Itk could provide a system for examining T cell development and lineage commitment in cells with increased Itk kinase activity.
The quantitative effects of modulating Itk self-association on T cell receptor signaling events are similar to those seen in studies of the Src family kinase, Lck. Src kinases, including Lck, are negatively-regulated by an intramolecular association between the SH2 domain and a phosphorylated tyrosine in the C terminal tail that is absent in the Tec kinases. A comprehensive examination of Lck autoinhibition was recently reported in which this intramolecular interaction is either disrupted or strengthened by sequence changes close to the phosphotyrosine (39). Lck-deficient Jurkat T cells were reconstituted with wild-type Lck or Lck mutants, and T cell receptor signaling leading to Erk activation was examined. Similar to our findings for wild-type Itk and Itk(BtkSH3), modulation of the Lck autoinhibitory interaction led to detectable, but modest changes in Erk activation. Together these studies indicate that shifting the equilibrium between active and inactive kinase conformations alters the strength of TCR mediated signaling but does so within a limited range of outcomes. This is in contrast to mutations located in active sites that have pronounced, all or nothing, effects on downstream signaling.
In the current study, the sequence changes that we have introduced into full length Itk have been limited to the SH3/SH2 interface. As already mentioned, the isolated PH domain of Itk also forms intermolecular self-associated complexes (26) and might therefore play a significant role in Itk clustering in T cells. Itk membrane association occurs via PH domain interactions with phosphatidylinositol (3,4,5) trisphosphate in a T cell stimulation dependent fashion (40–43). Localization of Itk at the membrane could favor PH/PH intermolecular interactions resulting in further stabilization of self-associated Itk complexes. Indeed, the contribution of the PH domain to Itk self–association may be minimal in systems such as the NIH 3T3 cells used here for co-immunoprecipitation experiments, but significantly more pronounced in the context of the T cell membrane environment. Hence, the modest functional effects observed upon altering the SH3/SH2 interface could be due to a dominant role for the PH domain that has not been affected in the Itk(BtkSH3) mutant. When more detailed structural insights into the PH/PH domain interface become available, this region of the Itk regulatory structure can be probed for specific sequence changes that disrupt PH domain self-association but are silent with respect to the membrane binding function of the PH domain. It seems likely that such mutations, by themselves or in combination with mutations in the SH3/SH2 interface, may shift the equilibrium further away from the self-associated form of Itk leading to further enhancement of Itk signaling in T cells.
Specific intermolecular self-association has been characterized for a number of protein systems (44). Within the kinase superfamily this mechanism activates receptor kinases by promoting trans auto-phosphorylation both within and outside of the protein kinase domain (45). In another example of activation by intermolecular association, the anti-viral protein kinase PKR dimerizes via phosphotyrosine-dependent binding to double-stranded RNA (46, 47). Alternatively, inhibition by dimerization occurs for the receptor-like protein tyrosine phosphatase-α where an inhibitory ‘wedge’ on one molecule inserts into the catalytic site of another molecule (48). A crystal structure of the kinase domain from yeast Snf1 also reveals a dimeric arrangement that putatively impedes catalytic activity by steric means (49). Likewise, a crystal structure of Ca2+/calmodulin-dependent protein kinase II (CaMKII) reveals a regulatory segment that sterically blocks substrate binding to the catalytic site (50). The CaMKII kinase domain itself is intrinsically active and dimer formation brings the regulatory segment into position to inhibit activity.
In light of these examples, a simple mechanistic model for Itk autoinhibition by self-association would invoke steric blockage of the catalytic site. However, it has been well documented that the isolated Itk kinase domain exhibits little or no catalytic activity (28–30) suggesting the possibility of an alternative autoinhibition mechanism. We have previously reported a kinetic analysis of a series of Itk fragments and have shown that the SH2 domain and linker between SH2 and kinase domains are required to achieve wild-type levels of activity (28). Like the Csk kinase (51), the SH2 and SH2-kinase linker region make direct contact with the kinase domain to stabilize the active conformation of Itk. Thus, inhibition of Itk catalytic activity by SH3/SH2 mediated self-association could be explained if the intermolecular interactions between Itk regulatory domains (particularly those involving the SH2 domain) compete with activating interactions between the SH2 and kinase domains.
T cell activation itself produces signals that compete with the self-association equilibrium and would shift the population of Itk in the cell toward a monomeric state. Specifically, exogenous binding partners such as transiently produced phospholigands would compete directly with Itk self-association by interfering with the SH2/SH3 interaction interface (9, 18, 22, 36, 52, 53). If self-association is disfavored, the regulatory domains could then adopt the catalytically competent conformation (28). Conversely, in the absence of such activating factors, Itk might remain self-associated and autoinhibited.
It is also possible to envision a role for Itk autoinhibition following activation of Itk by TCR engagement; in this case, self-association would be one mechanism that could terminate Itk signaling (in addition to the activity of phosphatases). In binding studies of the Itk SH3 and SH2 domains we found that Y180 phosphorylation in the SH3 domain enhances affinity for the Itk SH2 domain (54). Together with the observations that autophosphorylation at Y180 in the SH3 domain has no effect on Itk kinase activity and occurs in cis (54) it is reasonable to suggest that autophosphorylation may be a first step toward turning off Itk-mediated signaling, by promoting intermolecular self-association leading to a drop in kinase activity. Indeed, a previous report suggests that Itk clustering in T cells occurs following membrane association (27).
While the precise mechanistic details of when and how intermolecular association of Itk modulates its activity remain a question, this mode of autoinhibition for a non-receptor tyrosine kinase suggests that reagents (or mutations) that shift the equilibrium toward the self-associated state could dampen T cell activation while disfavoring self-association will increase Itk activity and concomitant T cell activation.
Supplementary Material
Acknowledgements
The authors would like to thank Dr. Owen Witte for providing the anti-pY223 and anti-pY551 Btk antibodies.
This work is supported by grants from the National Institutes of Health (NIAID, AI43957 and AI075150 to A.H.A; NIAID, AI37584 and AI66118 to L.J.B.). University of Massachusetts Medical School core resources supported by the Diabetes Endocrinology Research Center grant DK32520 were also used.
References
- 1.Gibson S, Leung B, Squire JA, Hill M, Arima N, Goss P, Hogg D, Mills GB. Identification, cloning, and characterization of a novel human T-cell-specific tyrosine kinase located at the hematopoietin complex on chromosome 5q. Blood. 1993;82:1561–1572. [PubMed] [Google Scholar]
- 2.Heyeck SD, Berg LJ. Developmental regulation of a murine T-cell-specific tyrosine kinase gene, Tsk. Proc Natl Acad Sci U S A. 1993;90:669–673. doi: 10.1073/pnas.90.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siliciano JD, Morrow TA, Desiderio SV. itk, a T-cell-specific tyrosine kinase gene inducible by interleukin 2. Proc Natl Acad Sci U S A. 1992;89:11194–11198. doi: 10.1073/pnas.89.23.11194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka N, Asao H, Ohtani K, Nakamura M, Sugamura K. A novel human styrosine kinase gene inducible in T cells by interleukin 2. FEBS Lett. 1993;324:1–5. doi: 10.1016/0014-5793(93)81520-a. [DOI] [PubMed] [Google Scholar]
- 5.Yamada N, Kawakami Y, Kimura H, Fukamachi H, Baier G, Altman A, Kato T, Inagaki Y, Kawakami T. Structure and expression of novel protein-tyrosine kinases, Emb and Emt, in hematopoietic cells. Biochem Biophys Res Commun. 1993;192:231–240. doi: 10.1006/bbrc.1993.1404. [DOI] [PubMed] [Google Scholar]
- 6.Liu KQ, Bunnell SC, Gurniak CB, Berg LJ. T cell receptor-initiated calcium release is uncoupled from capacitative calcium entry in Itk-deficient T cells. J Exp Med. 1998;187:1721–1727. doi: 10.1084/jem.187.10.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perez-Villar JJ, Kanner SB. Regulated association between the tyrosine kinase Emt/Itk/Tsk and phospholipase-C gamma 1 in human T lymphocytes. J Immunol. 1999;163:6435–6441. [PubMed] [Google Scholar]
- 8.Schaeffer EM, Debnath J, Yap G, McVicar D, Liao XC, Littman DR, Sher A, Varmus HE, Lenardo MJ, Schwartzberg PL. Requirement for Tec kinases Rlk and Itk in T cell receptor signaling and immunity. Science. 1999;284:638–641. doi: 10.1126/science.284.5414.638. [DOI] [PubMed] [Google Scholar]
- 9.Wilcox HM, Berg LJ. Itk phosphorylation sites are required for functional activity in primary T cells. J Biol Chem. 2003;278:37112–37121. doi: 10.1074/jbc.M304811200. [DOI] [PubMed] [Google Scholar]
- 10.Palacios EH, Weiss A. Function of the Src-family kinases, Lck and Fyn, in T-cell development and activation. Oncogene. 2004;23:7990–8000. doi: 10.1038/sj.onc.1208074. [DOI] [PubMed] [Google Scholar]
- 11.Berg LJ, Finkelstein LD, Lucas JA, Schwartzberg PL. Tec family kinases in T lymphocyte development and function. Annu Rev Immunol. 2005;23:549–600. doi: 10.1146/annurev.immunol.22.012703.104743. [DOI] [PubMed] [Google Scholar]
- 12.Cooper JA. The when and how of Src regulation. Cell. 1993;73:1051–1054. doi: 10.1016/0092-8674(93)90634-3. [DOI] [PubMed] [Google Scholar]
- 13.Roskoski R., Jr Src protein-tyrosine kinase structure and regulation. Biochem Biophys Res Commun. 2004;324:1155–1164. doi: 10.1016/j.bbrc.2004.09.171. [DOI] [PubMed] [Google Scholar]
- 14.Xu W, Doshi A, Lei M, Eck MJ, Harrison SC. Crystal structures of c-Src reveal features of its autoinhibitory mechanism. Mol Cell. 1999;3:629–638. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- 15.Xu W, Harrison SC, Eck MJ. Three-dimensional structure of the tyrosine kinase c-Src. Nature. 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- 16.Severin AJR, Boyken S, Fulton DB, Andreotti AH. Proline isomerization preorganizes the Itk SH2 domain for binding to the Itk SH3 domain. J Mol Biol. 2009;387:726–743. doi: 10.1016/j.jmb.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andreotti AH, Bunnell SC, Feng S, Berg LJ, Schreiber SL. Regulatory intramolecular association in a tyrosine kinase of the Tec family. Nature. 1997;385:93–97. doi: 10.1038/385093a0. [DOI] [PubMed] [Google Scholar]
- 18.Brazin KN, Fulton DB, Andreotti AH. A specific intermolecular association between the regulatory domains of a Tec family kinase. J Mol Biol. 2000;302:607–623. doi: 10.1006/jmbi.2000.4091. [DOI] [PubMed] [Google Scholar]
- 19.Mallis RJ, Brazin KN, Fulton DB, Andreotti AH. Structural characterization of a proline-driven conformational switch within the Itk SH2 domain. Nat Struct Biol. 2002;9:900–905. doi: 10.1038/nsb864. [DOI] [PubMed] [Google Scholar]
- 20.Laederach A, Cradic KW, Brazin KN, Zamoon J, Fulton DB, Huang XY, Andreotti AH. Competing modes of self-association in the regulatory domains of Bruton's tyrosine kinase: intramolecular contact versus asymmetric homodimerization. Protein Sci. 2002;11:36–45. doi: 10.1110/ps.26702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laederach A, Cradic KW, Fulton DB, Andreotti AH. Determinants of intra versus intermolecular self-association within the regulatory domains of Rlk and Itk. J Mol Biol. 2003;329:1011–1020. doi: 10.1016/s0022-2836(03)00531-x. [DOI] [PubMed] [Google Scholar]
- 22.Morrogh LM, Hinshelwood S, Costello P, Cory GO, Kinnon C. The SH3 domain of Bruton's tyrosine kinase displays altered ligand binding properties when auto-phosphorylated in vitro. Eur J Immunol. 1999;29:2269–2279. doi: 10.1002/(SICI)1521-4141(199907)29:07<2269::AID-IMMU2269>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 23.Okoh MP, Vihinen M. Interaction between Btk TH and SH3 domain. Biopolymers. 2002;63:325–334. doi: 10.1002/bip.10049. [DOI] [PubMed] [Google Scholar]
- 24.Pursglove SE, Mulhern TD, Mackay JP, Hinds MG, Booker GW. The solution structure and intramolecular associations of the Tec kinase SRC homology 3 domain. J Biol Chem. 2002;277:755–762. doi: 10.1074/jbc.M108318200. [DOI] [PubMed] [Google Scholar]
- 25.Hansson H, Okoh MP, Smith CI, Vihinen M, Hard T. Intermolecular interactions between the SH3 domain and the proline-rich TH region of Bruton's tyrosine kinase. FEBS Lett. 2001;489:67–70. doi: 10.1016/s0014-5793(00)02438-8. [DOI] [PubMed] [Google Scholar]
- 26.Huang YH, Grasis JA, Miller AT, Xu R, Soonthornvacharin S, Andreotti AH, Tsoukas CD, Cooke MP, Sauer K. Positive regulation of Itk PH domain function by soluble IP4. Science. 2007;316:886–889. doi: 10.1126/science.1138684. [DOI] [PubMed] [Google Scholar]
- 27.Qi Q, Sahu N, August A. Tec kinase Itk forms membrane clusters specifically in the vicinity of recruiting receptors. J Biol Chem. 2006;281:38529–38534. doi: 10.1074/jbc.M609180200. [DOI] [PubMed] [Google Scholar]
- 28.Joseph RE, Min L, Andreotti AH. The Linker between SH2 and Kinase Domains Positively Regulates Catalysis of the Tec Family Kinases. Biochemistry. 2007;46:5455–5462. doi: 10.1021/bi602512e. [DOI] [PubMed] [Google Scholar]
- 29.Brown K, Long JM, Vial SC, Dedi N, Dunster NJ, Renwick SB, Tanner AJ, Frantz JD, Fleming MA, Cheetham GM. Crystal structures of interleukin-2 tyrosine kinase and their implications for the design of selective inhibitors. J Biol Chem. 2004;279:18727–18732. doi: 10.1074/jbc.M400031200. [DOI] [PubMed] [Google Scholar]
- 30.Hawkins J, Marcy A. Characterization of Itk tyrosine kinase: contribution of noncatalytic domains to enzymatic activity. Protein Expr Purif. 2001;22:211–219. doi: 10.1006/prep.2001.1447. [DOI] [PubMed] [Google Scholar]
- 31.Heyeck SD, Wilcox HM, Bunnell SC, Berg LJ. Lck phosphorylates the activation loop tyrosine of the Itk kinase domain and activates Itk kinase activity. J Biol Chem. 1997;272:25401–25408. doi: 10.1074/jbc.272.40.25401. [DOI] [PubMed] [Google Scholar]
- 32.Ouyang W, Ranganath SH, Weindel K, Bhattacharya D, Murphy TL, Sha WC, Murphy KM. Inhibition of Th1 development mediated by GATA-3 through an IL-4-independent mechanism. Immunity. 1998;9:745–755. doi: 10.1016/s1074-7613(00)80671-8. [DOI] [PubMed] [Google Scholar]
- 33.Masuyama N, Oishi K, Mori Y, Ueno T, Takahama Y, Gotoh Y. Akt inhibits the orphan nuclear receptor Nur77 and T-cell apoptosis. J Biol Chem. 2001;276:32799–32805. doi: 10.1074/jbc.M105431200. [DOI] [PubMed] [Google Scholar]
- 34.Weng Z, Rickles RJ, Feng S, Richard S, Shaw AS, Schreiber SL, Brugge JS. Structure-function analysis of SH3 domains: SH3 binding specificity altered by single amino acid substitutions. Mol Cell Biol. 1995;15:5627–5634. doi: 10.1128/mcb.15.10.5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu H, Chen JK, Feng S, Dalgarno DC, Brauer AW, Schreiber SL. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell. 1994;76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]
- 36.Bunnell SC, Diehn M, Yaffe MB, Findell PR, Cantley LC, Berg LJ. Biochemical interactions integrating Itk with the T cell receptor-initiated signaling cascade. J Biol Chem. 2000;275:2219–2230. doi: 10.1074/jbc.275.3.2219. [DOI] [PubMed] [Google Scholar]
- 37.Miller AT, Berg LJ. New insights into the regulation and functions of Tec family tyrosine kinases in the immune system. Curr Opin Immunol. 2002;14:331–340. doi: 10.1016/s0952-7915(02)00345-x. [DOI] [PubMed] [Google Scholar]
- 38.Berg LJ. Signalling through TEC kinases regulates conventional versus innate CD8(+) T-cell development. Nat Rev Immunol. 2007;7:479–485. doi: 10.1038/nri2091. [DOI] [PubMed] [Google Scholar]
- 39.Nika K, Tautz L, Arimura Y, Vang T, Williams S, Mustelin T. A weak Lck tail bite is necessary for Lck function in T cell antigen receptor signaling. J Biol Chem. 2007;282:36000–36009. doi: 10.1074/jbc.M702779200. [DOI] [PubMed] [Google Scholar]
- 40.August A, Sadra A, Dupont B, Hanafusa H. Src-induced activation of inducible T cell kinase (ITK) requires phosphatidylinositol 3-kinase activity and the Pleckstrin homology domain of inducible T cell kinase. Proc Natl Acad Sci U S A. 1997;94:11227–11232. doi: 10.1073/pnas.94.21.11227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kojima T, F M, Watanabe Y, Hamazato F, Mikoshiba K. Characterization of the pleckstrin homology domain of Btk as an inositol polyphosphate and phosphoinositide binding domain. Biochem Biophys Res Commun. 1997;236:333–339. doi: 10.1006/bbrc.1997.6947. [DOI] [PubMed] [Google Scholar]
- 42.Rameh LE, A A, Carraway KL, Couvillon AD, Rathbun G, Crompton A, VanRenterghem B, Ravichandran KS, Burakoff SJ, Wang DS, Chen CS, Cantley LC. A comparative analysis of the phosphoinositide binding specificity of pleckstrin homology domains. J. Biol. Chem. 1997;272:22059–22066. doi: 10.1074/jbc.272.35.22059. [DOI] [PubMed] [Google Scholar]
- 43.Salim K, B MJ, Querfurth E, Zvelebil MJ, Gout I, Scaife R, Margolis RL, Gigg R, Smith CI, Driscoll PC, Waterfield MD, Panayotou G. Distinct specificity in the recognition of phosphoinositides by the pleckstrin homology domains of dynamin and Bruton's tyrosine kinase. EMBO J. 1996;15:6241–6250. [PMC free article] [PubMed] [Google Scholar]
- 44.Klemm JD, Schreiber SL, Crabtree GR. Dimerization as a regulatory mechanism in signal transduction. Annu Rev Immunol. 1998;16:569–592. doi: 10.1146/annurev.immunol.16.1.569. [DOI] [PubMed] [Google Scholar]
- 45.Lemmon MA, Schlessinger J. Transmembrane signaling by receptor oligomerization. Methods Mol Biol. 1998;84:49–71. doi: 10.1385/0-89603-488-7:49. [DOI] [PubMed] [Google Scholar]
- 46.Su Q, Wang S, Baltzis D, Qu LK, Wong AH, Koromilas AE. Tyrosine phosphorylation acts as a molecular switch to full-scale activation of the eIF2alpha} RNA-dependent protein kinase. Proc Natl Acad Sci U S A. 2006;103:63–68. doi: 10.1073/pnas.0508207103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lemaire PA, Lary J, Cole JL. Mechanism of PKR activation: dimerization and kinase activation in the absence of double-stranded RNA. J Mol Biol. 2005;345:81–90. doi: 10.1016/j.jmb.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 48.Jiang G, den Hertog J, Su J, Noel J, Sap J, Hunter T. Dimerization inhibits the activity of receptor-like protein-tyrosine phosphatase-alpha. Nature. 1999;401:606–610. doi: 10.1038/44170. [DOI] [PubMed] [Google Scholar]
- 49.Nayak V, Zhao K, Wyce A, Schwartz MF, Lo WS, Berger SL, Marmorstein R. Structure and Dimerization of the Kinase Domain from Yeast Snf1, a Member of the Snf1/AMPK Protein Family. Structure. 2006;14:477–485. doi: 10.1016/j.str.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 50.Rosenberg OS, Deindl S, Sung RJ, Nairn AC, Kuriyan J. Structure of the autoinhibited kinase domain of CaMKII and SAXS analysis of the holoenzyme. Cell. 2005;123:849–860. doi: 10.1016/j.cell.2005.10.029. [DOI] [PubMed] [Google Scholar]
- 51.Ogawa A, Takayama Y, Sakai H, Chong KT, Takeuchi S, Nakagawa A, Nada S, Okada M, Tsukihara T. Structure of the carboxyl-terminal Src kinase, Csk. J Biol Chem. 2002;277:14351–14354. doi: 10.1074/jbc.C200086200. [DOI] [PubMed] [Google Scholar]
- 52.Ching KA, Grasis JA, Tailor P, Kawakami Y, Kawakami T, Tsoukas CD. TCR/CD3-Induced activation and binding of Emt/Itk to linker of activated T cell complexes: requirement for the Src homology 2 domain. J Immunol. 2000;165:256–262. doi: 10.4049/jimmunol.165.1.256. [DOI] [PubMed] [Google Scholar]
- 53.Tsoukas CD, Grasis JA, Ching KA, Kawakami Y, Kawakami T. Itk/Emt: a link between T cell antigen receptor-mediated Ca2+ events and cytoskeletal reorganization. Trends Immunol. 2001;22:17–20. doi: 10.1016/s1471-4906(00)01795-6. [DOI] [PubMed] [Google Scholar]
- 54.Joseph RE, Fulton DB, Andreotti AH. Mechanism and functional significance of itk autophosphorylation. J Mol Biol. 2007;373:1281–1292. doi: 10.1016/j.jmb.2007.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.