Abstract
Dysregulation of apoptosis is associated with the development of human cancer and resistance to anticancer therapy. We have previously shown in tumor xenografts that DNA alkylating agents induce sporadic cell necrosis and regression of apoptosis-deficient tumors. Sporadic tumor cell necrosis is associated with extracellular release of cellular content such as the high mobility group box 1 (HMGB1) protein and subsequent recruitment of innate immune cells into the tumor tissue. It remained unclear whether HMGB1 and the activation of innate immunity played a role in tumor response to chemotherapy. In this study, we show that whereas DNA alkylating therapy leads to a complete tumor regression in an athymic mouse tumor xenograft model, it fails to do so in tumors deficient in HMGB1. The HMGB1-deficient tumors have an impaired ability to recruit innate immune cells including macrophages, neutrophils, and NK cells into the treated tumor tissue. Cytokine array analysis reveals that whereas DNA alkylating treatment leads to suppression of protumor cytokines such as IL-4, IL-10, and IL-13, loss of HMGB1 leads to elevated levels of these cytokines upon treatment. Suppression of innate immunity and HMGB1 using depleting Abs leads to a failure in tumor regression. Taken together, these results indicate that HMGB1 plays an essential role in activation of innate immunity and tumor clearance in response to DNA alkylating agents.
The ultimate goal of cancer therapy is to selectively kill cancer cells while sparing normal cells. A major burden to overcome during therapy is the impaired ability of cancer cells to undergo apoptosis, which contributes to both tumor development and drug resistance. Despite increasing efforts to design targeted therapeutic approaches toward specific molecular signatures of cancer, nonspecific chemotherapy by DNA alkylating damage remains among the most commonly prescribed in the clinic owing to its effectiveness in a wide spectrum of cancer types. However, despite its widespread use, the molecular mechanism for the antitumor activity of DNA alkylating agents remains largely unclear.
Although apoptosis has been intensively described to account for chemotherapy-induced cancer cell death, other types of cytocidal and cytostatic mechanisms have also gained attention. These include necrosis, mitotic catastrophe, senescence, and autophagy (1–4). Among these, necrosis has clinically been reported to occur in numerous cancers after chemotherapy and radiation therapy and has been used as one of the best prognostic factors for a positive outcome (5–8). We have demonstrated both in cell culture and in tumor xenografts that necrosis plays an important role in chemotherapy-induced cancer cell death, especially in apoptosis-deficient cells (9, 10).
An important biological consequence of cell death is the activation of the immune response. A fundamental difference between apoptosis and necrosis is their influence on the immune response. Apoptosis is often characterized by an anti-inflammatory response that is essential during development and for homeostasis in adult organisms to prevent chronic inflammation and autoimmune diseases. During apoptosis, the cell membrane remains intact. Apoptotic cells present “find-me” and “eat-me” signals on their surfaces and are quickly engulfed and cleared by phagocytes or neighboring cells (11, 12). In contrast, necrotic cells undergo a rapid loss of membrane integrity that allows the release of cellular constituents into the extracellular environment. This results in a proinflammatory response that is essential in defending against viral and pathogen infection and plays a critical role to warn the host of insult (13–15).
A number of molecules have been identified that elicit immune signaling during necrosis. These include S100 family molecules, high mobility group box 1 (HMGB1) protein, purine metabolites, uric acid, and heat shock proteins. HMGB1 is one of the most abundant proteins in the nucleus whose functions include binding and bending DNA to facilitate transcription factor binding and the assembly of site-specific DNA binding proteins including p53, stabilizing nucleosomes, and facilitating DNA repair processes (16, 17). In damaged or dying cells, HMGB1 dissociates from chromatin and is released into the extracellular environment due to loss of plasma membrane integrity (18). Outside the cell, HMGB1 acts as a damage-associated molecular pattern molecule to alert the host of damage by triggering immune responses. It binds with high affinity to receptors such as TLRs (TLR2 and TLR4) and the receptor for advanced glycation end products (RAGE), which are expressed on a wide variety of cells including immune cells and endothelial cells (19–22).
HMGB1 plays a complex role in cancer. The observed over-expression of HMGB1 in tumors, particularly in conjunction with its receptor RAGE, has been associated with the proliferation and metastasis of many tumor types (21, 22). Blockade of RAGE–HMGB1 signaling has been shown to inhibit tumor growth and metastasis in both xenograft and endogenously formed tumors (23). HMGB1 has recently been found to promote autophagy, a cell survival mechanism, both intrinsically (24) or in a paracrine fashion (25). In contrast, whereas animals injected with cells killed in vitro become resistant to the formation of newly inoculated xenograft tumors, those injected with HMGB1 knockdown cells fail to become tumor resistant (26, 27). HMGB1 released by dying tumor cells can stimulate Ag presentation on dendritic cells through its interaction with TLR2 or TLR4, which subsequently activates an adaptive anticancer immune response (26, 28). The ability for HMGB1 to modulate the immune response may be affected by its redox status, as HMGB1 has been reported to be oxidized in apoptotic cells and changes its binding patterns with cell surface receptors (29, 30). Although it remains to be determined whether conventional chemotherapy induces immune-stimulating cell death in vivo, these studies point to an antitumor role for therapy-induced HMGB1 release and the activation of the adaptive immune system.
In addition to the adaptive immune response, the innate immune response has been realized to play an important role in cancer biology, although its physiological consequences remain unclear and controversial (31, 32). Chronic inflammation has been shown to promote cancer cell survival and growth (33). Spontaneous tumor necrosis has been found to correlate with accelerated tumorigenesis (34). In contrast, activation of the innate immune system has been shown to contribute to the antitumor activity induced by the restoration of p53 in p53-null tumors (35). We have previously shown that DNA alkylating therapy can induce tumor regression in athymic nude mice that lack an adaptive immune response. This tumor regression is associated with sporadic tumor cell necrosis, HMGB1 release, and infiltration of innate immune cells into the treated tumor tissue (9). It remains to be determined whether HMGB1 plays a role in the innate immune response and tumor regression during DNA alkylating therapy. In the current study, we use tumor cells derived from hmgb1−/− cells to study the role of HMGB1 in therapy-induced activation of innate immunity and tumor response.
Materials and Methods
Cell lines
Immortalized wild-type murine embryonic fibroblasts (MEFs) were generated as previously described (36). Immortalized hmgb1−/− MEFs were a kind gift from Dr. Marco Bianchi (San Raffaele Scientific Institute, Milan, Italy).
Plasmids
Retroviral expression vectors for E1A (LPC–12S) and K-Ras (pBabe–Ras) were gifts of Dr. Scott Lowe (Cold Spring Harbor, NY). The Bcl–xL construct and GFP–HMGB1 (HMGB1 cloned into the pEGFP–C1 vector) were generated in Dr. Craig Thompson’s laboratory (University of Pennsylvania). Flag-tagged HMGB1 was cloned into the LPC vector using a pEF6–Flag–HMGB1 construct as previously described (37). pBabeMN–IRES–GFP (Gary Nolan, Stanford University) was used for GFP expression. The short hairpin RNA (shRNA) constructs were obtained from Sigma (TRCN0000018934).
Cell culture and tumor cell preparation
All cell lines were retrovirally transformed with E1A and K-Ras using 293T packaging cells via a calcium phosphate precipitation method in the presence of 10 µg/ml polybrene (Sigma). When generating Bcl-xL–expressing tumors, a retroviral Bcl–xL construct was used together with E1A and K-Ras plasmids. The cells were s.c. injected into the backs of 6-to 8-wk-old male athymic nude mice (Taconic Farms). When tumors formed, they were excised from the mice, minced, and digested with trypsin-EDTA (0.05%) and collagenase A (10 µg/ml in trypsin) for 21 min at 37°C with brief vortexing every 7 min. The tumor cells were then strained through a 70-µm nylon filter to obtain a single-cell suspension. The 293T cells, MDA-MB-231 cells, and MEFs were maintained in DMEM supplemented with 10% FBS, 2 mM l-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin at 37°C with 5.5% CO2.
Immunoblotting
Cells were lysed in RIPA buffer (1% sodium deoxycholine, 0.1% SDS, 1% Triton X-100, 10 mM Tris at pH 8, 0.14 M NaCl) with protease inhibitor mixture (G-Biosciences) plus EDTA. Sample concentration was determined by bicinchoninic acid protein analysis. To determine HMGB1 release from treated tumor cells, cell culture supernatant was collected from cells at indicated treatment points, centrifuged at low speed, and stored at −20°C. Twenty micrograms protein or 20 µl culture supernatant was resolved by SDS-PAGE, transferred to a nitrocellulose membrane, blocked for 1 h at room temperature with 5% milk in PBS containing 0.05% Tween 20, and probed overnight at 4°C with primary Abs diluted 1:1000, unless otherwise noted, in the blocking solution. Blots were washed with PBS containing 0.05% Tween 20, and membranes were probed with the appropriate secondary Abs diluted in block solution for 1 h at room temperature. Primary Abs against the following molecules were used: HMGB1 (ab18256; Abcam), Flag (F3165; Sigma), Bcl-xL (556361; BD Pharmingen), β-tubulin (T4026; Sigma), IL-4 (554386, 1:100; BD Pharmingen), IL-10 (551215, 1:100; BD Pharmingen), granzyme B (AF1865, 1:100; R&D Systems), caspase 3 (C76920; Transducation Laboratories), cleaved caspase 3 (9661; Cell Signaling Technology), and ICAD (sc-9066; Santa Cruz Biotechnology). Densitometric quantification of the immunoblot bands was performed with ImageJ software (National Institutes of Health).
Measurement of cell death
Propidium iodide (PI) exclusion was used to measure cell death. After treating cells for the indicated times, cells were harvested and resuspended in DMEM with PI (1 µg/ml), and cell viability was determined by flow cytometry using a FACSCalibur (BD Biosciences). Pictures of untreated and treated cells were taken with a Zeiss Axiovert S100 microscope using a 20× objective. Images were captured using an Infinity 3 camera (Lumenera Corporation) and analyzed with Infinity Analyze software.
Xenograft mouse tumor experiments
Male nude mice, age 6–8 wk, were obtained from Taconic Farms. Mice were housed and monitored at the Division of Laboratory Animal Resources at Stony Brook University. All experimental procedures and protocols were approved by the institutional animal care and use committee. Tumors were established by resuspending 1 × 106 tumor cells in 100 µl PBS and injecting the cells into the mid-flanks of mice using a 26-gauge needle. When palpable tumors formed (~100 mm3 in size), mice were placed randomly into treatment groups. Mice were either left untreated or treated via i.p. injections of 170 mg/kg cyclophosphamide monohydrate (CP) (C0768; Sigma-Aldrich) in sterile 0.9% sodium chloride every 5 d starting at day 9 or 10 after tumor inoculation. For each tumor, the tumor length (l) and width (w) was measured every 4–5 d with an electronic caliper. Tumor volume (v) was calculated using the formula v = (l × w2)/2 and plotted in cm3. The mice were imaged using the Maestro small animal imaging system (CRi). The animals bearing untreated tumors were sacrificed around day 30 post-tumor inoculation or when tumors reached 2 cm3 in size, in accordance with regulation of the IACUC.
Depletion experiments
To deplete circulating HMGB1 in mice, an HMGB1-neutralizing Ab (38, 39) was injected three times a week for the first 2 wk, starting 2 d before the first CP treatment, and two times a week thereafter at a concentration of 50 µg/mouse/injection. For neutrophil depletion, a neutrophil-neutralizing Ab (Ly-6G; 14-5931; eBioscience) was injected i.p. at a concentration of 150 µg/mouse/injection starting with the first CP treatment and continuing for the next three CP treatments for a total of four anti-neutrophil injections. For NK cell depletion, 25 µl of an anti-NK Ab (anti-Asialo GM1; 98-1000; Wako) was diluted in 75 µl saline and injected i.v. with the first CP treatment and continued twice a week for a total of six treatments.
Immunohistochemistry
Immunohistochemistry (IHC) was performed according to standard protocol unless otherwise noted. Briefly, for paraffin-embedded sections, tissue was fixed in 10% neutral buffered formalin overnight, dehydrated in gradually increasing concentrations of ethanol, perfused in paraffin at 60°C overnight, and embedded in paraffin the next day. For cryosections, the tissue was embedded directly in OCT medium and stored at −80°C. Tissue was sectioned into 6-µm sections, and paraffin tissue was dewaxed and rehydrated in decreasing concentrations of ethanol solutions. After rehydration of paraffin tissue sections, Ag retrieval was performed using citrate buffer at pH 6. Frozen sections were warmed to room temperature for 15 min and then fixed in 4% paraformaldehyde. For both paraffin and frozen sections, endogenous peroxidase quenching was then performed using 3% H2O2, tissue was incubated with a 5% animal serum blocking solution, and the tissue was incubated with primary Ab overnight at 4°C. Abs to the following markers were used on paraffin-embedded tissue: Flag (F3165, 1:100; Sigma), Mac-2 (1:2000; Cedarlane Laboratories), neutrophil allotypic marker (MCA771G, 1:50; AbD Serotec), granzyme B (AF1865, 1:10; R&D Systems), IL-1β (AF401-NA, 2 µg/ml; R&D Systems), and cleaved caspase 3 (9661, 1:50; Cell Signaling Technology). Perforin (sc-58643, 1:500; Santa Cruz Biotechnology) was used on frozen sections. Biotinylated secondary Abs were used at a concentration of 1:1000 for 1 h at room temperature. After a wash series, tissue was incubated with avidin/biotinylated HRP (ABC Elite kit; Vector Labs) according to the manufacturer’s instructions. Slides were submerged in diaminobenzidine/H2O2 substrate solution until the desired staining intensity was obtained, and slides were counterstained with hematoxylin. For quantitation, 5–10 randomly chosen microscope fields were counted. NK cell marker NKp46 (137601, 1:37; BioLegend) and dendritic cell marker CD11c (n418; AbD Serotec) were used on paraffin-embedded sections, and IHC analysis was performed by an automated processor (Discovery XT; Ventanna, Tuscon, AZ) using a protease Ag retrieval method.
Flow cytometry
Single tumor cell suspensions were prepared as previously mentioned, and RBCs were lysed using a buffered ammonium chloride (ACK) solution (0.15 M NH4Cl, 1.0 mM KHCO3, 0.1 mM Na2EDTA, pH 7.2–7.4). After adding ACK solution, the tube was briefly vortexed and centrifuged at 1000 rpm for 5 min. Lysing of RBCs was continued until the cells were visibly clear of RBCs. The cells were washed two times with PBS. After the last wash, the microcentrifuge tube was allowed to sit at room temperature for 5 min so that the large tumor pieces would descend to the bottom of the tube. The supernatant and its constituents were collected and used for flow cytometry analysis. To confirm neutrophil and NK cell depletion, blood was collected from mice via eye bleeds, and RBCs were lysed with ACK solution, washed twice with PBS plus 5% FBS, and subjected to flow cytometry. Flow cytometry was performed using a FACSCalibur (BD Biosciences). The following Abs were used: to detect macrophages, PE-conjugated anti-mouse CD11b (Mac-1; 553311; BD Biosciences) and allophycocyanin-conjugated anti-mouse F4/80 (BM8; 17–4801; eBioscience); to detect neutrophils, PE-conjugated anti-mouse Gr-1 (553128; BD Biosciences) and Alexa Fluor 647-conjugated antimouse neutrophil (MCA771A647T; AbD Serotec); and to detect NK cells, FITC-conjugated anti-mouse NK1.1 (553164; BD Biosciences) and NKp46 (13760; BioLegend).
Cytokine array
Tumors were extracted from untreated and treated animals and minced into small pieces. Tissue lysate was made with a glass tissue homogenizer, and protein concentration was determined by bicinchoninic acid assay. A mouse cytokine array from R&D Systems (ARY006) was used to assess anti-inflammatory and proinflammatory cytokines in the tumor tissue. All procedures were performed following the manufacturer’s instructions. The array blot was developed using enhanced chemiluminescent detection reagent, and the density of each cytokine spot was analyzed using ImageJ software. The amount of cytokines in the treated tumor was normalized to the untreated tumor and graphed.
Statistical analysis
Data were analyzed with Microsoft Excel and are represented as mean ± SEM. Statistical analysis to generate p values was performed using Graph-Pad Prism software with ANOVA.
Live cell imaging for supplemental videos
hmgb1−/− MEFs were transfected with GFP-HMGB1 and treated with 100 µg/ml mafosfamide (MAF) in the presence of 0.5 µg/ml PI and 0.5 µg/ml Hoechst 33342. Live cell imaging was captured with a Zeiss LSM 510 META NLO Two-Photon Laser Scanning Confocal Microscope System using the time series function and processed using Zeiss Confocal microscope imaging software.
Results
HMGB1 plays an essential role in tumor regression in response to chemotherapy in vivo
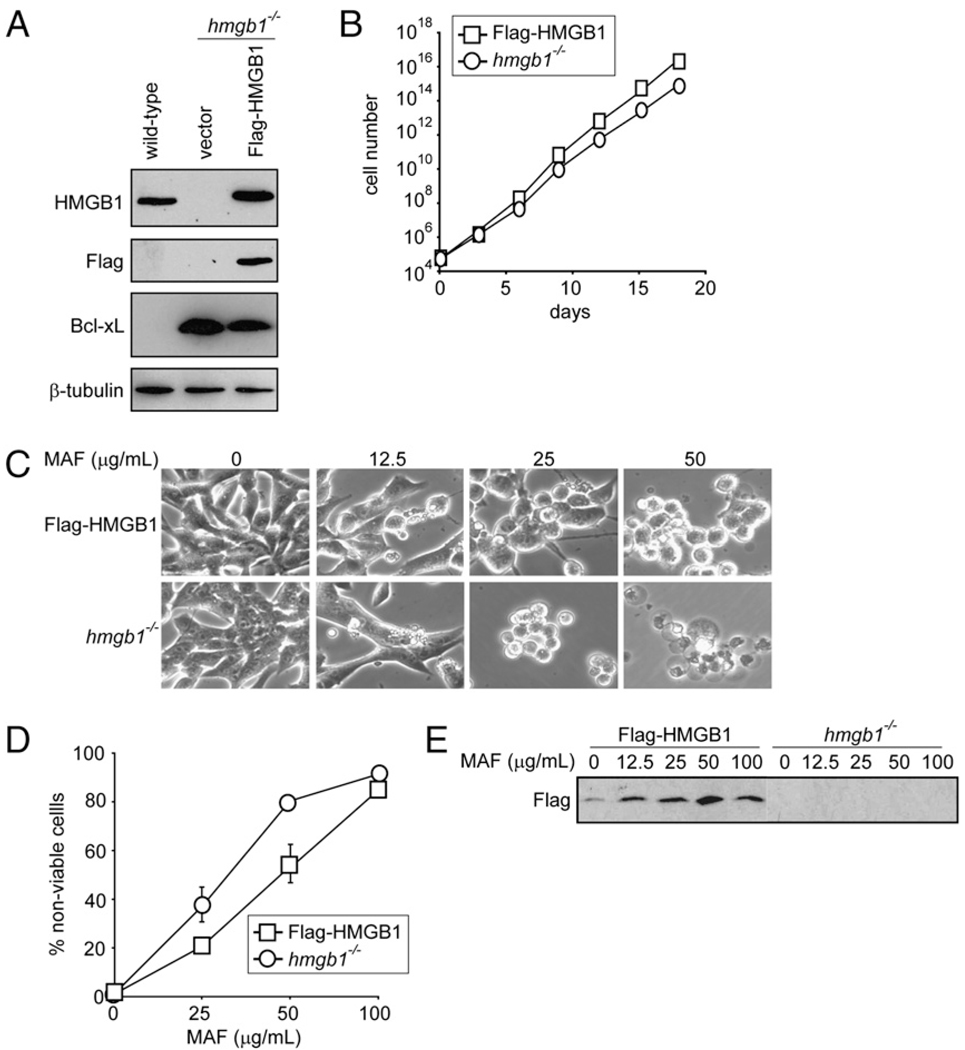
To determine the role of HMGB1 in tumor response to chemotherapy, hmgb1−/− MEFs (40) were transformed with E1A and K-Ras oncoproteins. To ensure a fair comparison of cells with an isogenic background, Flag-tagged HMGB1 (37) was introduced into the hmgb1−/− MEFs to restore HMGB1 at comparable levels to those of the wild-type cells (Fig. 1A). Bcl-xL was expressed in both the Flag-HMGB1 and hmgb1−/− cells to suppress apoptosis to mimic apoptosis-deficient human cancers (Fig. 1A). Additionally, the GFP was expressed in these cells to facilitate live tumor imaging. The cells were injected into athymic nude mice. After tumors formed, tumors were excised, and tumor cells were recovered and cultured as stable cell lines. The Flag-HMGB1 and hmgb1−/− tumor cells grew in culture at similar rates (Fig. 1B) and had comparable sensitivity to treatment with the DNA alkylating agent MAF (Fig. 1C, 1D). Upon MAF treatment, the Flag-HMGB1 tumor cells released HMGB1 into the culture media in a dose-dependent manner (Fig. 1E). Additionally, we performed time-lapse video imaging to capture the release of HMGB1 from the cell during necrosis. GFP-tagged HMGB1 was introduced into hmgb1−/− MEFs and treated with MAF in the presence of PI and a live cell permeant DNA dye, Hoechst 33342. Simultaneous decrease of cellular GFP-HMGB1 and loss of plasma membrane integrity indicated by PI-positive staining was observed (Supplemental Video 1). This is consistent with previous reports that DNA alkylating damage can induce tumor cell necrosis (9, 10) and that HMGB1 is released from necrotic cells (37, 18, 10). Hence, we have established transformed MEFs to study the role of HMGB1 during chemotherapy.
FIGURE 1.
Restoration of HMGB1 in hmgb1−/− tumor cells does not affect cell proliferation or sensitivity to chemotherapy in vitro. Wild-type and hmgb1−/− MEFs were transformed into apoptosis-deficient tumor cells using E1A, K-Ras, and Bcl-xL. Subsequently, a pool of these cells was transfected with Flag-tagged HMGB1 to restore HMGB1 in hmgb1−/− cells. A, Cell lysates were subjected to immunoblot analysis for HMGB1, Flag, Bcl-xL, and β-tubulin. B, The Flag-HMGB1 and hmgb1−/− cells were plated, and cell number was determined on the indicated days. Data shown are the average of four independent counts ± SEM (SEM is narrower than symbols so cannot be seen). C–E, Cells were treated with the DNA alkylating agent MAF at the indicated concentrations for 24 h. C, Cells were photographed under a phase-contrast filter using a 20× objective, and representative images are shown. D, Cell death was measured by PI exclusion and expressed as an average of three independent experiments ± SEM. E, Cell culture medium was collected from the untreated and treated cells and assessed for HMGB1 by immunoblot analysis using an Ab against Flag. Note that Flag-HMGB1 is found in the cell culture medium from Flag-HMGB1 but not hmgb1−/− cells.
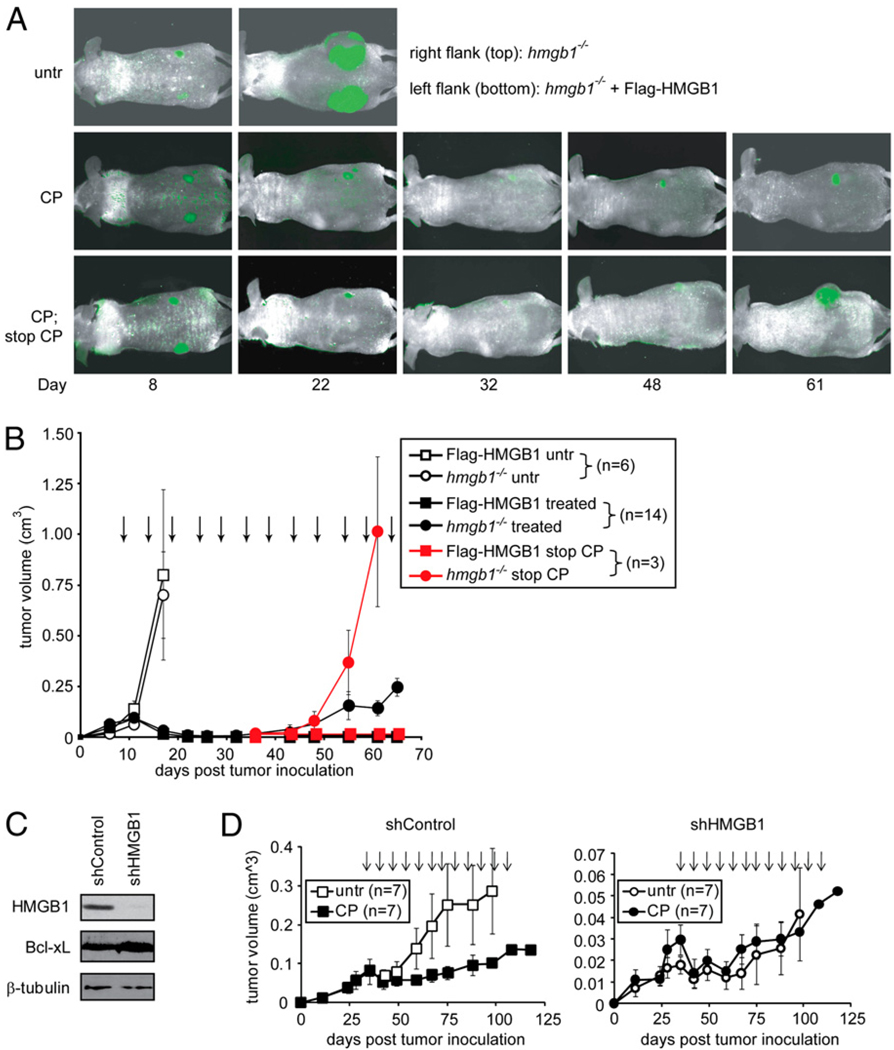
To study the effect of HMGB1 in response to chemotherapy in vivo, Flag-HMGB1 and hmgb1−/− tumor cells were injected bilaterally into the left flank and right flank, respectively, of 20 athymic nude mice. Both cell lines formed tumors that grew at similar rates (Fig. 2A, 2B). Fourteen of these animals were then compared side-by-side for their susceptibility to CP, a DNA alkylating agent that is commonly prescribed in the clinic. All 14 tumors with restored HMGB1 showed a virtually complete regression in response to CP treatment by day 22, as they were no longer detectable by either caliper measurement or fluorescence imaging (Fig. 2A, 2B). In sharp contrast, although hmgb1−/− tumors initially responded to treatment, all 14 tumors failed to regress fully. Instead, around day 30 these tumors began to increase in size. Furthermore, at day 35, 3 of the 14 mice were taken off of CP. Whereas the tumors with Flag-HMGB1 remained undetectable, the hmgb1−/− tumors recurred to form large tumors (Fig. 2A, bottom panel, and Fig. 2B, red line).
FIGURE 2.
CP-induced tumor regression in vivo is dependent on HMGB1. A and B, Wild-type and hmgb1−/− MEFs were transformed into apoptosis-deficient tumor cells using E1A, K-Ras, and Bcl-xL. Subsequently, a pool of these cells was transfected with Flag-tagged HMGB1 to restore HMGB1 in hmgb1−/− cells. Both Flag-HMGB1 and hmgb1−/− tumor cells were infected with GFP and injected bilaterally into the same mouse to allow tumors to form. When palpable tumors formed, the mice were left untreated or treated with 170 mg/kg CP every 5 d as indicated by arrows. Thirty days after the initial CP treatment, CP was discontinued on three mice (red symbols). A, Tumors were monitored by in vivo imaging, and representative images are shown. B, Tumor volume was determined by caliper measurement, and the average tumor volume was plotted with SEM. C, The antiapoptotic protein Bcl-xL was expressed in MDA-MB-231 human breast cancer cell line. A pool of these cells was lentivirally infected with an shRNA vector control or shRNA against HMGB1. Cell lysates were subjected to immunoblot for HMGB1, Bcl-xL, and β-tubulin. D, Fourteen athymic nude mice were injected with the MDA-MB-231 cells with shRNA vector control or HMGB1 knockdown. When tumors formed, mice were divided into two groups: untreated and those treated every 5 d with 170 mg/kg CP as indicated by the arrows. Tumor volume was determined by caliper measurement, and the average tumor volume was plotted with SEM.
To determine whether the dependence of HMGB1 for tumor regression exists in other tumor systems, we used a human breast cancer cell line, MDA-MB-231. Bcl-xL was expressed in MDAMB-231 cells, and subsequently HMGB1 expression was suppressed by shRNA knock-down (Fig. 2C). We compared the response to CP in HMGB1 knock-down cells versus parental cells using the mouse xenograft model. Whereas CP showed potent antitumor activity in mice grafted with parental tumor cells, the tumors with HMGB1 knock-down were significantly more resistant to CP treatment (Fig. 2D). It was noted that HMGB1 knock-down slowed tumor growth in vivo, and the tumor regression in CP-treated parental tumors was not as complete as that of the MEF tumors. These discrepancies may be due to the different tumor cell systems and/or due to that HMGB1 may epigenetically promote cell survival and proliferation in certain cancers (17, 41, 42). Nevertheless, these results, together with the MEF fibrosarcoma data (Fig. 2A, 2B), indicate that HMGB1 plays an essential role in tumor regression induced by DNA alkylating therapy.
HMGB1 released from chemotherapy-induced necrotic tumor cells is a chemoattractant
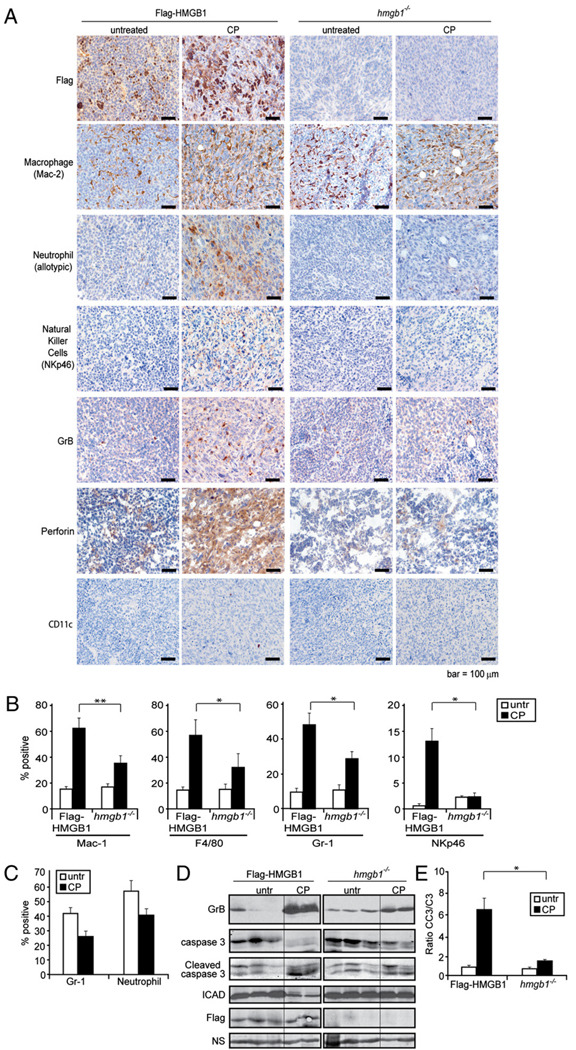
We have previously shown that CP treatment induces sporadic tumor cell necrosis and HMGB1 extracellular release in vivo (9). To determine if Flag-HMGB1 changed subcellular localization in response to CP treatment, tumors were extracted from untreated and CP-treated mice and subjected to IHC using an Ab against Flag. The hmgb1−/− tumors showed no positive staining for HMGB1. In tumors with restored HMGB1, the untreated tumor cells showed nuclear HMGB1 localization and the CP-treated tumors showed extracellular localization, indicating Flag-HMGB1 is released into the extracellular environment in response to CP treatment (Fig. 3A).
FIGURE 3.
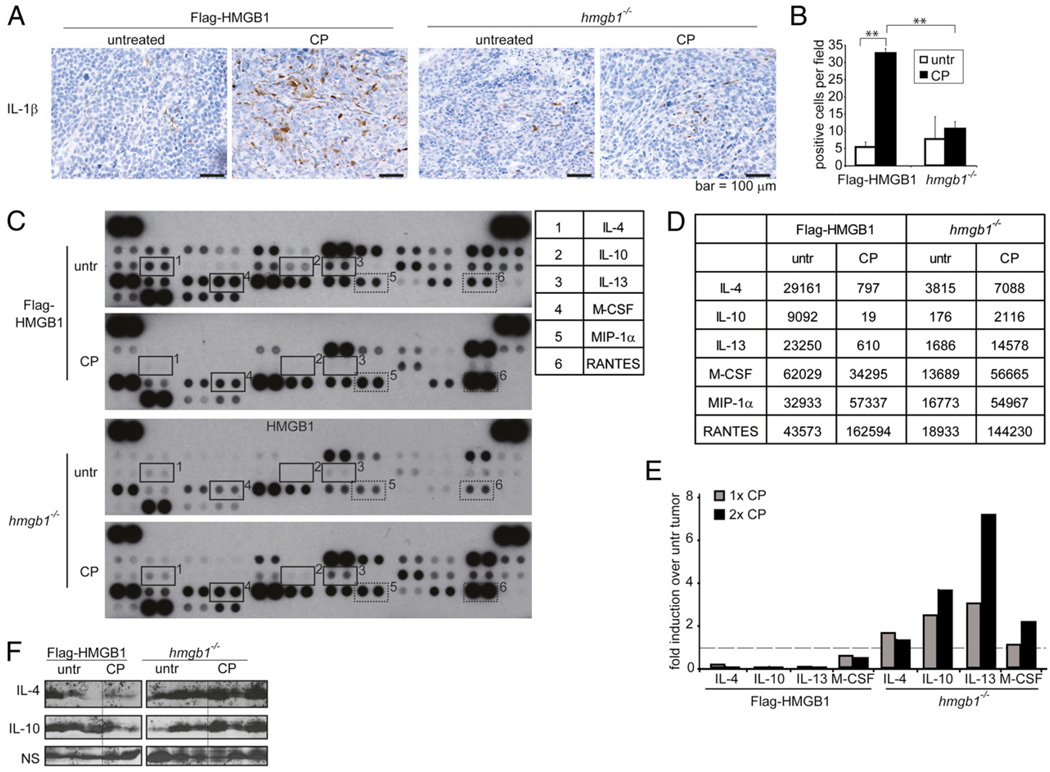
HMGB1 induces innate immune cell infiltration after chemotherapy. Mice bearing Flag-HMGB1 or hmgb1−/− tumors were either left untreated or treated twice with i.p. injections with 170 mg/kg CP. A, Tumors were excised and embedded in paraffin, sectioned, and subjected to IHC. Cellular localization of HMGB1 was assessed using a Flag Ab, and infiltration of macrophages, neutrophils, NK cells, and dendritic cells was assessed using Abs against Mac-2, an allotypic marker of neutrophils, NKp46, and CD11c, respectively. Activation of NK cells was examined using Abs against GrB and perforin. Scale bars, 100 µm. B, To quantitate the number of infiltrating macrophages and neutrophils, single-cell suspensions were prepared from untreated and treated tumor tissues and subjected to flow cytometry using anti-CD11b and anti-F4/80 Abs for macrophages, an anti–Gr-1 Ab for neutrophils, and an anti-NKp46 Ab for NK cells. The average from three independent experiments with two to three tumors in each group is shown. *p < 0.05, **p < 0.005. C, To determine if CP treatment results in decreased number of peripheral neutrophils, peripheral blood was collected and subjected to flow cytometry analysis using anti– Gr-1 or the allotypic anti-neutrophil Abs. Percentage of the positive cells is plotted. Data shown are the average of two to three mice per group. D, Immunoblot analysis was performed using the tumor lysates for GrB, caspase 3, cleaved caspase 3, ICAD, and Flag on three independent Flag-HMGB1 and hmgb1−/− untreated tumors and two independent Flag-HMGB1 and hmgb1−/− CP-treated tumors. E, Densitometry of the caspase 3 and cleaved caspase 3 immunoblot bands was determined by ImageJ software, and the ratio of activated caspase 3 (CC3) to full-length caspase 3 (C3) was plotted. *p < 0.05.
A prominent hallmark associated with HMGB1 released from damaged tissue or from immune cells in response to endotoxin exposure is leukocyte recruitment (41). As HMGB1 was released upon CP treatment, we next determined if leukocytes were recruited to CP-treated tumors. Similar to previous observations in wild-type tumors with endogenous HMGB1 (9), an increase in macrophage infiltration was observed in CP-treated Flag-HMGB1 tumors, whereas the increased infiltration of macrophages upon CP treatment was significantly lower in hmgb1−/− tumors shown by both IHC using an Ab against Mac-2 (Fig. 3A) and flow cytometry using Abs against CD11b and F4/80 (Fig. 3B). Strikingly, Flag-HMGB1 but not hmgb1−/− tumors had a significant infiltration of neutrophils detected by both IHC using an Ab against an allotypic marker of neutrophils (Fig. 3A) and by flow cytometry using an Ab against Gr-1 (Fig. 3B), despite a reduced number of neutrophils in the peripheral blood upon CP treatment as previously reported (Fig. 3C) (43). Similar to neutrophils, an anti-NK cell Ab (NKp46) revealed that whereas CP treatment induced a drastic infiltration of NK cells, the levels were markedly reduced in the CP-treated hmgb1−/− tumors compared with those in the HMGB1 restored tumors shown by both IHC (Fig. 3A) and flow cytometry (Fig. 3B). No obvious infiltration of dendritic cells was observed (Fig. 3A).
Granzymes, a molecular marker for activated NK cells, mediate NK cell killing activities via their delivery into target cells through perforin (44). The presence of granzyme B (GrB) and perforin was assessed by IHC. Upon CP treatment, there was a significant increase of both GrB and perforin in the Flag-HMGB1 tumors, whereas both markers were virtually absent in hmgb1−/− tumors (Fig. 3A, 3D). The enhanced GrB and perforin levels were associated with increased caspase 3 cleavage (Fig. 3D, 3E), which has been shown to be caused by proteolytic cleavage by GrB, independent of the mitochondrial apoptosis pathway (44, 45). Additionally, full-length ICAD, a proteolytic target of caspase 3 and GrB (46, 47), was significantly lower in the Flag-HMGB1 tumors compared with that in the hmgb1−/− tumors upon CP treatment (Fig. 3D). Taken together, these results indicate that HMGB1 released during chemotherapy can act as a chemo-attractant to activate the innate immune system including macrophages, neutrophils, and NK cells and that granzyme-mediated cell killing may be a mechanism for tumor clearance.
Activation of innate immunity plays an antitumor role in chemotherapy
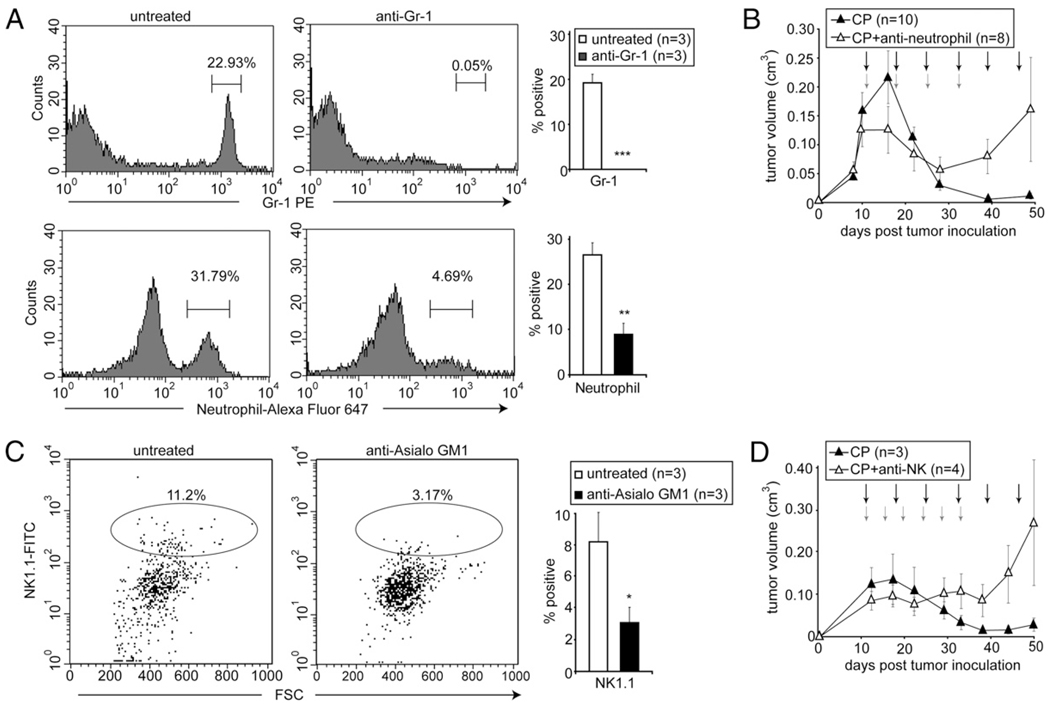
It has been shown that the activation of innate immunity contributes to tumor regression mediated by activation of p53 (35). We have previously shown that macrophages are required for complete tumor regression in response to CP treatment (9). As compromised infiltration of neutrophils and NK cells was observed in hmgb1−/− CP-treated tumors (Fig. 3A), we tested whether these innate immune cells also play an antitumor role in chemotherapy. An antineutrophil Ab (Ly-6G) and an anti-NK cell Ab (anti-Asialo GM1) were used to deplete neutrophils and NK cells, respectively, in athymic nude mice bearing apoptosis-deficient E1A/K-Ras–transformed tumors with endogenous HMGB1. Successful depletion was achieved, as indicated by lower counts of neutrophils and NK cells in peripheral blood (Fig. 4A, 4C). For neutrophil depletion, animals bearing tumors were treated with CP alone or CP in combination with i.p. injections of the neutrophil-depleting Ab. Whereas tumors in mice receiving CP alone showed complete regression by day 40, those in mice receiving CP in combination with the neutrophil-depleting Ab became resistant to the treatment despite an initial regression (Fig. 4B). For NK cell depletion, tumor-bearing mice were treated with CP alone or CP plus i.v. injections of the NK cell-depleting Ab. Similar to the neutrophil and macrophage depletion studies, mice with NK cell depletion could not achieve complete tumor regression, and tumors continued to grow after an initial delay (Fig. 4D).
FIGURE 4.
Depletion of neutrophils or NK cells leads to tumor resistance to chemotherapy. Mice bearing tumors with endogenous HMGB1 were treated by i.p. injections with 170 mg/kg CP every 5 d (black arrows) alone, or in combination with (A, B) i.p. injections with 150 µg of the anti–Gr-1 neutrophil-depleting Ab in combination with the first four CP treatments (red arrows), or (C, D) i.v. injections with 25 µl of the NK cell-depleting Ab every 3 d (red arrows). A and C, Peripheral blood was collected and analyzed by flow cytometry using indicated Abs. Representative histograms for neutrophils (A) or dot plot for NK cells (C) are shown. Shown on the right are the averages from three animals in each group + SEM. *p < 0.05, **p < 0.005, ***p < 0.0005. B and D, Tumors were measured by caliper, and the average volume of tumors with SEM was plotted.
HMGB1 suppresses an M2 immune response
Circulating immature monocytes are attracted by tumor-derived chemokines to migrate from the blood into the tumor tissue and differentiate into macrophages. Once inside the tumor, differentiation and polarization of the monocytes into activated (M1) or alternatively activated (M2) cells depend on the cytokines present in the tumor microenvironment (48). The M2 type cytokines have been shown to facilitate tissue repair and remodeling, and tumor progression therefore is characteristic of the tumor-associated macrophages, whereas the M1 type is usually associated with antitumor activity (49, 50). Therefore, switching tumor-associated macrophages from M2 to M1 may have significant therapeutic value. Because macrophage infiltration was enhanced in both Flag-HMGB1 and hmgb1−/− tumors upon CP treatment (Fig. 3A), and hmgb1−/− tumors failed to activate an antitumor immune response (Fig. 2), we determined whether HMGB1 could influence M1/M2 polarization. Indeed, IHC revealed that production of the proinflammatory cytokine IL-1β, an M1 cytokine, was stimulated by CP treatment in Flag-HMGB1 tumors but not in hmgb1−/− tumors (Fig. 5A, 5B). Notably, our cytokine array analysis revealed that in untreated tumors, the overall cytokine production is higher in Flag-HMGB1 tumors, consistent with the general function of HMGB1 as a transcriptional activator. However, upon CP treatment, although the production of certain cytokines such as MIP-1α and RANTES was elevated in both Flag-HMGB1 and hmgb1−/− tumors (Fig. 5C, 5D), levels of the M2-type immune-tolerant cytokines such as IL-4, IL-10, and IL-13 were enhanced in CP-treated hmgb1−/− tumors but decreased in Flag-HMGB1 tumors (Fig. 5C–F). Correlatively, the level of M-CSF, a well-established cytokine that drives macrophage M2 differentiation (51), was also enhanced in CP-treated hmgb1−/− tumors but decreased in Flag-HMGB1 tumors (Fig. 5C–E). These results indicate that the released HMGB1 induced by chemotherapy can modulate an antitumor immune response. This is consistent with previous reports showing that HMGB1 can activate innate immune cells to produce a number of proinflammatory cytokines but not anti-inflammatory cytokines (52, 53). These results indicate that HMGB1 may facilitate tumor regression in response to chemotherapy by suppressing the production of the protumor M2 cytokines in the tumor tissue.
FIGURE 5.
HMGB1 released during chemotherapy treatment mediates a proinflammatory response. Mice bearing Flag-HMGB1 or hmgb1−/− tumors were either left untreated or treated with i.p. injections of 170 mg/kg CP 5 d apart. A, Tumor tissue sections were subjected to IHC using an anti–IL-1β Ab. Scale bars, 100 µm. B, Ten randomly selected microscope fields were counted for IL-1β+ cells, and the average is shown on the right. **p < 0.005. C–F, Tumors were excised and made into lysates. The levels of cytokines were assessed with a mouse cytokine array panel in lysates from tumors in mice left untreated or treated one time or two times with CP. C and D, The (C) cytokine array blots and (D) raw OD readings using ImageJ are shown for selected cytokines. Numbered rectangles highlight the duplicate spots of anti-inflammatory cytokines (solid lines) and proinflammatory cytokines (dashed lines) that are altered by the status of HMGB1 presence, as summarized in the table. E, Results are shown as OD normalized to values for each cytokine in untreated tumor tissue. The dashed line represents the untreated tumor value set to one. F, Immunoblotting of IL-4 and IL-10 was used to confirm the cytokine array results in tumor cell lysates. Three independent Flag-HMGB1 and hmgb1−/− untreated tumors, three independent hmgb1−/− CP-treated tumors, and two Flag-HMGB1 CP-treated tumors were used.
HMGB1 is essential for activation of innate immunity and tumor regression
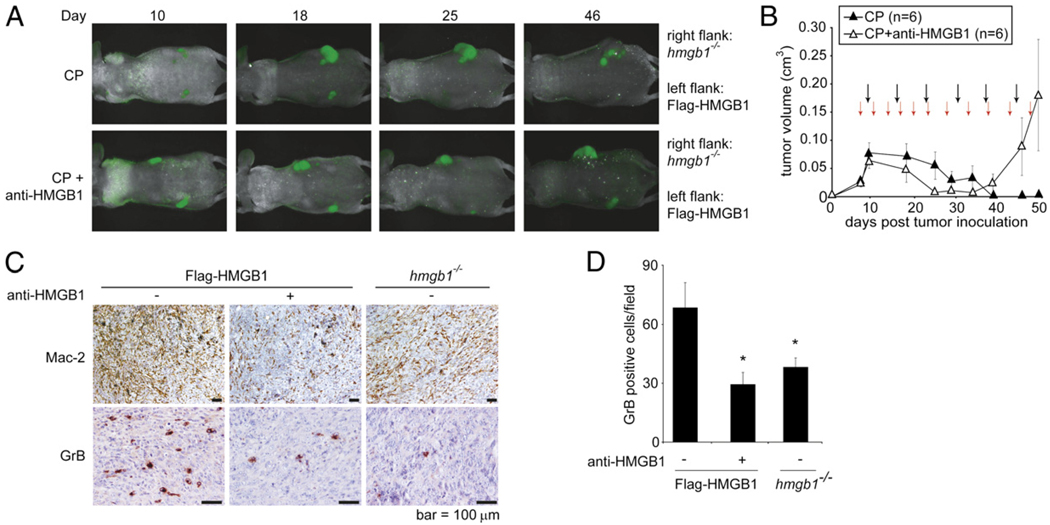
To confirm the importance of HMGB1 during chemotherapyinduced activation of the innate immune system and subsequent tumor regression, we used an HMGB1-neutralizing Ab (38, 39) in our tumor study. Flag-HMGB1 and hmgb1−/− tumor cells were injected bilaterally into the left and right flank of athymic nude mice, respectively. When tumors formed, mice were divided into two groups. One group received CP treatment alone, and the other received both CP and the HMGB1-neutralizing Ab. As anticipated, hmgb1−/− tumors failed to regress in response to both treatment regimens (Fig. 6A). The Flag-HMGB1 tumors initially responded to both treatments. However, the tumors in mice receiving CP in combination with the HMGB1-neutralizing Ab started to grow around day 35 (Fig. 6A, 6B). IHC analysis showed that the HMGB1-neutralizing Ab suppressed infiltration of Mac-2+ macrophages and GrB+ NK cells in CP-treated Flag-HMGB1 tumors to similar levels as in hmgb1−/− tumors (Fig. 6C, 6D). Taken together, these results demonstrate that HMGB1 plays an essential role in activating innate immunity and tumor regression during chemotherapy.
FIGURE 6.
Depletion of HMGB1 leads to tumor resistance and failure of innate immunity activation. A–D, Mice bearing Flag-HMGB1 or hmgb1−/− tumors were treated with i.p. injection of 170 mg/kg CP every 5 d (black arrows) alone or together with 50 mg of an HMGB1-neutralizing Ab twice a week (red arrows). A, Tumors were monitored by in vivo imaging, and representative images of the animals from each group are shown. B, Mice bearing tumors with endogenous HMGB1 were treated with CP alone or together with anti-HMGB1. Tumors were measured by caliper, and the average tumor volume was plotted with SEM. C, CP-treated tumors were excised and embedded in paraffin. Sections were subjected to IHC using an Ab against Mac-2 to assess the level of macrophage infiltration and an Ab against GrB to assess the level of activated NK cells. Note that similar to hmgb1−/− tumors, CP-treated tumors subjected to HMGB1 depletion (anti-HMGB1) had reduced levels of macrophage and activated NK cell infiltration compared with those of the Flag-HMGB1 CP-treated tumors. Scale bars, 100 µm. D, Ten randomly selected microscope fields were counted for GrB+ cells. Data shown are average + SEM. *p < 0.05.
Discussion
Although chemotherapy has long been known to induce cancer cell death, it is becoming appreciated that tumor response to chemotherapy is not simply cytotoxic or cytostatic (2). Both innate and adaptive immune responses can be activated by damaged or dying cells, pointing to an important question as to whether the immune response also plays a significant role in therapy. We have previously found that apoptosis is dispensable for tumor regression in response to DNA alkylating treatment. Alternative cell death pathways including necrosis, senescence, and autophagy have been implicated in cancer therapy. These cytotoxic/stress responses have been shown to cause release/secretion of intracellular contents that may contribute to immune stimulation. HMGB1 has been shown to be one of the molecules that can be induced to release via necrosis and autophagy (9, 27, 18, 24, 54). We find here that although tumor cells deficient in HMGB1 do not have survival advantages in response to DNA alkylating treatment in cell culture, they are resistant to chemotherapy in vivo. This resistance correlates with impaired innate immunity, indicated by lower levels of macrophage, neutrophil, and NK cell infiltration into the HMGB1-deficient tumor tissues upon treatment.
Our findings point to an important role of the innate immune response in therapy-induced tumor clearance. It has been shown that cells killed by chemotherapeutic agents in vitro can protect animals against newly inoculated tumors via Ag cross-presentation to activate cytolytic T cells in an HMGB1–TLR4–dependent manner (26). Here, by using immune-compromised athymic nude mice, we show that the innate immune response is important in tumor clearance in response to chemotherapy. This is consistent with a previous report showing that innate immunity is essential for tumor clearance in a p53 restoration mouse model (35). As we observed that depleting each of the macrophage, neutrophil, and NK cell populations led to defective tumor regression, it remains to be determined whether these innate immune cell populations function in a linear fashion or in concert to mediate the antitumor activity. Nevertheless, in clinical practice, CP has been shown to affect both the T cell and B cell compartments and to block Ab production (55, 56). Our findings provide an explanation for why CP, which is often used as an immune suppressant, works efficiently in cancer treatment by blocking adaptive immunity and allowing innate immunity to predominate.
The mechanism by which the innate immune system kills cancer cells remains to be determined. Our leukocyte depletion results show that neutrophils and NK cells play an antitumor role (Fig. 4), similar to macrophages as shown previously (9, 35). Consistent with the impaired NK cell recruitment, markedly lower levels of perforin and GrB were detected in hmgb1−/− tumors, suggesting that perforin/granzyme-mediated cytotoxicity may contribute to tumor cell death. This can also explain why tumor cells deficient in mitochondrial apoptosis are susceptible to alkylating therapy, as perforin and granzymes are known to mediate cell death independent of the mitochondrial apoptotic pathway (44, 45).
Our results also suggest that HMGB1 may contribute to tumor regression by regulating monocyte polarization. Macrophages have been shown to facilitate both cell death and tumor growth. Macrophages that mediate these opposing functions have been classified as M1 (antitumor) and M2 (protumor). Whereas M1 macrophages are characterized by classic cytotoxic cytokines, the protumor M2 macrophages are classified by elevated production of cytokines such as IL-4, IL-10, and IL-13. Notably, our cytokine array analysis revealed that upon CP treatment, the levels of the M2 cytokines IL-4, IL-10, and IL-13 are higher in hmgb1−/− tumors but are markedly suppressed in the presence of HMGB1 (Fig. 5C–F). The elevated levels of M2 cytokines correlate with the tumor regrowth observed in hmgb1−/− tumors (Fig. 2A, 2B). Hence, HMGB1 released by dying tumor cells may suppress M2 polarization of macrophages and prevent tumor regrowth and resistance to treatment. Modalities aimed at inducing tumor cell necrosis may likely result in final eradication of tumors as a consequence of nonspecific activation of the innate immune system. Although elevated levels of HMGB1 have been detected in various types of cancers (21, 22), boosting the innate immune response by inducing necrotic release of HMGB1 may be a promising approach for adjuvant treatment of cancer.
Supplementary Material
Acknowledgments
We thank Dr. Kevin Tracey for the HMGB1-depleting Ab, Dr. Myriam Malet-Martino for providing MAF, and Dr. Marco Bianchi for providing hmgb1−/− MEFs. We thank Drs. Scott Lowe, Jim Bliska, and Jorge Benach for providing reagents and suggestions. We thank Dr. Guowei Tian for help with live cell imaging and Dr. Christopher Czura and David Habiel for technical assistance. We also thank Drs. Kevin Tracey, Ute Moll, Nancy Reich, and Howard Fleit for discussion.
R.K. was supported by the National Institutes of Health (GM063769). J.M.C. was supported by National Institutes of Health Training Grants T32GM00796427 and T32CA009176. H.C.C. was supported by a Veterans Administration merit award. W.-X.Z. was supported by the National Cancer Institute (CA129536 and CA098092), Susan Komen for the Cure (KG081538), and the Carol Baldwin Breast Cancer Research Foundation.
Abbreviations used in this article
- CP
cyclophosphamide monohydrate
- GrB
granzyme B
- HMGB1
high mobility group box 1
- IHC
immunohistochemistry
- MAF
mafosfamide
- MEF
murine embryonic fibroblast
- PI
propidium iodide
- RAGE
receptor for advanced glycation end products
- shRNA
short hairpin RNA
Footnotes
The online version of this article contains supplemental material.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Brown JM, Attardi LD. The role of apoptosis in cancer development and treatment response. Nat. Rev. Cancer. 2005;5:231–237. doi: 10.1038/nrc1560. [DOI] [PubMed] [Google Scholar]
- 2.Havelka AM, Berndtsson M, Olofsson MH, Shoshan MC, Linder S. Mechanisms of action of DNA-damaging anticancer drugs in treatment of carcinomas: is acute apoptosis an “off-target” effect? Mini Rev. Med. Chem. 2007;7:1035–1039. doi: 10.2174/138955707782110196. [DOI] [PubMed] [Google Scholar]
- 3.Melino G, Knight RA, Nicotera P. How many ways to die? How many different models of cell death? Cell Death Differ. 2005;12(Suppl. 2):1457–1462. doi: 10.1038/sj.cdd.4401781. [DOI] [PubMed] [Google Scholar]
- 4.Okada H, Mak TW. Pathways of apoptotic and non-apoptotic death in tumour cells. Nat. Rev. Cancer. 2004;4:592–603. doi: 10.1038/nrc1412. [DOI] [PubMed] [Google Scholar]
- 5.Bacci G, Mercuri M, Longhi A, Ferrari S, Bertoni F, Versari M, Picci P. Grade of chemotherapy-induced necrosis as a predictor of local and systemic control in 881 patients with non-metastatic osteosarcoma of the extremities treated with neoadjuvant chemotherapy in a single institution. Eur. J. Cancer. 2005;41:2079–2085. doi: 10.1016/j.ejca.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 6.Delanian S, Lefaix JL. Current management for late normal tissue injury: radiation-induced fibrosis and necrosis. Semin. Radiat. Oncol. 2007;17:99–107. doi: 10.1016/j.semradonc.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Linder S, Havelka AM, Ueno T, Shoshan MC. Determining tumor apoptosis and necrosis in patient serum using cytokeratin 18 as a biomarker. Cancer Lett. 2004;214:1–9. doi: 10.1016/j.canlet.2004.06.032. [DOI] [PubMed] [Google Scholar]
- 8.Raymond AK, Chawla SP, Carrasco CH, Ayala AG, Fanning CV, Grice B, Armen T, Plager C, Papadopoulos NE, Edeiken J, et al. Osteosarcoma chemotherapy effect: a prognostic factor. Semin. Diagn. Pathol. 1987;4:212–236. [PubMed] [Google Scholar]
- 9.Guerriero JL, Ditsworth D, Fan Y, Zhao F, Crawford HC, Zong WX. Chemotherapy induces tumor clearance independent of apoptosis. Cancer Res. 2008;68:9595–9600. doi: 10.1158/0008-5472.CAN-08-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zong WX, Ditsworth D, Bauer DE, Wang ZQ, Thompson CB. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes Dev. 2004;18:1272–1282. doi: 10.1101/gad.1199904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lauber K, Blumenthal SG, Waibel M, Wesselborg S. Clearance of apoptotic cells: getting rid of the corpses. Mol. Cell. 2004;14:277–287. doi: 10.1016/s1097-2765(04)00237-0. [DOI] [PubMed] [Google Scholar]
- 12.Nagata S, Hanayama R, Kawane K. Autoimmunity and the clearance of dead cells. Cell. 2010;140:619–630. doi: 10.1016/j.cell.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 13.Fadok VA, Bratton DL, Rose DM, Pearson A, Ezekewitz RA, Henson PM. A receptor for phosphatidylserine-specific clearance of apoptotic cells. Nature. 2000;405:85–90. doi: 10.1038/35011084. [DOI] [PubMed] [Google Scholar]
- 14.Golstein P, Kroemer G. Cell death by necrosis: towards a molecular definition. Trends Biochem. Sci. 2007;32:37–43. doi: 10.1016/j.tibs.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 15.Zong WX, Thompson CB. Necrotic death as a cell fate. Genes Dev. 2006;20:1–15. doi: 10.1101/gad.1376506. [DOI] [PubMed] [Google Scholar]
- 16.Bianchi ME, Manfredi AA. High-mobility group box 1 (HMGB1) protein at the crossroads between innate and adaptive immunity. Immunol. Rev. 2007;220:35–46. doi: 10.1111/j.1600-065X.2007.00574.x. [DOI] [PubMed] [Google Scholar]
- 17.Ellerman JE, Brown CK, de Vera M, Zeh HJ, Billiar T, Rubartelli A, Lotze MT. Masquerader: high mobility group box-1 and cancer. Clin. Cancer Res. 2007;13:2836–2848. doi: 10.1158/1078-0432.CCR-06-1953. [DOI] [PubMed] [Google Scholar]
- 18.Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418:191–195. doi: 10.1038/nature00858. [DOI] [PubMed] [Google Scholar]
- 19.Andersson UG, Tracey KJ. HMGB1, a pro-inflammatory cytokine of clinical interest: introduction. J. Intern. Med. 2004;255:318–319. doi: 10.1111/j.1365-2796.2003.01304.x. [DOI] [PubMed] [Google Scholar]
- 20.Palumbo R, Bianchi ME. High mobility group box 1 protein, a cue for stem cell recruitment. Biochem. Pharmacol. 2004;68:1165–1170. doi: 10.1016/j.bcp.2004.03.048. [DOI] [PubMed] [Google Scholar]
- 21.Sims GP, Rowe DC, Rietdijk ST, Herbst R, Coyle AJ. HMGB1 and RAGE in inflammation and cancer. Annu. Rev. Immunol. 2010;28:367–388. doi: 10.1146/annurev.immunol.021908.132603. [DOI] [PubMed] [Google Scholar]
- 22.Tang D, Kang R, Zeh HJ, III, Lotze MT. High-mobility group box 1 and cancer. Biochim. Biophys. Acta. 2010;1799:131–140. doi: 10.1016/j.bbagrm.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taguchi A, Blood DC, del Toro G, Canet A, Lee DC, Qu W, Tanji N, Lu Y, Lalla E, Fu C, et al. Blockade of RAGE-amphoterin signalling suppresses tumour growth and metastases. Nature. 2000;405:354–360. doi: 10.1038/35012626. [DOI] [PubMed] [Google Scholar]
- 24.Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, Loughran P, Hoppe G, Bianchi ME, Tracey KJ, Zeh HJ, III, Lotze MT. Endogenous HMGB1 regulates autophagy. J. Cell Biol. 2010;190:881–892. doi: 10.1083/jcb.200911078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu L, Yang M, Kang R, Wang Z, Zhao Y, Yu Y, Xie M, Yin X, Livesey KM, Lotze MT, et al. HMGB1-induced autophagy promotes chemotherapy resistance in leukemia cells. Leukemia. 2011;25:23–31. doi: 10.1038/leu.2010.225. [DOI] [PubMed] [Google Scholar]
- 26.Apetoh L, Ghiringhelli F, Tesniere A, Obeid M, Ortiz C, Criollo A, Mignot G, Maiuri MC, Ullrich E, Saulnier P, et al. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat. Med. 2007;13:1050–1059. doi: 10.1038/nm1622. [DOI] [PubMed] [Google Scholar]
- 27.Rovere-Querini P, Capobianco A, Scaffidi P, Valentinis B, Catalanotti F, Giazzon M, Dumitriu IE, Müller S, Iannacone M, Traversari C, et al. HMGB1 is an endogenous immune adjuvant released by necrotic cells. EMBO Rep. 2004;5:825–830. doi: 10.1038/sj.embor.7400205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Curtin JF, Liu N, Candolfi M, Xiong W, Assi H, Yagiz K, Edwards MR, Michelsen KS, Kroeger KM, Liu C, et al. HMGB1 mediates endogenous TLR2 activation and brain tumor regression. PLoS Med. 2009;6:e10. doi: 10.1371/journal.pmed.1000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kazama H, Ricci JE, Herndon JM, Hoppe G, Green DR, Ferguson TA. Induction of immunological tolerance by apoptotic cells requires caspase-dependent oxidation of high-mobility group box-1 protein. Immunity. 2008;29:21–32. doi: 10.1016/j.immuni.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang D, Kang R, Cheh CW, Livesey KM, Liang X, Schapiro NE, Benschop R, Sparvero LJ, Amoscato AA, Tracey KJ, et al. HMGB1 release and redox regulates autophagy and apoptosis in cancer cells. Oncogene. 2010;29:5299–5310. doi: 10.1038/onc.2010.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zitvogel L, Apetoh L, Ghiringhelli F, Kroemer G. Immunological aspects of cancer chemotherapy. Nat. Rev. Immunol. 2008;8:59–73. doi: 10.1038/nri2216. [DOI] [PubMed] [Google Scholar]
- 33.Vakkila J, Lotze MT. Inflammation and necrosis promote tumour growth. Nat. Rev. Immunol. 2004;4:641–648. doi: 10.1038/nri1415. [DOI] [PubMed] [Google Scholar]
- 34.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gélinas C, Fan Y, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zong WX, Lindsten T, Ross AJ, MacGregor GR, Thompson CB. BH3-only proteins that bind pro-survival Bcl-2 family members fail to induce apoptosis in the absence of Bax and Bak. Genes Dev. 2001;15:1481–1486. doi: 10.1101/gad.897601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ditsworth D, Zong WX, Thompson CB. Activation of poly (ADP)-ribose polymerase (PARP-1) induces release of the pro-inflammatory mediator HMGB1 from the nucleus. J. Biol. Chem. 2007;282:17845–17854. doi: 10.1074/jbc.M701465200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H, Bloom O, Zhang M, Vishnubhakat JM, Ombrellino M, Che J, Frazier A, Yang H, Ivanova S, Borovikova L, et al. HMG-1 as a late mediator of endotoxin lethality in mice. Science. 1999;285:248–251. doi: 10.1126/science.285.5425.248. [DOI] [PubMed] [Google Scholar]
- 39.Yang H, Ochani M, Li J, Qiang X, Tanovic M, Harris HE, Susarla SM, Ulloa L, Wang H, DiRaimo R, et al. Reversing established sepsis with antagonists of endogenous high-mobility group box 1. Proc. Natl. Acad. Sci. USA. 2004;101:296–301. doi: 10.1073/pnas.2434651100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Calogero S, Grassi F, Aguzzi A, Voigtländer T, Ferrier P, Ferrari S, Bianchi ME. The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat. Genet. 1999;22:276–280. doi: 10.1038/10338. [DOI] [PubMed] [Google Scholar]
- 41.Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat. Rev. Immunol. 2005;5:331–342. doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- 42.Völp K, Brezniceanu ML, Bösser S, Brabletz T, Kirchner T, Göttel D, Joos S, Zörnig M. Increased expression of high mobility group box 1 (HMGB1) is associated with an elevated level of the antiapoptotic c-IAP2 protein in human colon carcinomas. Gut. 2006;55:234–242. doi: 10.1136/gut.2004.062729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hattori K, Shimizu K, Takahashi M, Tamura M, Oheda M, Ohsawa N, Ono M. Quantitative in vivo assay of human granulocyte colony-stimulating factor using cyclophosphamide-induced neutropenic mice. Blood. 1990;75:1228–1233. [PubMed] [Google Scholar]
- 44.Chowdhury D, Lieberman J. Death by a thousand cuts: granzyme pathways of programmed cell death. Annu. Rev. Immunol. 2008;26:389–420. doi: 10.1146/annurev.immunol.26.021607.090404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.MacDonald G, Shi L, Vande Velde C, Lieberman J, Greenberg AH. Mitochondria-dependent and -independent regulation of granzyme B-induced apoptosis. J. Exp. Med. 1999;189:131–144. doi: 10.1084/jem.189.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomas DA, Du C, Xu M, Wang X, Ley TJ. DFF45/ICAD can be directly processed by granzyme B during the induction of apoptosis. Immunity. 2000;12:621–632. doi: 10.1016/s1074-7613(00)80213-7. [DOI] [PubMed] [Google Scholar]
- 47.Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- 48.Lamagna C, Aurrand-Lions M, Imhof BA. Dual role of macrophages in tumor growth and angiogenesis. J. Leukoc. Biol. 2006;80:705–713. doi: 10.1189/jlb.1105656. [DOI] [PubMed] [Google Scholar]
- 49.Mantovani A, Sica A. Macrophages, innate immunity and cancer: balance, tolerance, and diversity. Curr. Opin. Immunol. 2010;22:231–237. doi: 10.1016/j.coi.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 50.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fleetwood AJ, Lawrence T, Hamilton JA, Cook AD. Granulocyte-macrophage colony-stimulating factor (CSF) and macrophage CSF-dependent macrophage phenotypes display differences in cytokine profiles and transcription factor activities: implications for CSF blockade in inflammation. J. Immunol. 2007;178:5245–5252. doi: 10.4049/jimmunol.178.8.5245. [DOI] [PubMed] [Google Scholar]
- 52.Andersson U, Wang H, Palmblad K, Aveberger AC, Bloom O, Erlandsson-Harris H, Janson A, Kokkola R, Zhang M, Yang H, Tracey KJ. High mobility group 1 protein (HMG-1) stimulates proinflammatory cytokine synthesis in human monocytes. J. Exp. Med. 2000;192:565–570. doi: 10.1084/jem.192.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rao DA, Tracey KJ, Pober JS. IL-1alpha and IL-1beta are endogenous mediators linking cell injury to the adaptive alloimmune response. J. Immunol. 2007;179:6536–6546. doi: 10.4049/jimmunol.179.10.6536. [DOI] [PubMed] [Google Scholar]
- 54.Thorburn J, Horita H, Redzic J, Hansen K, Frankel AE, Thorburn A. Autophagy regulates selective HMGB1 release in tumor cells that are destined to die. Cell Death Differ. 2009;16:175–183. doi: 10.1038/cdd.2008.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lerman SP, Weidanz WP. The effect of cyclophosphamide on the ontogeny of the humoral immune response in chickens. J. Immunol. 1970;105:614–619. [PubMed] [Google Scholar]
- 56.Stockman GD, Heim LR, South MA, Trentin JJ. Differential effects of cyclophosphamide on the B and T cell compartments of adult mice. J. Immunol. 1973;110:277–282. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.