Abstract
The human RAD52 protein, which exhibits a heptameric ring structure, has been shown to bind resected double strand breaks (DSBs), consistent with an early role in meiotic recombination and DSB repair. In this work, we show that RAD52 binds single-stranded and tailed duplex DNA molecules via precise interactions with the terminal base. When probed with hydroxyl radicals, ssDNA–RAD52 complexes exhibit a four-nucleotide repeat hypersensitivity pattern. This unique pattern is due to the interaction of RAD52 with either a 5′ or a 3′ terminus of the ssDNA, is sequence independent and is phased precisely from the terminal nucleotide. Hypersensitivity is observed over ∼36 nucleotides, consistent with the length of DNA that is protected by RAD52 in nuclease protection assays. We propose that RAD52 binds DNA breaks via specific interactions with the terminal base, leading to the formation of a precisely organized ssDNA–RAD52 complex in which the DNA lies on an exposed surface of the protein. This protein–DNA arrangement may facilitate the DNA–DNA interactions necessary for RAD52-mediated annealing of complementary DNA strands.
Keywords: hydroxyl radicals/hypersensitivity/RAD52/single-stranded DNA
Introduction
In simple eukaryotes, the RAD52 epistasis group of genes is required for normal resistance to ionizing radiation (Petes et al., 1991; Game, 1993). A key member of this group, RAD52, plays an important role in genetic recombination and the repair of double strand breaks (DSBs).
The Saccharomyces cerevisiae RAD52 protein has been purified and shown to promote the annealing of complementary single strands (Mortensen et al., 1996; Shinohara et al., 1998; Sugiyama et al., 1998). It also stimulates in vitro recombination reactions catalysed by RAD51 (Sung, 1997; New et al., 1998; Shinohara and Ogawa, 1998), a structural and functional homologue of the Escherichia coli RecA protein (Aboussekhra et al., 1992; Shinohara et al., 1992;Ogawa et al., 1993; Sung, 1994). These properties of RAD52 are consistent with roles in both RAD51-independent and RAD51-dependent recombination, as indicated by genetic studies (Petes et al., 1991; Paques and Haber, 1999).
The human RAD52 protein shares many properties with its yeast counterpart, including DNA strand annealing and stimulation of RAD51-mediated recombination reactions in vitro (Reddy et al., 1997; Benson et al., 1998; Baumann and West, 1999). Both yeast and human RAD52 proteins form ring-like structures (Shinohara et al., 1998; Van Dyck et al., 1998), and a low-resolution three-dimensional structure of human RAD52 protein has been determined (Stasiak et al., 2000). Human RAD52 consists of seven subunits that are organized in the form of a heptameric ring with a large central channel, a structure similar to that exhibited by hexameric DNA helicases. The RAD52 rings exhibit a pinwheel appearance in projection, related to that of bacteriophage T7 gp4 protein (Yu et al., 1996a), E.coli DnaB (Yu et al., 1996b) and SV40 large T antigen (San Martin et al., 1997). However, while it is generally accepted that the hexameric helicases bind DNA within the central channel, the binding site of RAD52 is unknown. The evidence that is available presently appears to favour the view that DNA may wrap around the outside of the RAD52 ring, an arrangement similar to that proposed for the P22 essential recombination function (Erf) (Poteete et al., 1983) and β protein from bacteriophage λ (Passy et al., 1999).
RAD52 has been shown to bind specifically to single-stranded tails present at sites of resected DSBs (Van Dyck et al., 1999), consistent with a role in the early stages of recombination and DSB repair. In this work, we extend these observations by showing that RAD52 binds to the terminal nucleotide of single-stranded (ss) and tailed duplex DNA, leading to the precise wrapping of ssDNA around or within the RAD52 protein. The structure formed by the ssDNA–RAD52 complex is uniquely sensitive to attack by hydroxyl radicals, in that the DNA exhibits a phased four-nucleotide repeat pattern of hypersensitivity. From these data, we propose that the ssDNA lies in a groove that is exposed on the surface of RAD52, an arrangement that could facilitate single strand annealing.
Results
Analysis of ssDNA–RAD52 complexes
Hydroxyl radicals (OH•) generated in vitro during the Fenton reaction have been used extensively to probe protein–DNA interactions. Hydroxyl radicals promote the sequence-independent cleavage of single- or double-stranded DNA by abstracting a hydrogen atom from ribose, leading to the breakdown of the sugar. Decreased reactivity at defined sites is usually indicative of protein binding in this region, which blocks access to the radicals. In some instances, however, protein binding can enhance cleavage by increasing the accessibility of the phosphodiester backbone or by channelling the OH• into a volume adjacent to the DNA strand.
When ssDNA–RAD52 complexes, formed by interaction of RAD52 with a 5′-32P-labelled 68 base synthetic oligonucleotide, were probed with OH•, we observed that the DNA was hypersensitive to OH• attack (Figure 1, lane d). A ladder of bands was observed with hypersensitivity located every four nucleotides along the DNA. This repeat pattern was seen along the entire length of the DNA (except for the 4n fragment, which could not be ethanol precipitated). In the absence of RAD52, the ssDNA was cleaved uniformly at each position along the phosphodiester backbone (lane c), indicating that the 4n hypersensitivity pattern was a consequence of RAD52 binding.
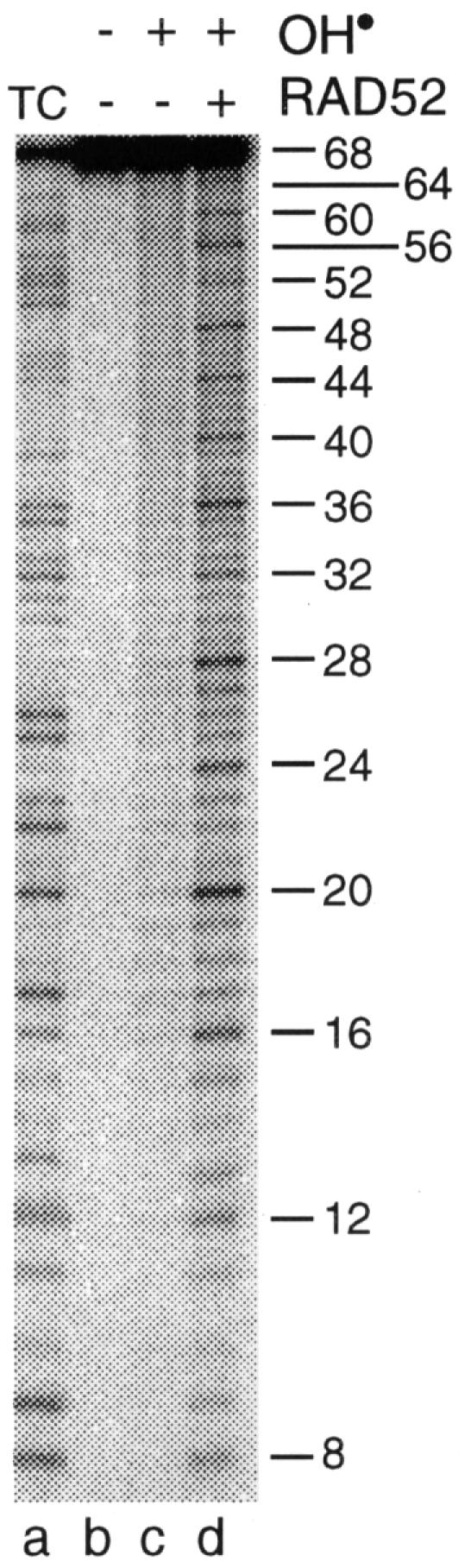
Fig. 1. Hypersensitivity of single-stranded DNA bound by RAD52 to hydroxyl radicals. 5′-32P-labelled oligo 1 (68n) was incubated in binding buffer in the absence (lane c) or presence (lane d) of RAD52 (40 nM) as described in Materials and methods. Reactions were supplemented with hydrogen peroxide and iron/EDTA and incubated for a further 10 min. 32P-labelled products were analysed by denaturing PAGE and detected by autoradiography. Lane a: T + C sequence ladders.
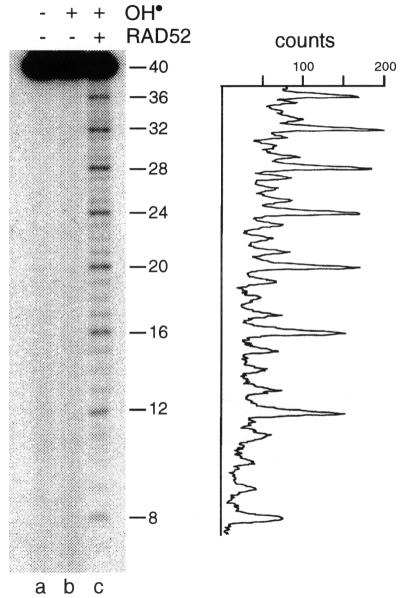
To determine whether the 4n repeat pattern was sequence dependent, a second oligonucleotide, poly(dT)40, was used. The pattern observed with the poly(dT)40–RAD52 complex was even more striking (Figure 2, lane c), and a quantification of the hypersensitivity (shown to the right of Figure 2) indicated that each fourth base was cleaved much more efficiently than other sites.
Fig. 2. Sequence-independent phasing from the DNA terminus. 5′-32P-labelled poly(dT)40 was incubated with RAD52 (20 nM) and probed with hydroxyl radicals as described in the legend to Figure 1. PhosphorImager profile of the hydroxyl radical pattern shown in lane c.
These results show that RAD52 binds ssDNA in a sequence-independent fashion to produce a complex in which the DNA lies exposed to OH• attack. The regular 4n pattern of hypersensitivity may be due to a distortion of the DNA as it wraps around or within RAD52.
Interaction of RAD52 with 5′- and 3′-tailed duplex DNA
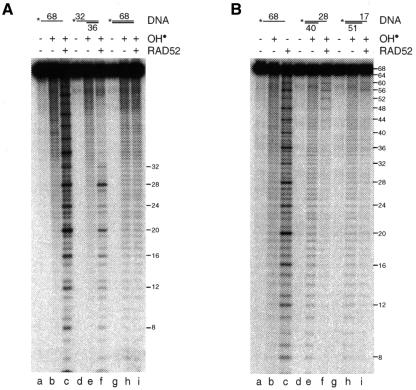
Since recombination and DSB repair are thought to be initiated by resected DSBs, and RAD52 interacts specifically with such substrates in vitro (Van Dyck et al., 1999), we next investigated whether complexes formed between tailed duplex DNA and RAD52 exhibited the unusual OH• sensitivity. To do this, we first compared the effect of OH• on ssDNA, 5′-tailed DNA and linear duplex DNA. The substrates, all 5′-32P-labelled in oligo 1, are indicated schematically in Figure 3A.
Fig. 3. Substrate specificity of RAD52. (A) 5′-32P-labelled oligo 1 (lanes a–c), 5′-tailed DNA with a 32 nucleotide overhang (lanes d–f) or duplex DNA (lanes g–i) was incubated in the absence or presence of RAD52 (40 nM) as indicated. The complexes were then probed with hydroxyl radicals and the products analysed by denaturing PAGE. The 5′-overhang and duplex substrates were made by annealing 5′-32P-labelled oligo 1 with oligo 3 and oligo 2, respectively. (B) 5′-32P-labelled oligo 1 (lanes a–c) or 3′-tailed DNA with overhangs of 28 (lanes d–f) or 17 nucleotides (lanes g–i) was incubated in the absence or presence of RAD52 (80 nM). The complexes were then probed with hydroxyl radicals and the products analysed as described in (A). The 28n and 17n overhang substrates were produced by annealing 32P-labelled oligo 1 with oligo 4 and oligo 6, respectively.
Whereas the ssDNA–RAD52 complex showed its characteristic four-nucleotide repeat pattern (Figure 3A, lane c), OH• hypersensitivity was not observed with duplex DNA (lane i). The tailed DNA molecule, however, was found to be sensitive to OH• (Figure 3A, lane f), but not along the full length of the DNA substrate. Indeed, the 4n pattern was only seen in the region corresponding to the single-stranded tail.
The experiments described above indicate that RAD52 binds to the terminal base of single-stranded or 5′-tailed DNA, and that the OH• repeat pattern is phased from the terminus. To determine whether RAD52 could also bind 3′ termini, we analysed OH• patterns on 3′-tailed duplex substrates carrying overhangs that were 28 or 17 nucleotides in length (Figure 3B). In this experiment, we found that the ssDNA tails were also hypersensitive to OH•, with the substrate containing a 28 base tail giving rise to six hypersensitive bands (Figure 3B, lane f), whereas that with a 17 base tail produced only three hypersensitive sites (lane i).
The results obtained with the 5′- and 3′-overhang substrates demonstrate that RAD52 is binding to the end of the substrate and that the OH• repeat pattern is phased in from the terminal base. The hypersensitivity pattern stops close to the transition from single-stranded to double-stranded DNA. The results also demonstrate that RAD52 can bind DNA termini without regard for their polarity.
In the case of ssDNA, the results show that the 4n hypersensitivity occurs throughout the length of the DNA substrate. Due to the ability of RAD52 to bind 5′ or 3′ ssDNA termini, this 4n repeat pattern was only observed with oligonucleotides possessing a length that is divisible by 4n. Other DNA lengths were found to exhibit over lapping patterns (data not shown).
DNA binding by RAD52
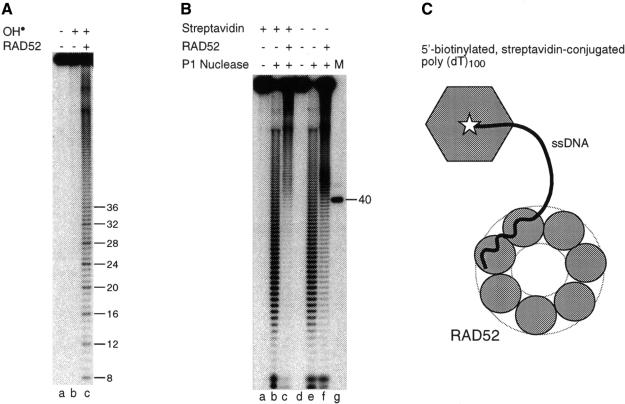
To determine the length of DNA bound by RAD52, it was necessary to block binding to one end of the ssDNA, and this was achieved by synthesizing 5′-biotinylated poly(dT)100 to which streptavidin could be conjugated (indicated schematically in Figure 4C). Whereas a biotinylated 5′ terminus could be bound by RAD52, the presence of streptavidin limited RAD52 to 3′-terminal interactions. When the 5′-biotinylated streptavidin-conjugated 3′-32P-end-labelled poly(dT)100 was incubated with RAD52 and probed with OH•, we found that the hypersensitivity extended 36 nucleotides from the 3′ terminus (Figure 4A, lane c).
Fig. 4. Chemical and nuclease footprinting of the ssDNA–RAD52 complex. (A) 5′-biotinylated streptavidin-conjugated poly(dT)100, labelled at the 3′ terminus with 32P, was incubated in the absence or presence of RAD52 (26 nM) and probed with hydroxyl radicals as indicated. The products were separated by denaturing PAGE and visualized by autoradiography. (B) 5′-biotinylated poly(dT)100, 32P-labelled at the 3′-terminus and conjugated with streptavidin as indicated, was incubated with or without RAD52 (26 nM) for 20 min at 37°C. The ssDNA–RAD52 complexes were then treated with P1 nuclease as described in Materials and methods. 32P-labelled products were analysed by denaturing PAGE and autoradiography. Lane g: 32P-labelled poly(dT)40 marker. (C) Schematic diagram indicating the binding of RAD52 to the 3′ terminus of 5′-biotinylated streptavidin-conjugated poly(dT)100. The RAD52 heptameric ring is indicated binding to the 3′ terminus of the ssDNA, which has biotin (star) conjugated to streptavidin at the 5′ end. The path taken by the ssDNA when bound to the RAD52 heptamer is unknown.
To confirm that RAD52 binds only 36 nucleotides, we also performed P1 nuclease assays on the poly(dT)100–RAD52 complex. Under the conditions used, P1 was expected to act as both an endo- and exonuclease. When the biotinylated streptavidin-conjugated oligonucleotide was incubated with RAD52 and P1 nuclease added, a protein ‘footprint’ was obtained that extended ∼40 nucleotides from the 3′ terminus (Figure 4B, lane c). In the absence of RAD52, the DNA was digested more uniformly by the nuclease (Figure 4B, lane b).
In the experiment of Figure 4B, both DNA ends were blocked by protein binding (streptavidin at the 5′ terminus and RAD52 at the 3′ terminus), allowing P1 to act only as an endonuclease. Omission of the streptavidin, however, permitted entry of the P1 from the 5′ terminus, enabling us to define the size of the DNA-binding site of RAD52 a third way. In this case, we again observed that ∼40 nucleotides at the 3′ end of the ssDNA substrate were protected by RAD52 (Figure 4B, lane f). Under these conditions, a region of increased nuclease cleavage was observed at sites located 40–50 nucleotides from the 3′ terminus, where the exonuclease was blocked by bound RAD52. Taken together, these results are consistent with the OH• data and indicate that RAD52 protein binds ∼36 nucleotides of ssDNA.
DNA cross-linking
To determine whether it is possible to detect direct interactions between RAD52 and ssDNA, the protein was incubated with 32P-labelled poly(dT)40 and the resulting complexes were fixed by photo-cross-linking. Protein subunits cross-linked to DNA were then resolved by denaturing SDS–PAGE, which revealed the presence of two distinct product bands (Figure 5, lane d). At the present time, it is not known whether these bands represent the cross-linking of DNA to one or two RAD52 subunits of the heptameric ring, or whether two DNA molecules become cross-linked to the same RAD52 monomer. In the absence of RAD52 (Figure 5, lane b), or when the products of complete reactions were deproteinized prior to SDS–PAGE (lane e), products of reduced mobility were not observed.
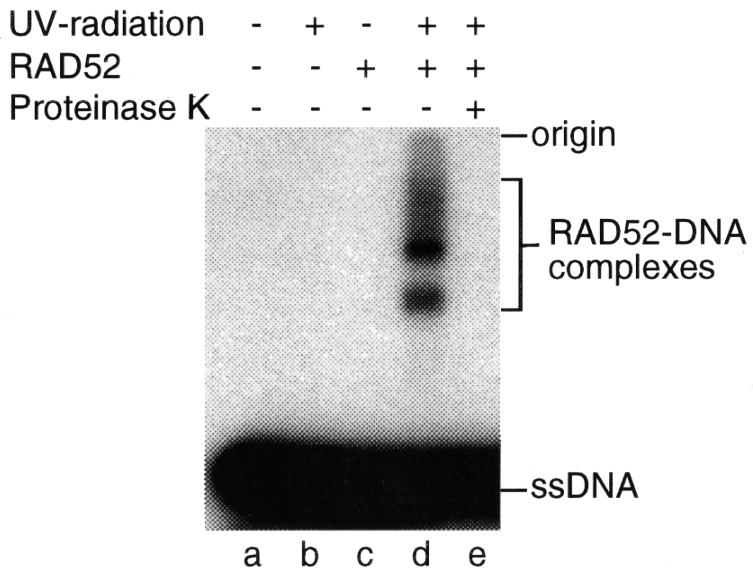
Fig. 5. Protein–DNA cross-linking. 5′-32P-labelled poly(dT)40, incubated with or without RAD52, was UV irradiated as indicated. Aliquots were analysed directly or after deproteinization by incubation for 30 min at 37°C with 2 mg/ml proteinase K in 0.5% SDS. 32P-labelled DNA was visualized by autoradiography following SDS–PAGE. The RAD52–ssDNA complexes are indicated.
Relationship between OH• sensitivity and single strand annealing
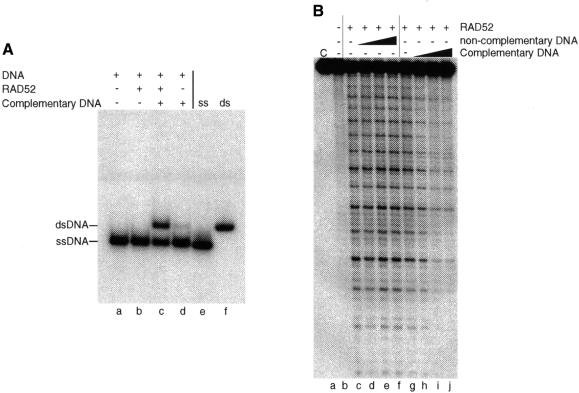
It is known that RAD52 promotes DNA annealing reactions in vitro (Mortensen et al., 1996; Reddy et al., 1997; Shinohara et al., 1998; Sugiyama et al., 1998). As shown in Figure 6A, RAD52 promotes the re-annealing of a 32P-labelled oligo with its complement to form duplex DNA products (lane c). We therefore investigated whether the OH• hypersensitivity pattern was altered by the presence of a complementary strand of DNA. To do this, 5′-32P-end-labelled oligo 1 (68 nucleotides) was incubated with RAD52 for 20 min. At this time, complementary (oligo 2) or non-complementary (oligo 1) unlabelled ssDNA was added and incubation was continued for 10 min. The resulting complexes were then probed with hydroxyl radicals.
Fig. 6. Effect of single strand annealing on hydroxyl radical sensitivity. (A) Single strand annealing by RAD52. Complexes formed between 32P-labelled oligo 1 and RAD52 (15 nM) were supplemented with unlabelled complementary (oligo 2) or non-complementary (oligo 1) DNA as indicated. Reactions were stopped and, following deproteinization, were analysed by neutral PAGE. Lanes e and f: ssDNA (oligo 1) and dsDNA (oligos 1 + 2) markers. (B) 5′-32P-labelled oligo 1 was incubated with RAD52 (15 nM) for 20 min. Increasing amounts (0, 0.5, 1.0 and 2.0 ng) of the same (oligo 1) or complementary (oligo 2) DNA (both unlabelled) were then added and incubation continued for 10 min. Reactions were supplemented with hydrogen peroxide and iron/EDTA and incubated for a further 10 min. The products were denatured and analysed by denaturing PAGE. Lane a: control lane showing oligo 1 without OH• treatment.
As shown in Figure 6B (lanes g–j), the presence of the complementary single strand resulted in a significant decrease in OH• sensitivity of the 32P-labelled strand. A loss of OH• sensitivity was not observed in control reactions to which non-complementary DNA was added (Figure 6B, lanes c–f). These results indicate that the hypersensitivity to hydroxyl radicals is lost upon RAD52-mediated single strand annealing.
Discussion
In this work, we have shown that RAD52 interacts with single-stranded and tailed duplex DNA to form complexes that, when probed with hydroxyl radicals, exhibit an unusual four-nucleotide repeat hypersensitivity pattern. To our knowledge, such a pattern, which is sequence independent and appears to be phased by precise interactions with either a 5′- or a 3′-terminal base, has not been observed with any other protein.
The binding of RAD52 to DNA termini is consistent with recent electron microscopic observations of the way in which RAD52 interacts with resected DSBs (Van Dyck et al., 1999). In these studies, it was also shown that RAD52 could protect DSBs from exonuclease attack. Taken together, the data indicate that RAD52 is capable of ‘capping’ DNA termini, a function that may be important for the stability and repair of DSBs in vivo.
Although it is known that RAD52 exhibits a heptameric ring structure in vitro (Stasiak et al., 2000), there is presently very little evidence to indicate the path taken by the DNA when bound by the ring. Although RAD52 rings contain a large central channel and show a distinct chiral arrangement of subunits, similar to the structure formed by a family of hexameric DNA helicases (Egelman, 1996), there is currently no evidence to suggest that the DNA passes though this central channel. Indeed, electron microscopic observations of complexes formed between RAD52 and circular DNA lend support to the view that the DNA passes along the outside of the RAD52 ring (Van Dyck et al., 1998), an arrangement that may be similar to that proposed for the P22 Erf protein and the β protein of bacteriophage λ (Poteete et al., 1983; Passy et al., 1999).
The OH• hypersensitivity data, together with nuclease protection assays, indicate that RAD52 binds ∼36 nucleotides, a length that is insufficient to allow the passage of DNA around the entire ring, unless the DNA wraps around the crown of the RAD52 heptamer. It is more likely that DNA is bound by one or two subunits, consistent with results obtained in our cross-linking studies. Although we cannot at present exclude the possibility that the DNA passes through the central channel of RAD52, we interpret the OH• hypersensitivity patterns to indicate that the ssDNA lies in an exposed groove on the surface of the protein, and that the phosphodiester backbone is highly distorted and exposed to solvent. Such an arrangement would make the bases accessible for interaction with complementary DNA, and thus facilitate single strand annealing. Alternatively, if each monomer in RAD52 is capable of binding the terminus of a ssDNA molecule, the ability of the RAD52 heptamer to bind multiple DNA termini could increase the local concentration of DNA ends and thereby facilitate the terminal interactions required for single strand annealing. Such an arrangement would also permit single strand annealing reactions to take place between homologous DNA molecules containing heterologous terminal sequences.
Consistent with the notion that the OH• hypersensitivity represents a measure of the way in which DNA lies exposed on the surface of RAD52, we find that the hypersensitivity pattern is lost when the bound strand anneals with its complement. This change may reflect an alteration in accessibility of the phosphodiester backbone, which occurs upon DNA–DNA pairing, or may be a consequence of the release of paired duplex DNA by the RAD52 heptamer.
Materials and methods
Proteins
Recombinant human RAD52 protein was purified from baculovirus-infected Sf9 cells as described (Van Dyck et al., 1999).
DNA
All oligonucleotides were synthesized by phosphoramidite chemistry on an Applied Biosystems 380B DNA synthesizer and used fully deprotected. Following ethanol precipitation, each oligonucleotide was resuspended in 10 mM Tris–HCl pH 8.0, 1 mM EDTA at a concentration of 5 µg/µl. The DNA sequences are as follows: oligo 1 (68n), 5′-TATCGAATCCGTCTAGTCAACGCTGCCGAATTCTACCAGTG AGGTTTGGGCTCCTCAACCTGCAGGTT-3′; oligo 2 (68n), 5′-AAC CTGCAGGTTGAGGAGCCCAAACCTCACTGGTAGAATTCGGCA GCGTTGACTAGACGGATTCGATA-3′; oligo 3 (36n), 5′-AACCTG CAGGTTGAGGAGCCCAAACCTCACTGGTAG-3′; oligo 4 (40n), 5′-ACTGGTAGAATTCGGCAGCGTTGACTAGACGGATTCGATA- 3′; oligo 5 (58n), 5′-TTGAGGAGCCCAAACCTCACTGGTAGAA TTCGGCAGCGTTGACTAGACGGATTCGATA-3′; oligo 6 (51n), 5′-GCCCAAACCTCACTGGTAGAATTCGGCAGCGTTGACTAGA CGGATTCGATA-3′; oligo 7: poly(dT)40; oligo 8, 5′-biotinylated poly(dT)100 (purchased from Oswel DNA service).
DNA was 5′-32P-end-labelled by treatment with T4 polynucleotide kinase (USB) and [γ-32P]ATP, or 3′-32P-end-labelled by treatment with terminal transferase (NEB) in the presence of [α-32P]ddATP. Proteins were inactivated by incubation at 65°C for 15 min and the DNA purified by passage through a G25 spin column (Pharmacia) to remove the unincorporated nucleotides.
Duplex and tailed substrates were produced by annealing 32P-end-labelled 68mer (oligo 1) with excess complementary oligonucleotide as described (Parsons et al., 1990).
Hydroxyl radical footprinting
32P-labelled DNA (∼1 ng) was incubated with or without RAD52 in binding buffer [50 mM triethanolamine–HCl pH 7.5, 1 mM dithiothreitol (DTT), 100 µg/ml bovine serum albumin (BSA)] for 20 min at 37°C in a total volume of 100 µl. Hydroxyl radicals were generated using hydrogen peroxide and iron/EDTA (Tullius and Dombroski, 1986; Bennett et al., 1993) and after 5 min incubation the reactions were stopped by addition of thiourea. The DNA products were then ethanol precipitated, denatured and electrophoresed on an 8% denaturing polyacrylamide gel. 32P-labelled DNA products were detected by autoradiography after drying the gels onto Whatman 3 mm paper followed by exposure to Biomax MR film. OH• sensitivity patterns were quantified using a Molecular Dynamics PhosphorImager with ImageQuant software. Maxam–Gilbert TC sequencing ladders were made as described (Maxam and Gilbert, 1980).
In some reactions containing 5′-biotinylated poly(dT)100, the 3′-32P-end-labelled DNA was incubated for 5 min at 37°C with 10 ng of streptavidin (Sigma) prior to the addition of RAD52.
Protein–DNA cross-linking
32P-labelled poly(dT)40 was incubated with or without RAD52 in binding buffer for 20 min at 37°C. The reactions were then divided into two, such that one half could be irradiated with 4.5 kJ UV254 light and analysed for the presence of protein–DNA complexes by SDS–PAGE on a 4–20% gradient gel. Non-irradiated samples were analysed in a similar way. 32P-labelled DNA was detected by autoradiography.
DNA annealing assays
Reaction mixtures (100 µl) containing 32P-end-labelled oligo 1 (∼1 ng) were incubated in the absence or presence of RAD52 in binding buffer for 20 min at 37°C. Complementary (oligo 2) or non-complementary (oligo 1) unlabelled DNA was then added and incubation continued for 10 min. Reactions were stopped and deproteinized by addition of 25 µl of stop buffer (100 mM Tris–HCl pH 7.5, 5% SDS, 250 mM EDTA, 20 mg of proteinase K/ml) followed by a further incubation for 10 min. DNA products were analysed by electrophoresis through 10% polyacrylamide gels using a Tris-borate buffer system, followed by autoradiography.
P1 nuclease assays
Biotinylated poly(dT)100, bound or unbound by streptavidin (10 ng), was incubated with or without RAD52 in binding buffer for 20 min at 37°C. Reactions were then supplemented with 1.5 U of P1 nuclease (BRL) and incubation was continued for 5 min. DNA products were ethanol precipitated, denatured and electrophoresed on an 8% denaturing polyacrylamide gel.
Acknowledgments
Acknowledgements
We thank Ed Egelman, Andrzej Stasiak, Dale Wigley and our colleagues at Clare Hall for their interest and comments. The Imperial Cancer Research Fund and the Human Frontiers Science Program supported this work. E.V.D. was supported in part by an EC fellowship.
References
- Aboussekhra A., Chanet,R., Adjiri,A. and Fabre,F. (1992) Semidominant suppressors of srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to prokaryotic RecA proteins. Mol. Cell. Biol., 12, 3224–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P. and West,S.C. (1999) Heteroduplex formation by human Rad51 protein: effects of DNA end-structure, hRP-A and hRad52. J. Mol. Biol., 291, 363–374. [DOI] [PubMed] [Google Scholar]
- Bennett R.J., Dunderdale,H.J. and West,S.C. (1993) Resolution of Holliday junctions by RuvC resolvase: cleavage specificity and DNA distortion. Cell, 74, 1021–1031. [DOI] [PubMed] [Google Scholar]
- Benson F.E., Baumann,P. and West,S.C. (1998) Synergistic actions of Rad51 and Rad52 in genetic recombination and DNA repair. Nature, 391, 401–404. [DOI] [PubMed] [Google Scholar]
- Egelman E.H. (1996) Homomorphous hexameric helicases: tales from the ring cycle. Structure, 4, 759–762. [DOI] [PubMed] [Google Scholar]
- Game J.C. (1993) DNA double-strand breaks and the RAD50–RAD57 genes in Saccharomyces.Semin. Cancer Biol., 4, 73–83. [PubMed] [Google Scholar]
- Maxam A.M. and Gilbert,W. (1980) Sequencing end-labelled DNA with base specific chemical cleavages. Methods Enzymol., 65, 499–560. [DOI] [PubMed] [Google Scholar]
- Mortensen U.H., Bendixen,C., Sunjevaric,I. and Rothstein,R. (1996) DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl Acad. Sci. USA, 93, 10729–10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New J.H., Sugiyama,T., Zaitseva,E. and Kowalczykowski,S.C. (1998) Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein-A. Nature, 391, 407–410. [DOI] [PubMed] [Google Scholar]
- Ogawa T., Yu,X., Shinohara,A. and Egelman,E.H. (1993) Similarity of the yeast Rad51 filament to the bacterial RecA filament. Science, 259, 1896–1899. [DOI] [PubMed] [Google Scholar]
- Paques F. and Haber,J.E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev., 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons C.A., Kemper,B. and West,S.C. (1990) Interaction of a four-way junction in DNA with T4 endonuclease VII. J. Biol. Chem., 265, 9285–9289. [PubMed] [Google Scholar]
- Passy S.I., Yu,X., Li,Z., Radding,C.M. and Egelman,E.H. (1999) Rings and filaments of β protein from bacteriophage λ suggest a superfamily of recombination proteins. Proc. Natl Acad. Sci. USA, 96, 4279–4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes T.D., Malone,R.E. and Symington,L.S. (1991) Recombination in yeast. In The Molecular and Cellular Biology of the Yeast Saccharomyces: Genome Dynamics, Protein Synthesis and Energetics, Vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 407–521. [Google Scholar]
- Poteete A.R., Sauer,R.T. and Hendrix,R.W. (1983) Domain structure and quaternary organisation of the bacteriophage P22 Erf protein. J. Mol. Biol., 171, 401–418. [DOI] [PubMed] [Google Scholar]
- Reddy G., Golub,E.I. and Radding,C.M. (1997) Human Rad52 protein promotes single-strand DNA annealing followed by branch migration. Mutat. Res., 377, 53–59. [DOI] [PubMed] [Google Scholar]
- San Martin M.C., Gruss,C. and Carazo,J.M. (1997) Six molecules of SV40 large T antigen assemble in a propeller-shaped particle around a channel. J. Mol. Biol., 268, 15–20. [DOI] [PubMed] [Google Scholar]
- Shinohara A. and Ogawa,T. (1998) Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature, 391, 404–407. [DOI] [PubMed] [Google Scholar]
- Shinohara A., Ogawa,H. and Ogawa,T. (1992) Rad51 protein involved in repair and recombination in S.cerevisiae is a RecA-like protein. Cell, 69, 457–470. [DOI] [PubMed] [Google Scholar]
- Shinohara A., Shinohara,M., Ohta,T., Matsuda,S. and Ogawa,T. (1998) Rad52 forms ring structures and cooperates with RPA in single-strand DNA annealing. Genes Cells, 3, 145–156. [DOI] [PubMed] [Google Scholar]
- Stasiak A.Z., Larquet,E., Stasiak,A., Müller,S., Engel,A., Van Dyck,E., West,S.C. and Egelman,E.H. (2000) The human Rad52 protein exists as a heptameric ring. Curr. Biol., 10, 337–340. [DOI] [PubMed] [Google Scholar]
- Sugiyama T., New,J.H. and Kowalczykowski,S.C. (1998) DNA annealing by Rad52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc. Natl Acad. Sci. USA, 95, 6049–6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P. (1994) Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast Rad51 protein. Science, 265, 1241–1243. [DOI] [PubMed] [Google Scholar]
- Sung P. (1997) Function of yeast Rad52 protein as a mediator between replication protein-A and the Rad51 recombinase. J. Biol. Chem., 272, 28194–28197. [DOI] [PubMed] [Google Scholar]
- Tullius T.D. and Dombroski,B.A. (1986) Hydroxyl radical footprinting: high resolution information about DNA–protein contacts and application to λ repressor and Cro protein. Proc. Natl Acad. Sci. USA, 83, 5469–5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyck E., Hajibagheri,N.M.A., Stasiak,A. and West,S.C. (1998) Visualisation of human Rad52 protein and its complexes with hRad51 and DNA. J. Mol. Biol., 284, 1027–1038. [DOI] [PubMed] [Google Scholar]
- Van Dyck E., Stasiak,A. and West,S.C. (1999) Binding of double-strand breaks in DNA by human Rad52 protein. Nature, 398, 728–731. [DOI] [PubMed] [Google Scholar]
- Yu X., Hingorani,M.M., Patel,S.S. and Egelman,E.H. (1996a) DNA is bound within the central hole to one or two of the six subunits of the T7 DNA helicase. Nature Struct. Biol., 3, 740–743. [DOI] [PubMed] [Google Scholar]
- Yu X., Jezewska,M.J., Bujalowski,W. and Egelman,E.H. (1996b) The hexameric Escherichia coli DnaB helicase can exist in different quaternary states. J. Mol. Biol., 259, 7–14. [DOI] [PubMed] [Google Scholar]