Abstract
John Nash showed that within a complex system individuals are best off if they make the best decision that they can, taking into account the decisions of the other individuals. Here, we investigate if similar principles influence the evolution of signaling networks in multicellular animals. Specifically, by analyzing a set of metazoan species, we observe a striking negative correlation of genomically encoded tyrosine content with biological complexity (as measured by the number of cell types in each organism). We discuss how this observed tyrosine loss correlates with the expansion of tyrosine kinases in the evolution of the metazoan lineage and how it may relate to the optimization of signaling systems in multi-cellular animals. We propose that this phenomenon illustrates genome-wide adaptive evolution to accommodate beneficial genetic perturbation.
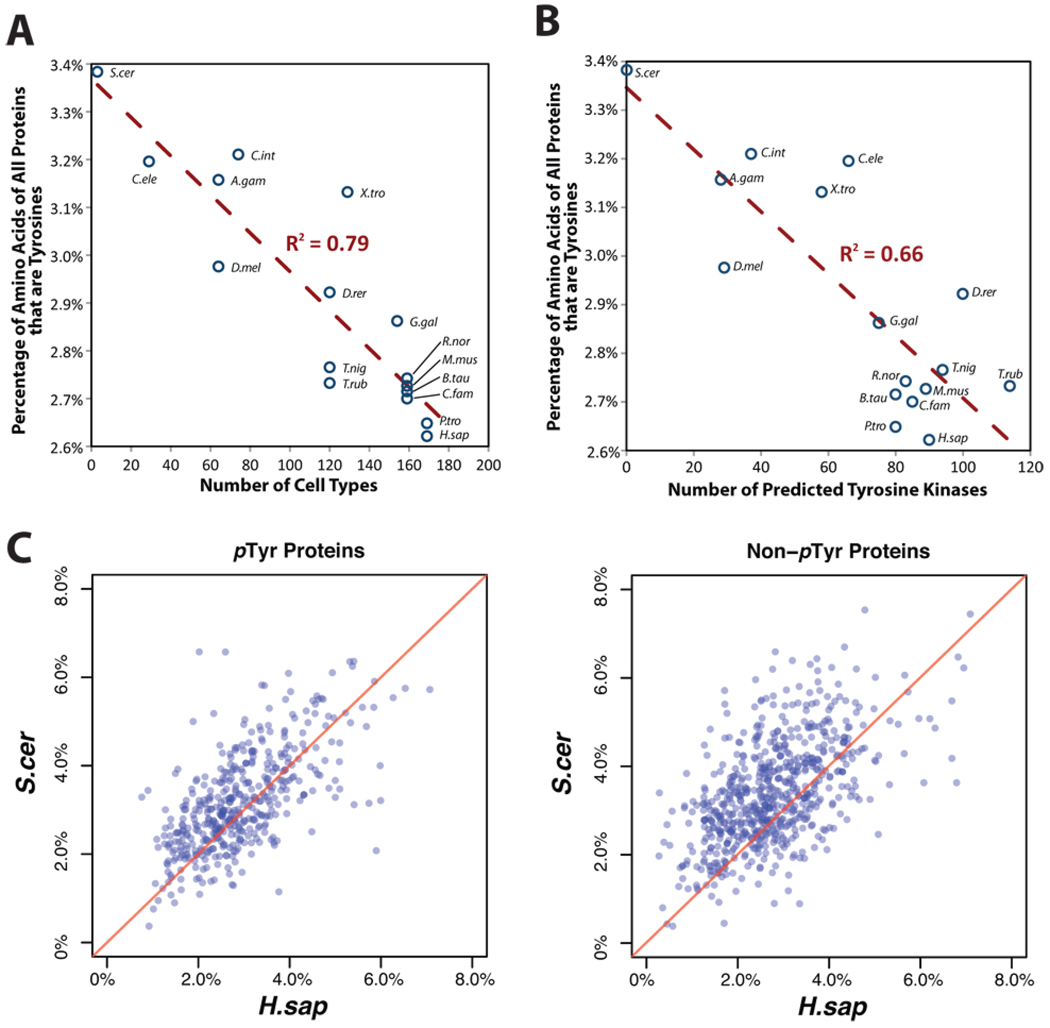
It is a biological paradox that organism complexity shows limited correlation with gene repertoire size (1). However, some protein families (2) have expanded with organism complexity as measured by number of cell types (3), especially those involved in regulation, such as tyrosine kinases in signaling, cell-cell communication and tissue boundary formation (4, 5). We observe a striking negative correlation of genomically-encoded tyrosine content with the number of distinct cell types in metazoan species (Spearman’s ρ = −0.89, approximate P-value = 3.0 × 10−6; Pearsons ρ = −0.89, approximate P-value = 4.0 × 10−6 , Fig. 1A). Thus, metazoans with more cell types have proportionally less potential tyrosine phospho-sites. Similarly, we observed that the number of tyrosine kinase domains correlates negatively with genomic tyrosine content (Spearman’s ρ = −0.68, approximate P-value = 3.7 × 10−3; Pearson’s ρ = −0.81, approximate P-value = 1.3 × 10−4, Fig. 1B). Including dual-specificity MLK and MEK kinases revealed a similar pattern (fig. S1A).
Fig. 1.
Correlation of expansion of phospho-tyrosine signaling systems with loss of genome encoded tyrosine residues. A, The genomically-encoded tyrosine content in metazoan organisms and yeast correlate negatively and significantly with organism complexity as measured by distinct cell types (2). Bakers yeast (S. cerevisiae) is included as a unicellular eukaryote for comparison. The species analyzed are yeast (S. cerevisiae), worm, (C. elegans), sea squirt (C. intestinalis), fly (D. melanogaster), mosquito (A. gambiae), zebrafish (D. rerio), tetraodon pufferfish (T. nigroviridis), Japanese pufferfish (T. rubripes), frog (X. tropicalis), chicken (G. gallus), dog (C. familiaris), cow (B. taurus), mouse (M. musculus), rat (R. norvegicus), chimpanzee (P. troglodytes) and human (H. sapiens). B, The number of tyrosine kinase domains in metazoans and yeast correlates negatively and significantly with the number of distinct cell types. C, The fraction of tyrosines in human-yeast ortholog protein pairs. Every point in the scatter plot represents a human-yeast ortholog protein pair where the (x, y) values denote the tyrosine content in human and yeast proteins, respectively. For simplicity, only proteins with an inferred one-to-one orthologous relationship between human and yeast are analyzed (for example, to avoid accelerated sequence divergence due to functional redundancy of paralogs). Orthologous protein pairs lying above the red diagonal (x = y) lines have higher tyrosine composition in yeast than human. The left scatter plot is for 437 human proteins conserved in yeast and known to be tyrosine-phosphorylated and the right plot is for 647 human proteins conserved in yeast not known to be tyrosine-phosphorylated.
These observations suggests an evolutionary model where the acquisition of a tyrosine kinase results in systems-level adaptation to remove deleterious phosphorylation events that cause aberrant cellular behavior and diseases (4). Assuming that a cell begins with a single tyrosine kinase, which is subsequently duplicated, it follows that the kinases may functionally diverge, as a result of relaxation in evolutionary constraints, to phosphorylate new substrates. Emerging kinase specificities could be retained if new substrates confer selection advantage. However, it is unlikely that every new phosphorylation event is beneficial. We hypothesize that optimization of newly emerged signaling networks would follow (6) through elimination of detrimental phosphorylation events by tyrosine-removing mutations. Even if many new phosphorylation sites are not deleterious, an organism with minimized noisy signaling systems is likely to have a fitness advantage. This scenario is repeated with the subsequent duplication of tyrosine kinases leading to more tyrosine residues lost (see SOM).
Despite several recent systematic phospho-proteomic studies (7), many human proteins have no observed phosphotyrosines. Our model suggests tyrosine loss had occurred predominantly in these proteins to minimize tyrosine phosphorylation. To test this hypothesis, we investigated differences in tyrosine loss between these proteins (Non-pTyr) and those that are tyrosine-phosphorylated (pTyr). Comparing members of these two groups to their orthologous proteins in S. cerevisiae (see SOM), which lack conventional tyrosine kinases, enabled us to assess the degree of tyrosine loss that may be triggered by the onset of phospho-tyrosine signaling in metazoans.
A significantly smaller fraction of amino acids are tyrosines in human proteins than in their yeast orthologs (approximate P = 3.5 × 10−4, paired Wilcoxon signed rank test, Fig. 1C). However, this phenomenon was statistically more pronounced in Non-pTyr proteins than in pTyr proteins (approximate P = 5.1 × 10−9, Mann-Whitney test, Fig. 1C). A similar trend was observed based on absolute tyrosine residue counts (approximate P = 2.0 × 10−7, Mann-Whitney test, fig. S1B), and on a higher confidence subset of pTyr proteins that either have multiple phospho-tyrosines or have sites observed in multiple studies (approximate P = 1.3 × 10−7, Mann-Whitney test).
Thus, tyrosine loss was strongly favored in human protein evolution, most notably in protein subsets that are not known to be tyrosine-phosphorylated. Genetic drift (8) is unlikely to account for these differences observed in a large number of evolutionarily distant human-yeast protein orthologs. As tyrosine is an essential and the most expensive amino acid to biosynthesize (9) after tryptophan and phenylalanine, essentiality and biosynthetic cost could be major factors in the observed loss. This is unlikely however, because we observed a strong positive correlation of number cell types with tryptophan and a weaker negative correlation for phenylalanine (table S1). Instead, we propose that positive selection of tyrosine-removing mutations occurred in the metazoan lineage to reduce adventitious tyrosine phosphorylation, at least in part. This optimization process likely shaped signaling networks crucial for the development of multi-cellular animals. Additionally, this could provide a mechanism to prevent unspecific phosphorylation events, that operates with evolution of domains and contextual factors to co-localize kinases with their substrates (10, 11). Tyrosine phosphorylation typically exerts its functional effects through allosteric regulation, or by creating binding sites for phosphobinding domains like SH2 and PTB (12). In agreement, we observed a slightly stronger negative correlation of genomically-encoded tyrosine content with the number of inferred phospho-tyrosine binding domains than tyrosine kinase domain count (Spearman’s ρ = −0.81, Pearson’s ρ = −0.88, see fig. S1A).
We note that the choanoflagellate Monosiga brevicollis, which is a member of the only known unicellular lineage with canonical tyrosine kinases (13), is an outlier in the cell type correlation studied above (data not shown). This observation is consistent with the emerging picture that choanoflagellates represent a distinct evolutionary branch from metazoans in which phospho-tyrosine signaling systems have been used for divergent functions (14, 15). Nevertheless, the Monosiga analysis is still consistent with optimization of phosphotyrosine signaling in this lineage – compared to metazoans analyzed here, Monosiga has higher numbers of tyrosine kinases and lower genomically-encoded tyrosine content (data not shown).
Other factors, such as tyrosine sulfation, could have contributed to the observed tyrosine loss, which raises the question whether other post-translational modifications and regulatory mechanisms are under similar evolutionary selection. We observed strong negative correlation of number of cell types with amino acids that can be methylated or glycosylated (table S1). The numbers of genomically-encoded threonine showed strong negative correlations with serine/threonine kinase and cell type numbers, although these trends were not observed with serine (fig. S2), suggesting possible coarse-grained functional differences between serine-and threonine-phosphorylation in metazoans.
Our findings suggest that the implementation of tyrosine kinase signaling, as a biological innovation that likely assisted the development of multi-cellular organisms, required system-level adaptive mutations. Analogous to the arguments by John Nash in his dissertation (16), this phenomenon highlights a general principle of adaptive evolution pertaining to the introduction of new components into a complex system, and parallels evolution of some human societies where the local populations have to adjust and adapt to the influx of immigrants contributing to the societies’ economic development. This principle may serve as an important framework when considering the evolution and fidelity of complex biological systems. Finally, this work raises the possibility that complex regulatory diseases, such as cancer, might result from systems-wide adaptive changes in human genomes and signaling systems.
Supplementary Material
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/1174301
Materials and Methods
Figs. S1 and S2
Table S1
References and Notes
References and Notes
- 1.Szathmry E, Jordn F, Pl C. Science. 2001;292:1315. doi: 10.1126/science.1060852. [DOI] [PubMed] [Google Scholar]
- 2.Vogel C, Chothia C. PLoS Comput Biol. 2006;2:e48. doi: 10.1371/journal.pcbi.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonner JT. Integ. Biol. 1998;1:28. [Google Scholar]
- 4.Hunter T. Curr Opin Cell Biol. 2009 doi: 10.1016/j.ceb.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Songyang Z, Cantley LC. Trends Biochem Sci. 1995;20:470. doi: 10.1016/s0968-0004(00)89103-3. [DOI] [PubMed] [Google Scholar]
- 6.Zarrinpar A, Park S-H, Lim WA. Nature. 2003;426:676. doi: 10.1038/nature02178. [DOI] [PubMed] [Google Scholar]
- 7.Jørgensen C, Linding R. Brief Funct Genomic Proteomic. 2008;7:17. doi: 10.1093/bfgp/eln001. [DOI] [PubMed] [Google Scholar]
- 8.Wright S. Genetics. 1931;16:97. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raiford DW, et al. J Mol Evol. 2008;67:621. doi: 10.1007/s00239-008-9162-9. [DOI] [PubMed] [Google Scholar]
- 10.Linding R, et al. Cell. 2007;129:1415. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller ML, et al. Sci Signal. 2008;1:ra2. doi: 10.1126/scisignal.1159433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seet BT, Dikic I, Zhou MM, Pawson T. Nat. Rev. Mol. Cell Biol. 2006;7:473. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- 13.King N, et al. Nature. 2008;451:783. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pincus D, Letunic I, Bork P, Lim WA. Proc Natl Acad Sci U S A. 2008;105:9680. doi: 10.1073/pnas.0803161105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manning G, Young SL, Miller WT, Zhai Y. Proc Natl Acad Sci U S A. 2008;105:9674. doi: 10.1073/pnas.0801314105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nash J. Ph.D. thesis. Princeton University; 1950. Non-cooperative games. [Google Scholar]
- 17.We thank Claus Jørgensen, Jiangzhi Zhang, Karen Colwill, Jing Jin and Kresten Lindorff-Larsen for suggestions and fruitful discussions. This project was in part supported by Genome Canada through the Ontario Genomics Institute and the Canadian Institutes of Health Research (MOP-84324). C.S.H.T. conceived the project. C.S.H.T., W.A.L., G.D.B., T.P. and R.L. designed the experiments. C.S.H.T., R.L. and A.P. performed the experiments. C.S.H.T., G.D.B., W.A.L., T.P. and R.L. wrote the paper. R.L. managed the project.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.