Abstract
Livers are comprised of maturational lineages of cells beginning extrahepatically in the hepato-pancreatic common duct near the duodenum and intrahepatically in zone 1 by the portal triads. The extrahepatic stem cell niches are the peribiliary glands deep within the walls of the bile ducts; those intrahepatically are the canals of Hering in postnatal livers and that derive from ductal plates in fetal livers. Intrahepatically, there are at least 8 maturational lineage stages from the stem cells in zone 1 (periportal), through the midacinar region (zone 2), to the most mature cells and apoptotic cells found pericentrally in zone 3. Those found in the biliary tree are still being defined. Parenchymal cells are closely associated with lineages of mesenchymal cells, and their maturation is coordinated. Each lineage stage consists of parenchymal and mesenchymal cell partners distinguishable by their morphology, ploidy, antigens, biochemical traits, gene expression, and ability to divide. They are governed by changes in chromatin (e.g. methylation), gradients of paracrine signals (soluble factors and insoluble extracellular matrix components), mechanical forces, and feedback loop signals derived from late lineage cells. Feedback loop signals, secreted by late lineage stage cells into bile, flow back to the periportal area and regulate the stem cells and other early lineage stage cells, in mechanisms dictating the size of the liver mass. Recognition of maturational lineage biology and its regulation by these multiple mechanisms offers new understandings of liver biology, pathologies, and strategies for regenerative medicine.
Keywords: hepatic stem cells, hepatoblasts, hepatic maturational lineages, feedback loop signaling, regenerative medicine
I. The Liver’s Maturational Lineages
Hepatic stem cells and their mesenchymal partners, angioblasts, give rise to daughter cells maturing into lineages of parenchymal and mesenchymal cells with stepwise changes in cell size, morphology, ploidy, gene expression, growth potential and signaling(1–4). Currently, there is evidence for at least 8 intrahepatic lineage stages (Figures 1 and 2)(5–6). Continued efforts to characterize the liver’s lineage biology should result in recognition of additional stages. This overview focuses on early intrahepatic lineage stages in human livers and includes aspects of their regulation. Information on later lineage stages of cells, additional background and references is included in the online supplement.
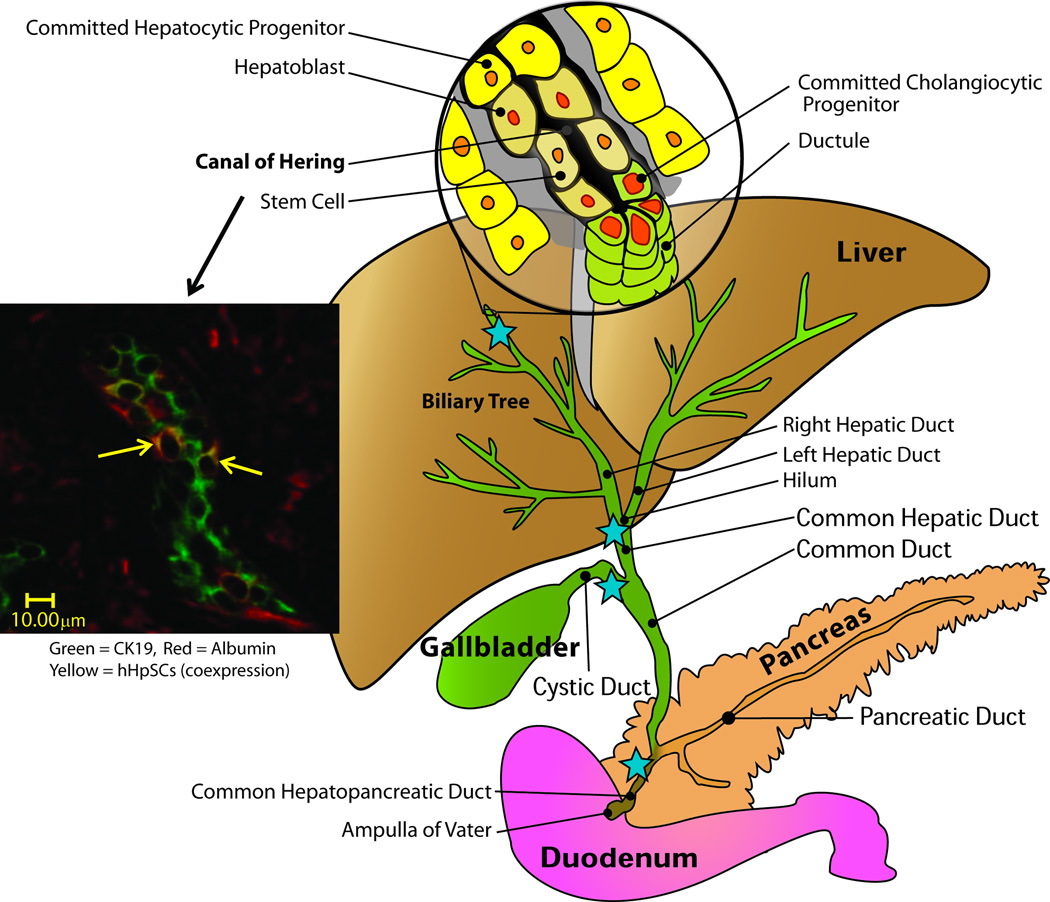
Figure 1.
Schematic image of liver, the biliary tree and panceas and their connections with the duodenum. The blue stars indicate sites at which there are high numbers of peribiliary glands, the stem cell niches of the biliary tree.
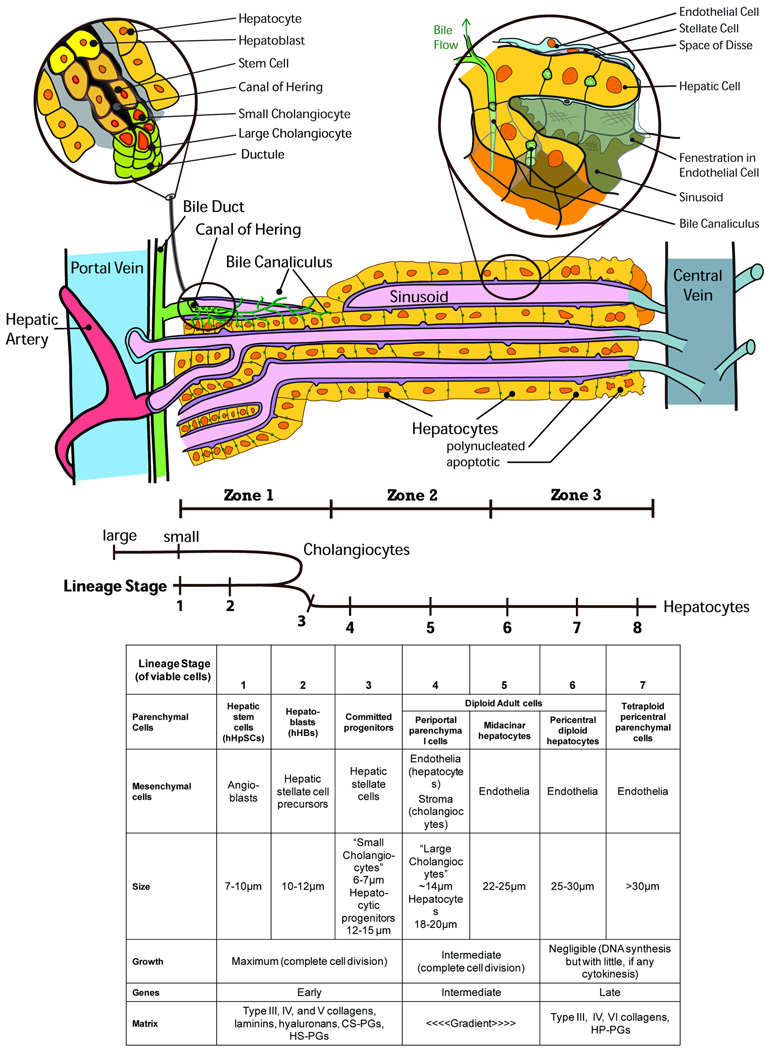
Figure 2.
Schematic image of intrahepatic maturational lineages.
Stage 1.Hepatic stem cells (hHpSCs) are multipotent stem cells located within the liver’s stem cell compartment, the ductal plates of fetal and neonatal livers, and canals of Hering in pediatric and adult livers(5,7–12). The compartment represents the anatomic and physiological link between the intralobular canalicular system of hepatocytes and the biliary tree and resides along sites that project starlike from the portal tracts. They constitute ~0.5–2% of the parenchyma of livers of all age donors. The hHpSCs cells range in size from 7–10µm in diameter and have a high nucleus-to-cytoplasm ratio. Tolerant of ischemia, they can remain viable in cadaveric livers for up to ~6 days after asystolic death(11–12).
The hHpSC phenotypic profile includes epithelial cell adhesion molecule (EpCAM), neural cell adhesion molecule(NCAM), CD133,CXCR4,SOX9,SOX17,FOXA2, cytokeratins(CK) 8/18/19, Hedgehog proteins (Sonic and Indian), intranuclear telomerase protein, claudin 3, MDR1, weak expression of albumin and MHC antigens. They do not express α-fetoprotein (AFP), intercellular adhesion molecule (ICAM-1), P450s, or markers for hemopoietic (e.g. CD34/38/45/90, glycophorin), endothelial (e.g. VEGFr, CD31, von Willebrand factor) or mesenchymal cells (e.g. CD146, desmin, vitamin A, CD105)(6–7,13). It remains unclear whether C-kit(CD117), expressed in the liver’s stem cell niches(8,14–15), is on hHpSCs or associated angioblasts, as CD117+ flow cytometry selects for angioblasts(7,13).
Some proteins, such as CK19, are synthesized and found in punctuate form, but not converted to filaments, as seen in hepatoblasts(7). Similarly, little albumin is synthesized but not packaged as in later lineage stages, implicating lineage-dependent distinctions in post-transcriptional and translational protein processing.
The hHpSCs are isolated by dual immunoselection for EpCAM+/NCAM+ cells from livers of all donor ages. In adult livers, which have scarce hepatoblast populations, EpCAM+ selection alone results in predominant hHpSCs isolation(7,16).
In culture, the hHpSCs form colonies capable of self-replication(17) and of differentiation to mature cells in culture and in vivo(7,18). Cells expand ex vivo if cultured in Kubota’s Medium, a serum-free medium containing only insulin, transferrin/fe, lipids, no copper, and low calcium (19–20) or if co-cultured with angioblasts. These feeders are replaceable with purified type III collagen substrates, low cross-linking hyaluronan hydrogel embedding or a mixture of both (13,21). If transplanted in vivo, they yield mature liver tissue. If cultured under distinct conditions (see below) they lineage-restrict into hepatoblasts(13).
Stage 2.Hepatoblasts(hHBs) are diploid bipotent cells giving rise to hepatocytic and cholangiocytic lineages, associated with precursors of both endothelia and hepatic stellate cells, and the liver’s probable transit amplifying cells(13). They reside throughout parenchyma of fetal and neonatal livers or as single cells and small cell aggregates tethered to the ends of canals of Hering in adult livers(8). With donor age, hHBs decline to <0.01% of the parenchymal cells in postnatal livers(7–8). They expand during regenerative processes associated with certain diseases such as cirrhosis. Previously, hHBs were referred to as “intermediate hepatobiliary cells of the ductular reactions”(22); extensive characterization enabled us to update their nomenclature with hepatoblasts(8). They can be isolated by dual immunoselection for EpCAM+/ICAM-1+. They have enormous expansion potential cultured in Kubota’s Medium, especially if supplemented with EGF and HGF, or on feeders of stellate cell precursors replaceable by substrata of type IV collagen, laminin, hyaluronans or mixtures of these, albeit without proven self-replication(13,23–24).
The hHBs, larger(10–12µm) and with higher amounts of cytoplasm than hHpSCs, have an antigenic profile that overlaps with hHpSCs(6–7,15). Shared phenotypic traits include CXCR4, CD133, SOX17, MDR1, cytokeratins(CK) 8/18 and 19, Hedgehog proteins (Sonic and Indian), and null expression of late P450s (e.g. P450-3A) or markers for hemopoietic, endothelia or mesenchymal cells (as in hHpSCs). Protein expression changes include reduction in EpCAM levels with primary localization to plasma membrane surfaces; filamentous CK14 and CK19(8, 15, 25); elevated albumin levels with discrete cytoplasmic packaging(7); switch from NCAM to ICAM-1; expression of early P450s (e.g. P450-A7) and CK7; and strong positive expression of hepatic-specific AFP, distinct from a hemopoietic progenitor variant form with alternative splicing of exon 1, a probable clue of mesendoderm to endoderm differentiation(26). They have approximately 5X the telomerase activity of hHpSCs and telomerase protein localized both in the nucleus and in the cytoplasm(27). A comparison of the phenotypic profiles of HpSCs and HBs can be found in Table 1 and in Figures 3,4.
Table 1.
Phenotypic Profiles of Multipotent Cell Populations in Human Livers
| Property | Human Hepatic Stem Cells (hHpSCs) |
Human Hepatoblasts (hHBs) |
|---|---|---|
| Average diameter (measured by forward scatter in flow cytometric analyses of isolated cells) | 7–9 µm | 10–12 µm |
| Nucleus to cytoplasmic ratio | Highest observed of all parenchymal progenitor subpopulations evaluated | Intermediate between that in hHpSCs and mature parenchymal cells |
| Percentage of parenchymal cells(7) | 0.5–1.5 % in livers of all donor ages and with minimal ischemia; percentages higher in ischemic livers | >80% (fetal livers) |
| ~50% (neonatal livers) [percentages change rapidly day by day postnatally] | ||
| <0.01% (adult livers) | ||
| Survival after cardiac arrest (tolerance for ischemia)(7) | Viable cells for several days after cardiac arrest | Viable for more than a day, but not as long as hHpSCs |
| Morphology of colonies in vitro (7) | Uniform; densely packed; look similar to ES cell colonies | Cord-like colonies interspersed with clear channels that are presumptive canaliculi |
| Evidence for Self-renewal(17) | Clonogenic expansion with stability of phenotype; doubling times of ~36 hours on plastic; can be passaged repeatedly; fastest doubling times (~20–24 hours) for hHpSCs on substrata of type III collagen | Significant expansion potential but not yet evidence for self-replication (under the conditions tested to date). Probable transit amplifying cells |
| Pluripotency(7) | Multipotent | Bipotent |
| Anaerobic metabolism (metabolomic studies)(21) | +++ | +++ |
| Conditions for clonogenic expansion(19) | Kubota’s Medium plus feeders of angioblasts replaceable with type III collagen (monolayers) or hyaluronans into which is mixed type III collagen (3-D) | Kubota’s Medium plus feeders of hepatic stellate cells replaceable with type IV collagen/laminin (monolayers) or hyaluronans into which is mixed type IV collagen/laminin (3-D) |
| CD44 (hyaluronan receptor)(24) | High concentrations | High concentrations |
| Claudin 3(16) | +++ | Negative |
| Indian Hedgehog(5) | +++ Highest level in cells in the center of the colonies | ++ Lower levels, but pattern of distribution is the same |
| Sonic Hedgehog(5) | ++ Located at edge of cells; concentrated in cells at edge of colonies at sites of high concentration of angioblasts | + Lower levels, but pattern of distribution is the same |
| Patched (Hedgehog receptor)(5) | +++ Found in all cells and in colonies throughout the colony | ++ Levels lower, but still evident |
| Telomerase(27) | + mRNA encoding telomerase and the protein found in nucleus. No telomerase protein in the cytoplasm | +++ mRNA encoding telomerase and the protein found in nucleus; with differentiation, increasing numbers of the cells have it in the cytoplasm; 5X higher activity than in hHpSCs |
| P450s(16) | Negative for all assayed | P450 A7 but not late forms of P450s |
| CK 8 and 18(16) | ++ | ++ |
| CK 19 (7, 16) | ++ (not in filament form) | ++ (filaments evident) |
| E-cadherin(7) | ++ | ++ |
| EpCAM(7–8) | +++ (throughout the cells) | ++ (plasma membrane) |
| NCAM /ICAM-1(7–8) | ++ /− | −/++ |
| Albumin(7) | ± | ++ |
| α-fetoprotein(7–8) | Negative | +++ |
| *Mesenchymal Markers | Negative | Negative |
| **Angioblasts/Endothelial cell Markers | Negative | Negative |
| ***Hemopoietic markers | Negative | Negative |
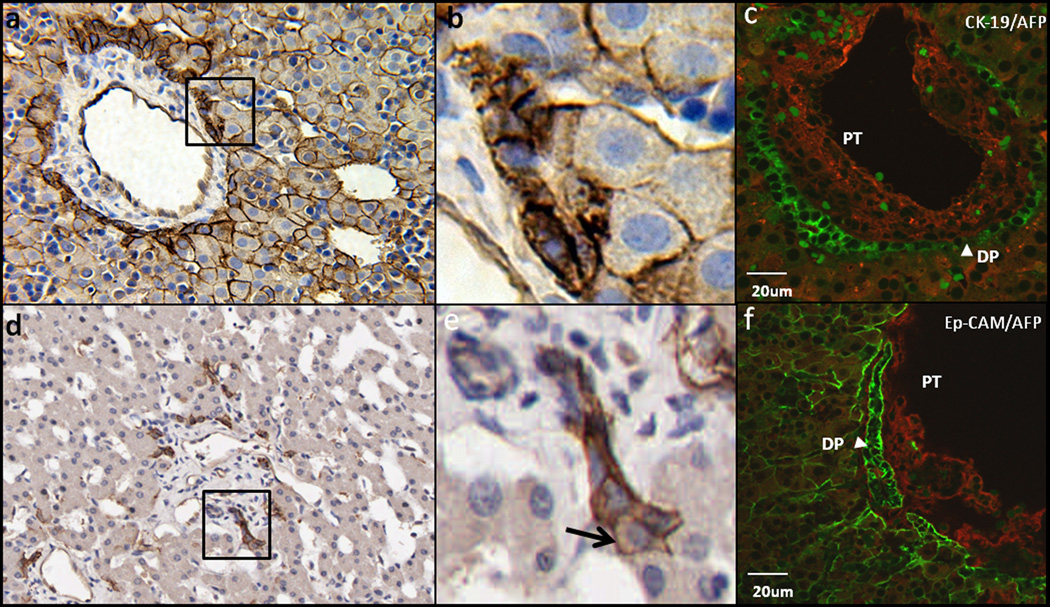
Figure 3.
Human hepatic stem cell and hepatoblast phenotypes in vivo. a,b) EpCAM expression in fetal liver: EpCAM expressed in the ductal plate is not only at the cell surface but also in the cytoplasm. EpCAM expressed in the hepatoblasts is specific to the cell surface. d,e) EpCAM expression in adult liver: One end of the Canal of Hering connects to the bile duct, the other end connects to hepatoblasts (arrow), indicating that the hepatoblasts are derived from primitive hepatic stem cells harbored in Canals of Hering. c) Double staining for CK-19/AFP and f) Ep-CAM/AFP of human fetal liver in the portal triad area and analyzed by confocal microscopy. CK-19 (c, green) is expressed not only by remodeling ductal plate but faintly expressed by some of the hepatoblasst. Ep-CAM (f, green) is detected in all the parenchymal cells and biliary epithelial cells forming bile duct and ductal plate (DP). AFP(red) is expressed by hepatoblasts throughout the fetal liver and undetectable in the ductal plate. (PT: Portal triad; DP: Ductal Plate)
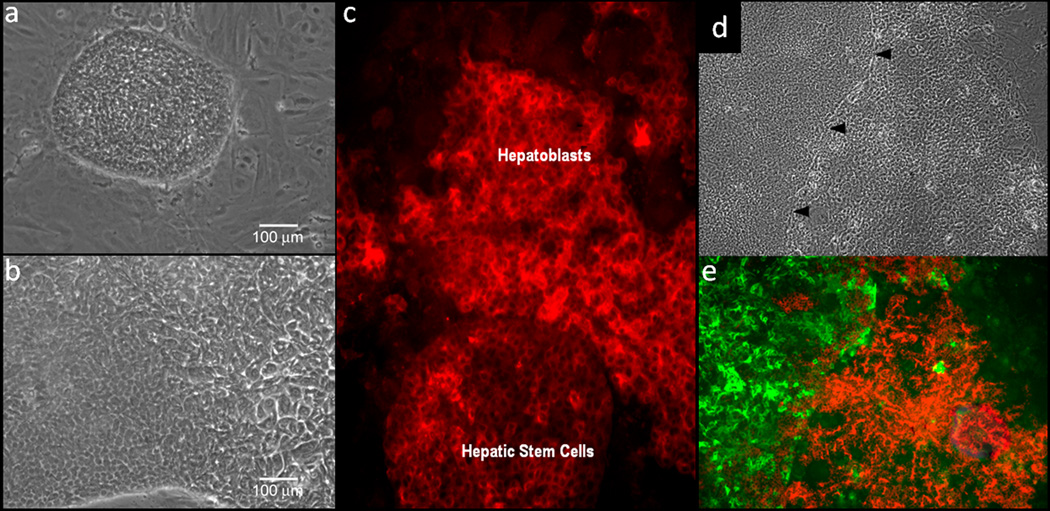
Figure 4.
Human hepatic stem cell and hepatoblast phenotypes in culture. a) Morphology of human hepatic stem cells and b) human hepatoblasts in culture on plastic. c) Albumin staining of human hepatic stem cells, transitioning to hepatoblasts. d) hepatic stem cells stained with NCAM (green) and hepatoblasts stained with ICAM (red).
Stage 3.Committed progenitors are ~12–15µm diploid, unipotent, immature cells. These precursors give rise to only one adult cell type. They lose most stem cell gene expression (e.g. NCAM, Hedgehog proteins), express either hepatocytic or biliary markers, and abound in fetal and neonatal tissues or chronic liver diseases (viral, alcoholic and non-alcoholic fatty liver diseases, autoimmune hepatitis, cholangiopathies), unlike normal adult tissues(28).
Committed hepatocytic progenitors, also called intermediate hepatocytes, express albumin, enzymes associated with glycogen synthesis (e.g. glucose-6-phosphate), and lack biliary markers (e.g.CK19) and AFP. They are associated with endothelial cell precursors and are located in vivo in the liver plates between the HBs and the diploid adult hepatocytes.
“Small cholangiocytes” are diploid biliary cells, 6–8µm with cuboidal shape, a high nucleus-to-cytoplasm ratio, small endoplasmic reticulum(29–30), and are associated with hepatic stellate cell precursors(13). They co-localize with hHpSCs in the stem cell niche, lining the canals of Hering, intrahepatic bile ducts and bile ductules with internal diameters below 15µm. Direct links between the canals of Hering and bile ductules, which may traverse the limiting plate and thus may have an intralobular segment (periportal) in addition to their intraportal location, support current hypotheses that point to small cholangiocytes as committed biliary progenitors(31). In human and rodent livers, they express high levels of the anti-apoptotic proteins annexin V and bcl2 (B-cell lymphoma 2 protein). At a functional level, they express endothelin receptors type A (EDNRA) and type B (EDNRB), endogenous opioid peptides, insulin, histamine (H1), acetylcholine (M3), and α-1-adrenergic agonists, aquaporin 4; they are negative for the Cl−/HCO3− exchanger and receptors for secretin or somatostatin. During chronic feeding with bile salts taurocholate and taurolithocholate, small cholangiocytes express otherwise negative Na+-dependent apical bile acid transporter (ABAT) de novo, suggesting a role in the cholehepatic recirculation of bile salts in conditions of overload(32). Finally, cystic fibrosis transmembrane conductance regulator (CFTR) is present in human, but not rodent, small cholangiocytes(31).
Stages 4–6.Diploid adult cells are the only parenchymal cells with significant proliferative capacity under all known in vitro or in vivo conditions. Exceptions are in conditions potentially involving genetic reprogramming through chromatin demethylation, the only known mechanism for restoring cytokinensis, as occurs in tyrosinemia(33) or with massive loss of mature parenchymal cells (e.g. >80%) due to a transgene(34). Diploid adult hepatocytes (“small hepatocytes”), partnered with endothelia, can undergo 6–7 rounds of division within 3 weeks in culture but have limited subcultivation capacity(19). Large cholangiocytes, partnered with stellate cells, are columnar shape, display a small nucleus and conspicuous cytoplasm, an abundant Golgi apparatus between the apical pole and the nucleus, and rough endoplasmic reticulum more abundant than small cholangiocytes(30,35–36). Large cholangiocytes line interlobular ducts located in the portal triads. The connections of hHpSCs in canals of Hering to the septal and segmental bile ducts has not yet been investigated, and markers in septal ducts, segmental ducts and larger ducts are found also in cells in peribiliary glands, the stem cell niches of the biliary tree(37). Large cholangiocytes express CFTR and Cl−/HC03− exchanger, aquaporin 4 and aquaporin 8, secretin and somatostatin receptors other than receptors for hormones and neuropeptides. In addition, they express the Na+-dependent bile acid transporter ABAT (apical bile acid transporter), MDR (multidrug transporter) and MRP (multidrug resistance associated proteins). When large cholangiocytes are damaged by acute carbon tetrachloride (CCl4) or GABA administration, small cholangiocytes proliferate, and acquire phenotypical and functional features of large cholangiocytes(38–39), suggesting that the population of small cholangiocytes lining the canals of Hering and ductules may represent precursors of large cholangiocytes lining larger ducts. The integrated differential gene expression between small and large normal cholangiocytes demonstrate through microarray that the proteins related to cell proliferation tend to be highly expressed by small cholangiocytes, whereas large cholangiocytes express functional and differentiated genes(36). This is consistent with studies showing, either with bile duct injury due to CCl4 and GABA administration or with bile duct regrowth following partial hepatectomy, that small cholangiocyte proliferation is activated presumably to repopulate bile ducts. These findings suggest that small cholangiocytes are less mature, have a high resistance to apoptosis, and have marked proliferative activities, while large cholangiocytes are more differentiated contributing mainly to ductal bile secretion and absorption. Therefore, while hepatocytic cell lineages proceed from periportal areas toward the central vein, cholangiocytes proceed in the opposite direction from canals of Hering/ductules toward larger ducts. See the online supplement for further information.
II. Regulation of the Parenchymal Cell Lineages
A. Paracrine Signaling between Epithelial-Mesenchymal Partners
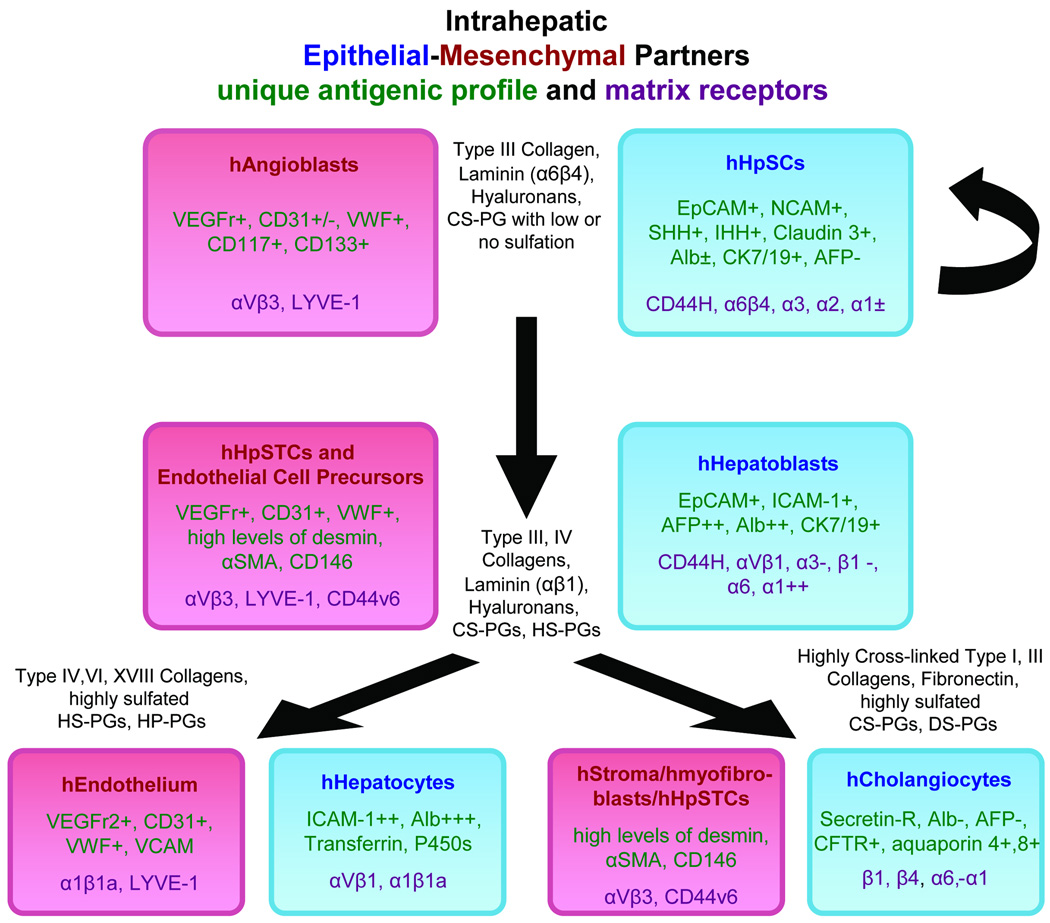
Paracrine signaling is the primary form of regulation between parenchymal cells and partnering mesenchymal cells and represents classic epithelial-mesenchymal relationships widely described in developmental biology since the 1930s. A new facet is that coordinate maturation of these [parenchymal]:[mesenchymal] cell associations, starting with [hHpSCs]:[angioblasts] and splitting into lineages of [hepatocyte]:[endothelia] and [cholangiocyte]:[stellate cells], gives rise to lineage-dependent gradients of paracrine signals(13) that govern the biological responses of cells at each lineage stage. Defined subsets of these lineage-dependent paracrine signals, soluble and insoluble matrix ones, can be used to establish cells at a specific lineage stage in culture (Figure 5).
Figure 5.
Schematic image indicating the Coordinate Maturation of the Epithelia (Parenchymal cells) and their Mesenchymal Partners and some of the identified extracellular matrix components found at the particular lineage stages. Not shown in the figure are the soluble signals that also are lineage dependent. Some of those identified and that are lineage dependent are: hepatic stem cells are LIF, IL-6, IL-11, and acetylcholine; hepatoblasts are HGF, EGF, bFGF, IL-6, IL-11, and acetylcholine; hepatocytes are HGF, EGF, bFGF, T3, glucagon, and hydrocortisone; cholangiocytes are VEGF, HGF, bFGF. and acetylcholine.
The intrahepatic stem cell niche contains type III collagen, α6β4 integrin-binding form of laminin, hyaluronans and a minimally sulfated chondroitin sulfate proteoglycan (CS-PG)(13). Transition to [hHBs]:[endothelia and stellate cell precursors] results in changes to type IV collagen, αVβ1 integrin-binding laminin, hyaluronans, more sulfated CS-PGs and forms of heparan sulfate-PGs (HS-PGs). The [hepatocyte]:[endothelia] lineages are associated with network collagens (e.g. type IV and VI) and increasingly sulfated forms of HS-PGs ending, in zone 3, in heparin-PGs (HP-PGs). The [cholangiocyte]:[stellate cell] lineages are associated with fibrillar collagens (e.g. types I and III) and progression from CS-PGs towards highly sulfated PGs, including dermatan sulfate-PGs (DS-PGs)(13,17,24). Many soluble signals bind to and work synergistically with matrix components to regulate cells, particularly PGs and their glycosaminoglycan chains (GAGs). Matrix-bound soluble signals are biphasic, yielding mitogenic versus differentiative responses depending on the specific matrix chemistry with which they are associated.
B. Feedback Loop Signaling
Late lineage stage cells produce positive and negative signaling regulators, including bile salts, various soluble factors and matrix components(40). Positive regulators include hepatopoietin, released by dying zone 3 cells that stimulate stem/progenitors expansion (M. Roach, J. Hambor, unpublished observations). Negative regulators include ecto-nucleotidases expressed by portal hepatoblasts like NTPDase2, which inhibits purinergic activation of basolateral P2Y receptors in periportal cholangiocytes under homeostatic conditions. Conversely, loss of NTPDase2 expression after experimental cholestasis in portal hepatoblasts allows activation of periportal P2Y receptors and increases cholangiocyte proliferation(41).
Another facet of regulation is mediated by acetylcholine. It stimulates proliferation of stem/progenitor cells and cholangiocytes expressing M3 acetylcholine receptors(42). In normal liver and even after partial hepatectomy, late lineage stage hepatocytes lacking M3 receptors release acetyl cholinesterase into the bile that delivers it to zone 1 where it destroys acetylcholine in the stem cell niche, thus blocking proliferation of stem/progenitor cells and cholangiocytes. In contrast, during conditions of pericentral damage, the acetyl cholinesterase is not released, resulting secondarily in induction by acetylcholine of stem/progenitor cell expansion. Denervated transplanted livers lack acetylcholine modulation of proliferation of cells lining the canals of Hering. Hepatitis-injured transplanted livers also exhibit lower numbers of progenitor and reactive ductular cells than innervated matched controls. Experiments in rats with galactosamine-damaged livers confirm that vagotomy induces impaired regeneration of progenitors and ductal reaction in cholangiocytes(43).
Mechanotransduction mechanisms are another major set affecting lineage biology, most involving cytoskeletal rearrangements. The cytoskeleton is a ubiquitous cellular component with characteristics of amplification systems and connections with matrix. Some of these connections allow cells to sense microenvironment rigidity through non-muscle myosin II, which directs stiffness-dependent differentiation in mesenchymal stem cells(44). Germ layer organization and cell sorting depends on cell adhesion forces and cortex tension relying on actomyosin network activity(45). Integrins connect the cytoskeleton to matrix substrata, recruit focal adhesions that adapt cells to mechanical stresses, bind ligands and regulate intracellular signaling(46). Mechanical stretch in liver cells induces activation and synthesis of morphogens in the TGF-β family of Activin/Nodal signaling(47). SMAD transcription factors regulate TGF-β signaling pathways and regulate gene expression through kinesin-mediated nucleocytoplasmic shuttling along intact microtubules(48–49).
Primary cilia in cells from soft organs also participate in mechanotransduction by probing and amplifying the effects of intraluminal flow above cell apical surfaces. They mediate polarized signal transduction pathways that use the cytoskeleton to ensure specific and non-diffusable signal trafficking to the nucleus(50). PDGRα and Hedgehog signaling take place in primary cilia(51–52) in livers of all ages(5) through dynein-mediated shuttling of Gli transcription factors(53). Some chromatin targets of Gli transcription factors include PTCH, WNT and BMP genes, all involved in embryonic development and differentiation mechanisms(54–56). Hedgehog expression gradients also demarcate the extension of endodermal organs during development(52,57). In conjunction, this information suggests primary cilia are relevant participants in endoderm maturation and differentiation.
Bile secretion is an important mechanism for homeostatic control of tissue mass, operating as an inductor in mechano-transduction. Bile is a Newtonian fluid in normal physiological conditions with salt concentration-dependent viscosity(58). Bile tonicity increases while flowing in the pericentral-to-periportal direction as hepatic parenchyma perform secretory functions. Abnormal bile tonicity is characteristic of pathological conditions(59). Shear forces from bile flow, proportional to bile viscosity, can function as long-range mechanical signals communicating states of hepatic function across the entire liver maturational gradient to cholangiocytes in the proximal biliary tree through primary cilia bending. This bending triggers stress-induced Ca2+, cAMP signaling cascades and receptor-mediated PDGRα and Hedgehog signaling, which makes bile a mechanical probe for liver homeostatic control(42).
Liver Regeneration. Two distinct forms of liver regeneration take place after: a) partial hepatectomy, and b) selective loss of pericentral cells. After partial hepatectomy, feedback loop signaling is essentially intact. DNA synthesis occurs in cells across the liver plates but only a portion of the cells undergo cytokinesis, yielding in increased numbers of polyploid cells, higher numbers of apoptotic cells, and more rapid turnover of the liver with restoration of the normal ploidy profiles within weeks(60). Feedback loop signaling is the explanation for liver cells in culture in which secreted signals from late lineage stage cells inhibit the growth of any early lineage stage cells(20).
Selective loss of pericentral cells with toxic injury to zone 3 cells (and sometimes also to zone 2) results in muting of the feedback loop signaling that activates rapid cell division of early lineage stage cells(12,61). In response, periportal cells undergo rapid hyperplastic growth (complete cell division) followed by differentiation. These phenomena, the classic “oval cell response” in rodents and the “ductular reactions” seen in human massive hepatic necrosis (e.g. acetaminophen toxicity, acute hepatotropic viral infection), have long been recognized to involve extensive expansion of the stem/progenitor cell populations(12). Chronic injury to the liver, as occurs with repeated drug exposures, radiation, or certain viral infections like hepatitis B or C, result in loss of late lineage stage cells, eliciting chronic regenerative responses.
C. Relevance to Clinical Programs
Hepatic lineage biology and mechanisms of its regulation will have relevance for many clinical programs. Examples include tissue sourcing for clinical programs, strategies for liver cell therapies, immunological issues, and most profoundly an understanding of liver tumors and logical strategies by which to treat liver cancers.
Sourcing of tissue for any clinical therapy is dictated by the proportion of cells at the different lineage stages in tissue of a given donor age. Fetal and neonatal tissues with lineages skewed towards early stages will be ideal for stem/progenitor cell therapies, whereas adult livers will be ideal for programs requiring rapid need for late lineage stage functions.
Liver cell therapies for inborn errors of metabolism will be affected by feedback loop signaling, since there will be no selection for the transplanted cells over endogenous cells, necessitating higher numbers of cells to be transplanted. By contrast, patients with liver failure due to virus, drugs, or radiation (involving a loss of feedback loop signals) can be transplanted with smaller numbers of cells given the strong selective pressure for transplanted cells to expand quickly to reconstitute liver mass.
Concerns regarding a need for immunosuppression will be affected by lineage biology. Non-immunogenic stem/progenitors can acquire immunogenicity with maturation that potentially can be managed by use of stellate cells, known to produce immunomodulatory signals. Liver cell therapies should also use grafting methods that optimize liver engraftment and prevent cell loss to ectopic sites unlike vascular route delivery, especially for stem/progenitors(62).
D. Liver Cancer Stem Cells
The idea that cancers are transformed stem/progenitor cells originated with the pioneering work of Van Potter in the 1960s, who proposed that hepatomas contain cells undergoing “blocked ontogeny”(63). This idea was further elucidated for all types of cancers by Barry Pierce and Stewart Sell(64) who clarified that many functions thought to be related to cancer (e.g. AFP expression) are normal functions of an expanded stem/progenitor cell population and that identification of key distinctions must involve comparison of cancer cells to their normal stem cell(61,65). Indeed, normal stem/progenitor cells are strikingly similar to tumor cells in morphology, gene expression, and growth properties, and tumors can be identified as an expanded lineage stage(61,66). The clinical use of stem cells may come with an increased risk of tumors depending on the donors (e.g. if there are undiagnosed tumor cells among the endogenous stem cells) and on the patient’s medical condition (e.g. severe immunosuppression).
Strategies for cancer therapies will be revolutionized if revamped with lineage biology knowledge. Treatments with drugs or radiation are known to affect later lineage stages preferentially. If a specific treatment also targets the lineage stage(s) containing malignantly transformed cells, then the treatment can be curative. If they fail to target that stage, there will be a lethal rebound effect: the treatment kills cells in later lineage stages, mutes feedback loop signaling, and secondarily unhinges early lineage stages where malignant cells reside. Therefore, future cancer therapies should involve strategies identifying the lineage stage of the tumor and whether the treatment targets that stage or, alternatively, uses lineage mechanism regulation, such as feedback loop signals, to control the rate of growth of tumor cells.
III. Conclusions
The intrahepatic maturational lineages begin within the stem cell compartments, located periportally, and progress through the midacinar region and ending near the central vein. The parenchymal cells, along with their mesenchymal cell partners, are governed by gradients of paracrine signals, including sets of soluble factors and insoluble extracellular matrix components, and by specific mechanical forces. Feedback loop signals regulate the stem/progenitors, controlling liver mass and tissue regeneration. Understanding stem cell and lineage biology in the liver and their regulation offers new considerations for basic and industrial investigations and for more biologically rational clinical program strategies.
Supplementary Material
ACKNOWLEDGEMENTS
UNC. Funding for the investigators at UNC derived from a sponsored research grant from Vesta Therapeutics (Research Triangle Park, NC), from NIH grants (DK52851, AA014243, IP30-DK065933), a Department of Energy Grant (DE-FG02-02ER-63477) and sponsored research grants from Vesta Therapeutics (Bethesda, MD), Vertex Pharmaceuticals (Cambridge, Mass), and Zenith Biotech (Guildford, CT).
Sapienza University. V. Cardinale received salary support with a scholarship from Sapienza University of Rome for studies done at UNC. D. Alvaro is supported by MIUR (Italian Minister of University and Research) grants: PRIN #2007, prot. 2007HPT7BA-003 and by Federate Atheneaum funds from the University “Sapienza” of Rome. Dr. Gaudio was supported by MIUR grants: PRIN#2007, prot. 2007HPT7BA_001 and Federate Atheneaum funds from the University “Sapienza” of Rome.
Scott & White and Texas A&M Health Science Center. Work was supported by the Dr. Nicholas C. Hightower Centennial Chair of Gastroenterology to Dr. Alpini from Scott & White Hospital, a VA Research Scholar Award, a VA Merit Award and the NIH grants DK062975, K76898 and DK58411 to Dr. Alpini.
Abbreviations
With respect to specific cellular subpopulations, the species of origin is indicated by a small letter in front of the abbreviation (r = rat; m=mouse; h= human);
- AFP
α-fetoprotein
- αSMA
α- smooth muscle actin
- GABA
γ-aminobutyric acid
- ALB
Albumin
- ABAT
Apical bile acid transporter
- bcl2 protein
B-cell lymphoma 2 protein
- CCl4
Carbon tetracholoride
- CS-PG
Chondroitin sulfate proteoglycan
- CFTR
Cystic fibrosis transmembrane conductance regulator
- CK
Cytokeratin
- DS-PG
Dermatan sulfate proteoglycan
- NTPDase2
Ectonucleotidase
- ENDRA and ENDRB
Endothelin receptors type A and type B
- EpCAM
Epithelial cell adhesion molecules
- HS-PG
Heparan sulfate proteoglycan
- HP-PG
Heparin proteoglycan
- hHBs versus rHBs
Hepatoblasts, human versus rat
- hHpSTCs versus rHpSTCs
Hepatic stellate cells, human versus rat
- hHpSCs versus rHpSCs
Hepatic stem cells, human versus rat
- ICAM
Intercellular adhesion molecules
- KDR
Kinase insert domain receptor
- MHC
Major histocompatibility complex
- MRP
Multidrug resistance associated proteins
- MDR
Multidrug transporter
- NCAM
Neural cell adhesion molecule
- PG
Proteoglycan
- SHH and IHH proteins
Sonic and Indian hedge hog proteins
- TGF-β
Transforming growth factor beta
- VCAM
Vascular cell adhesion molecule
- VEGFr
Vascular endothelial growth factor receptor
- vWF
von Willebrand factor
Footnotes
Author contributions:
R Turner and LM Reid did most of the writing of the sections on hepatic stem cells, hepatoblasts and maturational lineages of parenchymal cells and editing of the manuscript. O. Lozoya prepared the sections on mechanical effects on cells. Y. Wang helped with sections on the biliary tree stem cells and on regulation of the cells by paracrine signaling. C Barbier and E Wauthier helped to edit the review. The sections on the biliary tree, biliary tree stem cells, and cholangiocytes were written and edited by Drs. G. Alpini, D. Alvaro, E. Gaudio and V. Cardinale. The schematic figures were drawn by G. Mendel. The authors declare that they have no conflicts.
REFERENCES
- 1.Sigal SH, Brill S, Fiorino AS, Reid LM. The liver as a stem cell and lineage system. American Journal of Physiology. 1992;263:G139–G148. doi: 10.1152/ajpgi.1992.263.2.G139. [DOI] [PubMed] [Google Scholar]
- 2.Liu H, Di Cunto F, Imarisio S, Reid LM. Citron kinase is a cell cycle-dependent, nuclear protein required for G2/M transition of hepatocytes. Journal Biological Chemistry. 2003;278:2541–2548. doi: 10.1074/jbc.M210391200. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka M, Okabe M, Suzuki K, Kamiya Y, Tsukahara Y, Saito S, Miyajima A. Mouse hepatoblasts at distinct developmental stages are characterized by expression of EpCAM and DLK1: drastic change of EpCAM expression during liver development. Mechanisms of Development. 2009;126:665–676. doi: 10.1016/j.mod.2009.06.939. [DOI] [PubMed] [Google Scholar]
- 4.Okabe M, Tsukahara Y, Tanaka M, Suzuki K, S S, Kamiya Y, T T, et al. Potential hepatic stem cells reside in EpCAM+ cells of normal and injured mouse livers. Development. 2009;136:1951–1960. doi: 10.1242/dev.031369. Development 2009;136:1951–1960. [DOI] [PubMed] [Google Scholar]
- 5.Sicklick JK, Li YX, Melhem A, Schmelzer E, Zdanowicz M, Huang J, Caballero M, et al. Hedgehog signaling maintains resident hepatic progenitors throughout life. American Journal of Physiology.Gastrointestinal Liver Physiology. 2006;290:G859–G870. doi: 10.1152/ajpgi.00456.2005. [DOI] [PubMed] [Google Scholar]
- 6.Schmelzer E, Wauthier E, Reid LM. The phenotypes of pluripotent human hepatic progenitors. Stem Cells. 2006;24:1852–1858. doi: 10.1634/stemcells.2006-0036. [DOI] [PubMed] [Google Scholar]
- 7.*Schmelzer E, *Zhang L, *Bruce A, Wauthier E, Ludlow J, Yao H, Moss N, et al. Human hepatic stem cells from fetal and postnatal donors. Journal of Experimental Medicine. 2007;204:1973–1987. doi: 10.1084/jem.20061603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L, Theise N, Chua M, Reid LM. Human hepatic stem cells and hepatoblasts: Symmetry between Liver Development and Liver Regeneration. Hepatology. 2008;48:1598–1607. doi: 10.1002/hep.22516. [DOI] [PubMed] [Google Scholar]
- 9.Kuwahara R, Kofman AV, Landis CS, Swenson ES, Barendswaard E, Theise ND. The hepatic stem cell niche: identification by label retaining cell assay. Hepatology. 2008;47:1994–2002. doi: 10.1002/hep.22218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saxena R, Theise N. Canals of Hering: recent insights and current knowledge. Seminars in Liver Disease. 2004;24:43–48. doi: 10.1055/s-2004-823100. [DOI] [PubMed] [Google Scholar]
- 11.Stachelscheid H, Urbaniak T, Ring A, Spengler B, Gerlach JC, Zeilinger K. Isolation and characterization of adult human liver progenitors from ischemic liver tissue derived from therapeutic hepatectomies. Tissue Engineering, Part A. 2009;15:1633–1643. doi: 10.1089/ten.tea.2008.0291. [DOI] [PubMed] [Google Scholar]
- 12.Zhou H, Rogler LE, Teperman L, Morgan G, Rogler CE. Identification of hepatocytic and bile ductular cell lineages and candidate stem cells in bipolar ductular reactions in cirrhotic human liver. Hepatology. 2007;45:716–724. doi: 10.1002/hep.21557. [DOI] [PubMed] [Google Scholar]
- 13.Wang* Y, Yao* H-l, Barbier C, Wauthier E, Cui C-b, Moss N, Yamauchi M, et al. Lineage-Dependent Epithelial-Mesenchymal Paracrine Signals Dictate Growth versus Differentiation of Human Hepatic Stem Cells to Adult Fates. Hepatology. 2010;52:1443–1454. doi: 10.1002/hep.23829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crosby HA, Kelly DA, Strain AJ. Human hepatic stem-like cells isolated using c-kit or CD34 can differentiate into biliary epithelium. Gastroenterology. 2001;120:534–544. doi: 10.1053/gast.2001.21175. [DOI] [PubMed] [Google Scholar]
- 15.Theise ND, Saxena R, Portmann BC, Thung SN, Yee H, Chiriboga L, Kumar A, et al. The canals of Hering and hepatic stem cells in humans. Hepatology. 1999;30:1425–1433. doi: 10.1002/hep.510300614. [DOI] [PubMed] [Google Scholar]
- 16.Schmelzer E, Wauthier E, Reid LM. Phenotypes of pluripotent human hepatic progenitors. Stem Cell. 2006;24:1852–1858. doi: 10.1634/stemcells.2006-0036. [DOI] [PubMed] [Google Scholar]
- 17.McClelland R, Wauthier E, Zhang L, Barbier C, Melhem A, Schmelzer E, Reid LM. Ex vivo conditions for self-replication of human hepatic stem cells. Tissue Engineering. 2008;14:1–11. doi: 10.1089/ten.tec.2008.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Cui C, Miguez P, Yamauchi M, Costello J, Wauthier E, Gerber D, et al. Lineage restriction of hepatic stem cells to mature fates is made efficient by tissue-specific biomatrix scaffolds. Hepatology. 2010 doi: 10.1002/hep.24012. [DOI] [PubMed] [Google Scholar]
- 19.Kubota H, Reid LM. Clonogenic hepatoblasts, common precursors for hepatocytic and biliary lineages, are lacking classical major histocompatibility complex class I antigen. Proc Natl Acad Sci U S A. 2000;97:12132–12137. doi: 10.1073/pnas.97.22.12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wauthier E, Schmelzer E, Turner W, Zhang L, Lecluyse E, Ruiz J, Turner R, et al. Hepatic stem cells and hepatoblasts: identification, isolation, and ex vivo maintenance. Methods Cell Biol. 2008;86:137–225. doi: 10.1016/S0091-679X(08)00008-3. [DOI] [PubMed] [Google Scholar]
- 21.Turner WS, Seagle C, Galanko JA, Favorov O, Prestwich GD, Macdonald JM, Reid LM. Nuclear magnetic resonance metabolomic footprinting of human hepatic stem cells and hepatoblasts cultured in hyaluronan-matrix hydrogels. Stem Cells. 2008;26:1547–1555. doi: 10.1634/stemcells.2007-0863. [DOI] [PubMed] [Google Scholar]
- 22.Roskams TA, Theise ND, Balabaud C, Bhagat G, Bhathal PS, Bioulac-Sage P, Brunt EM, et al. Nomenclature of the Finer Branches of the biliary Tree: Canals, Ductules, and Ductular Reactions in Human Livers. Hepatology. 2004;39:1739–1745. doi: 10.1002/hep.20130. [DOI] [PubMed] [Google Scholar]
- 23.Kubota H, Yao H, Reid LM. Identification and characterization of vitamin A-storing cells in fetal liver. Stem Cell. 2007;25:2339–2349. doi: 10.1634/stemcells.2006-0316. [DOI] [PubMed] [Google Scholar]
- 24.Turner WS, Schmelzer E, McClelland R, Wauthier E, Chen W, Reid LM. Human hepatoblast phenotype maintained by hyaluronan hydrogels. J Biomed Mater Res B Appl Biomater. 2007;82:156–168. doi: 10.1002/jbm.b.30717. [DOI] [PubMed] [Google Scholar]
- 25.Haruna Y, Saito K, Spaulding S, Nalesnik MA, Gerber MA. Identification of bipotential progenitor cells in human liver development. Hepatology. 1996;23:476–481. doi: 10.1002/hep.510230312. [DOI] [PubMed] [Google Scholar]
- 26.Kubota H, Storms RW, Reid LM. Variant forms of alpha-fetoprotein transcripts expressed in human hematopoietic progenitors. Implications for their developmental potential towards endoderm. Journal of Biological Chemistry. 2002;277:27629–27635. doi: 10.1074/jbc.M202117200. [DOI] [PubMed] [Google Scholar]
- 27.Schmelzer E, Reid LM. Telomerase activity in human hepatic stem cells, hepatoblasts and hepatocytes from neonatal, pediatric, adult and geriatric donors. European Journal of Hepatology and Gastroenterology. 2009;21:1191–1198. doi: 10.1097/MEG.0b013e32832973fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan J, Hytiroglou P, Wieczorek R, Park YN, Thung SN, Arias B, Theise ND. Immunohistochemical evidence for hepatic progenitor cells in liver diseases. Liver. 2002;22:365–373. doi: 10.1034/j.1600-0676.2002.01622.x. [DOI] [PubMed] [Google Scholar]
- 29.Alvaro D, Mancino MG, Glaser S, Gaudio E, Marzioni M, Francis H, Alpini G. Proliferating cholangiocytes: a neuroendocrine compartment in the diseased liver. Gastroenterology. 2007;132:415–431. doi: 10.1053/j.gastro.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 30.Glaser S, Francis H, DeMorrow S, LeSage G, Fava G, Marzioni M, Venter J, et al. Heterogeneity of the intrahepatic biliary epithelium. World Journal of Gastroenterology. 2006;12:3523–3536. doi: 10.3748/wjg.v12.i22.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alpini G, Ulrich C, Roberts S, Phillips JO, Ueno Y, Podila PV, Colegio O, et al. Molecular and functional heterogeneity of cholangiocytes from rat liver after bile duct ligation. American Journal of Physiology. 1997;272:G289–G297. doi: 10.1152/ajpgi.1997.272.2.G289. [DOI] [PubMed] [Google Scholar]
- 32.Alpini G, Ueno Y, Glaser SS, Marzioni M, Phinizy JL, Francis H, Lesage G. Bile acid feeding increased proliferative activity and apical bile acid transporter expression in both small and large rat cholangiocytes. Hepatology. 2001;34:868–876. doi: 10.1053/jhep.2001.28884. [DOI] [PubMed] [Google Scholar]
- 33.Duncan A, Hickey RD, Paulk NK, Culberson AJ, Olson SB, Finegold MJ, Grompe M. Ploidy Reductions in Murine Fusion-derived Hepatocytes. Plos Genetics. 2009 doi: 10.1371/journal.pgen.1000385. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rhim JA, Sandgren EP, Degen JL, Palmiter RD, Brinster RL. Replacement of diseased mouse liver by hepatic cell transplantation. Science. 1994;263:1149–1152. doi: 10.1126/science.8108734. [DOI] [PubMed] [Google Scholar]
- 35.Alvaro D, Alpini G, Jezequel AM, Bassotti C, Francia C, Fraioli F, Romeo R, et al. Role and mechanisms of action of acetylcholine in the regulation of rat cholangiocyte secretory functions. J Clin Invest. 1997;100:1349–1362. doi: 10.1172/JCI119655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ueno Y, Alpini G, Yahagi K, Kanno N, Moritoki Y, Fukushima K, Glaser S, et al. Evaluation of differential gene expression by microarray analysis in small and large cholangiocytes isolated from normal mice. Liver International. 2003;23:449–459. doi: 10.1111/j.1478-3231.2003.00876.x. [DOI] [PubMed] [Google Scholar]
- 37.Terada T, Kato M, Horie S, Endo K, Kitamura Y. Expression of pancreatic alpha-amylase protein and messenger RNA in hilar primitive bile ducts and hepatocytes during human fetal liver organogenesis: an immunohistochemical and in situ hybridization study. Liver. 1998;18:313–319. doi: 10.1111/j.1600-0676.1998.tb00811.x. [DOI] [PubMed] [Google Scholar]
- 38.LeSage GD, Glaser SS, Marucci L, Benedetti A, Phinizy JL, Rodgers R, Caligiuri A, et al. Acute carbon tetrachloride feeding induces damage of large but not small cholangiocytes from BDL rat liver. American Journal of Physiology. 1999;276:G1289–G1301. doi: 10.1152/ajpgi.1999.276.5.G1289. [DOI] [PubMed] [Google Scholar]
- 39.Mancinelli R, Franchitto A, Gaudio E, Onori P, Glaser S, Francis H, Venter J, et al. After Damage of Large Bile Ducts by Gamma-Aminobutyric Acid, Small Ducts Replenish the Biliary Tree by Amplification of Calcium-Dependent Signaling and de Novo Acquisition of Large Cholangiocyte Phenotypes. American Journal of Pathology. 2010 doi: 10.2353/ajpath.2010.090677. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bishop JR, Schuksz M, Esko JD, Bishop JR, Schuksz M, Esko JD. Heparan sulfate proteoglycans fine-tune mammalian physiology. Nature. 2007;446:1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- 41.Jhandier MN, Kruglov EA, Lavoie EG, Sevigny J, Dranoff JA. Portal fibroblasts regulate the proliferation of bile duct epithelia via expression of NTPDase2. J Biol Chem. 2005;280:22986–22992. doi: 10.1074/jbc.M412371200. [DOI] [PubMed] [Google Scholar]
- 42.Woo K, Dutta AK, Patel V, Kresge C, Feranchak AP. Fluid flow induces mechanosensitive ATP release, calcium signalling and Cl− transport in biliary epithelial cells through a PKCzeta-dependent pathway. The Journal of Physiology. 2008;586:2779–2798. doi: 10.1113/jphysiol.2008.153015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cassiman D, Libbrecht L, Sinelli N, Desmet V, Denef C, Roskams T. The vagal nerve stimulates activation of the hepatic progenitor cell compartment via muscarinic acetylcholine receptor type 3. Am J Pathol. 2002;161:521–530. doi: 10.1016/S0002-9440(10)64208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 45.Krieg M, et al. Tensile forces govern germ-layer organization in zebrafish. Nature Cell Biology. 2008;10:429–436. doi: 10.1038/ncb1705. [DOI] [PubMed] [Google Scholar]
- 46.Schwartz MA, DeSimone DW. Cell adhesion receptors in mechanotransduction. Current Opinions in Cell Biology. 2008;20:551–556. doi: 10.1016/j.ceb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sakata R, Ueno T, Nakamura T, Ueno H, Sata M. Mechanical stretch induces TGF-beta synthesis in hepatic stellate cells. Eur J Clin Invest. 2004;34:129–136. doi: 10.1111/j.1365-2362.2004.01302.x. [DOI] [PubMed] [Google Scholar]
- 48.Batut J, Howell M, Hill CS. Kinesin-mediated transport of Smad2 is required for signaling in response to TGF-beta ligands. Developmental Cell. 2007;12:261–274. doi: 10.1016/j.devcel.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 49.Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 50.Praetorius HA, Spring KR. A physiological view of the primary cilium. Annu Rev Physiol. 2005;67:515–529. doi: 10.1146/annurev.physiol.67.040403.101353. [DOI] [PubMed] [Google Scholar]
- 51.Eggenschwiler JT, Anderson KV. Cilia and developmental signaling. Annual review of Cell and Developmental Biology. 2007;23:345–373. doi: 10.1146/annurev.cellbio.23.090506.123249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.King PJ, Guasti L, Laufer E. Hedgehog signalling in endocrine development and disease. Journal of Endocrinology. 2008;198:439–450. doi: 10.1677/JOE-08-0161. [DOI] [PubMed] [Google Scholar]
- 53.Kim J, Kato M, Beachy PA. Smoothened in the primary cilum to transcriptional activation in the nucleus. Proceedings of the National Academy of SCiences (USA) 2009;106:21666–21671. doi: 10.1073/pnas.0912180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McLin VA, Rankin SA, Zorn AM. Repression of Wnt/beta-catenin signaling in the anterior endoderm is essential for liver and pancreas development. Development. 2007;134:2207–2217. doi: 10.1242/dev.001230. [DOI] [PubMed] [Google Scholar]
- 55.Ober EA, Field HA, Stainier DY. From endoderm formation to liver and pancreas development in zebrafish. Mech Dev. 2003;120:5–18. doi: 10.1016/s0925-4773(02)00327-1. [DOI] [PubMed] [Google Scholar]
- 56.Cohen MM., Jr The hedgehog signaling network. American Journal of Medical Genetics. 2003:5–28. doi: 10.1002/ajmg.a.20495. [DOI] [PubMed] [Google Scholar]
- 57.Rivas-Carrillo JD, Okitsu T, Tanaka N, Kobayashi N. Pancreas development and beta-cell differentiation of embryonic stem cells. Current Medicinal Chemistry. 2007;14:1573–1578. doi: 10.2174/092986707780831096. [DOI] [PubMed] [Google Scholar]
- 58.Sangeetha NM, Bhat S, Choudhury AR, Maitra U, Terech P. Properties of Hydrogels Derived from Cationic Analogues of Bile Acid: Remarkably Distinct Flowing Characteristics. The Journal of Physical Chemistry B. 2004;108:16056–16063. [Google Scholar]
- 59.Luo X, Li W, Bird N, Chin SB, Hill NA, Johnson AG. On the mechanical behavior of the human biliary system. World J Gastroenterol. 2007;13:1384–1392. doi: 10.3748/wjg.v13.i9.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sigal SH, Rajvanshi P, Gorla GR, Sokhi RP, Saxena R, Gebhard DR, Jr, Reid LM, et al. Partial hepatectomy-induced polyploidy attenuates hepatocyte replication and activates cell aging events. American Journal of Physiology-Gastrointestinal and Liver Physiology. 1999;276:G1260–G1272. doi: 10.1152/ajpgi.1999.276.5.G1260. [DOI] [PubMed] [Google Scholar]
- 61.Roskams T. Liver stem cells and their implication in hepatocellular and cholangiocarcinoma. Oncogene. 2006;25:3818–3822. doi: 10.1038/sj.onc.1209558. [DOI] [PubMed] [Google Scholar]
- 62.Turner R, Gerber D, Reid L. The future of cell transplant therapies: a need for tissue grafting. Transplantation. 2010;90:807–810. doi: 10.1097/TP.0b013e3181f24ea2. [DOI] [PubMed] [Google Scholar]
- 63.Potter VR. The present status of the blocked ontogeny hypothesis of neoplasia: the thalassemia connection. Oncodevelopmental Biology & Medicine. 1981;2:243–266. [PubMed] [Google Scholar]
- 64.Sell S. On the stem cell origin of Cancer. American Journal of Pathology. 2010;176:1–11. doi: 10.2353/ajpath.2010.091064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamashita Y, Ji J, Budhu A, Forgues M, Jia H, Ye Q, Qin L, et al. Wnt/β-catenin signaling regulates cancer initiating cells (EpCAM+ AFP+) with stem cell features and metastatic activities in hepatocellular carcinoma. Gastroenterology. 2009;136:1012–1024. doi: 10.1053/j.gastro.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamashita T, Forgues M, Wang W, Kim JW, Ye Q, Jia H, Budhu A, et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Research. 2008;68:1451–1461. doi: 10.1158/0008-5472.CAN-07-6013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.