Abstract
Here we describe a protocol for hematopoietic differentiation of human pluripotent stem cells (hPSCs) and generation of mature myeloid cells from hPSCs through expansion and differentiation of hPSC-derived lin-CD34+CD43+CD45+ multipotent progenitors. The protocol is comprised of three major steps: (i) induction of hematopoietic differentiation by coculture of hPSCs with OP9 bone marrow stromal cells, (ii) short-term expansion of multipotent myeloid progenitors with a high dose of GM-CSF, and (iii) directed differentiation of myeloid progenitors into neutrophils, eosinophils, dendritic cells (DCs), Langerhans cells (LCs), macrophages, and osteoclasts. The generation of multipotent hematopoietic progenitors from hPSCs requires 9 days of culture, and an additional 2 days are needed to expand myeloid progenitors. Differentiation of myeloid progenitors into mature myeloid cells requires an additional 5–19 days of culture with cytokines, depending on the cell type.
Keywords: human embryonic stem cells, human induced pluripotent stem cells, myeloid progenitors, hematopoiesis, neutrophils, eosinophils, macrophages, osteoclasts, Langerhans cells
INTRODUCTION
Pluripotent stem cells are defined as cells capable of self-renewal and giving rise to derivatives of all three germ layers (ectoderm, endoderm, and mesoderm). The first expandable in vitro human pluripotent stem cells, embryonic stem cells (hESCs), were successfully derived from a blastocyst in 19981. Since then, hESCs have already become a valuable model for the study of early human development and in vitro analysis of lineage commitment and differentiation. In 2006, the Yamanaka group revealed that mouse fibroblasts could be reprogrammed to pluripotency using four transcription factors2. A year later induced pluripotent stem cells (iPSCs) were obtained from human somatic cells3,4. Because iPSCs can be generated from patients with various diseases, they can be used to obtain cells and progenitors carrying a disease-specific genetic trait and investigate the impact of this trait on the disease’s phenotypic development. In addition, iPSCs could potentially provide an unlimited supply of cells for drug testing.
Three major approaches have been developed to induce hematopoietic differentiation from hESCs. They include the embryoid body method5,6,7, coculture of hESCs with stromal cells lines 8,9,10,11 and two-dimensional (2D) culture systems12. The protocol described here uses the coculture of pluripotent stem cells on OP9 mouse bone marrow stromal cells to induce blood formation. The major advantage of the OP9 system is that efficient hematopoietic differentiation from hESCs can be achieved within a short period of time (8–9 days) and without added cytokines. OP9 coculture can be used to obtain multipotent hematopoietic progenitors and mature cells including T and B lymphocytes11,13,14 and megakaryocytes15,16 which are more difficult to obtain using embryoid body method or other feeder cell lines17. Although OP9 coculture was developed initially for differentiation of hESCs11, we found that this differentiation system is also very effective for generation of hematopoietic cells from hiPSCs18.
We demonstrated that all hematopoietic progenitors in hESC differentiation cultures can be identified by the expression of CD4319. CD43 is detected on all types of clonogenic blood progenitors and reliably separates the hematopoietic CD34+ cells from CD34+CD31+CD43− endothelial and CD34+CD31−CD43− mesenchymal cells. The early CD43+ cells co-express CD235a and CD41a and possess erythro-megakaryocytic differentiation potential. Cells with broader lymphomyeloid differentiation potential and lin-CD34+CD43+CD45− phenotype appear next. Finally, a lin-CD34+CD43+CD45+ population highly enriched in myeloid progenitors emerges. This population of cells can be expanded with GM-CSF and used to generate almost all types of mature myeloid cells using a particular cytokine combination20. Recently, we showed that hiPSCs can be differentiated into blood and endothelial cells as well. While we observed significant variation in the differentiation efficiency of seven tested hiPSC lines, the pattern of hiPSC differentiation was very similar to that of hESCs18,20.
Herein, we provide the protocol for the generation of hematopoietic progenitors and mature myeloid cells from hESCs and hiPSCs. This protocol is optimized from our earlier reports and is comprised of three major steps: (i) induction of hematopoietic differentiation by coculture of hPSCs with OP9 bone marrow stromal cells, (ii) short-term expansion of multipotent myeloid progenitors with a high dose of GM-CSF20,21, and (iii) directed differentiation of myeloid progenitors into neutrophils, eosinophils, dendritic cells (DCs), Langerhans cells (LCs), macrophages, and osteoclasts20. Although other methods for the generation of neutrophils, DCs, monocyte/macrophages, and osteoclasts from hESCs have been described22,23,24,25,26,27, the progenitors which give rise to mature myeloid cells in these differentiation systems remain obscure. Our method for the generation of myelomonocytic cells is based on the generation and expansion of prospectively identifiable lin-CD34+CD43+CD45+ multipotent hematopoietic progenitors from hPSCs. Applying this approach when differentiating hPSCs which have been genetically engineered to conditionally express specific genes, or hPSCs obtained from patients with a specific genetic abnormality will make it possible to analyze the effects of genes and oncogenes on myeloid development, lineage specification, and leukemic transformation. In addition, this method can provide an unlimited supply of homogenous populations of myeloid progenitors and mature myeloid cells for functional studies and drug testing. However, the described protocol has limited utility for studies of hPSC responses to specific growth factors, since it employs a serum and xenogeneic feeders to induce hematopoietic differentiation.
Experimental design
Generation, expansion and isolation of myeloid progenitors
When placed on OP9 feeders, hPSCs undergo a series of changes leading to the formation of a whole spectrum of myeloid progenitors. Lin−CD34+CD43+CD45+ cells with robust myeloid potential arise on day 6 of differentiation and expand over the following 2–3 days. These multipotent progenitors are highly enriched in myeloid colony-forming cells (CFCs) and can be further expanded with 200 ng/ml of GM-CSF. To maximize expansion, we dissociate the hPSC/OP9 cocultures and place the cells in nonadherent conditions where they spontaneously reaggregate. Because hematopoietic cells are released from the aggregates into suspension, they can easily be isolated by Percoll-based density gradient centrifugation without an additional enzymatic digestion. Although the majority (>60%) of the CD45+ cells collected after Percoll separation are lin−CD34+CD43+CD45+ cells enriched in myeloid colony-forming cells (CFCs), approximately 20% of the cells are CD235a+CD41a+/− erythroid and megakaryocytic progenitors, with few CD235a−CD41a−CD43+CD45− cells. To isolate lin−CD34+CD43+CD45+cells with more than 95% purity, we first deplete erythroid and megakaryocytic progenitors by magnet-activated cell sorting (MACS) using anti-human CD235a and CD41a antibodies. Subsequently, CD45+ cells are isolated from the CD235a−CD41a− fraction by positive selection with anti-human CD45 antibody (Fig. 1).
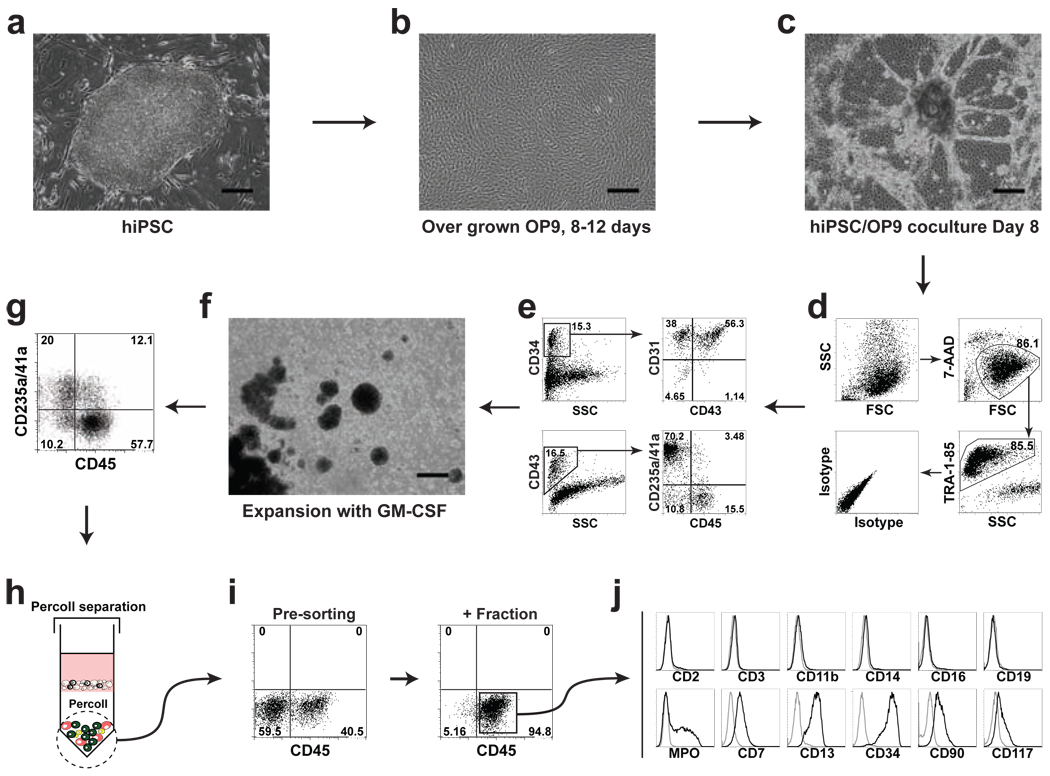
Figure 1.
Schematic diagram of the protocol used to obtain multipotent lin−CD34+CD43+CD45+ progenitors from human pluripotent stem cells. (a) Undifferentiated hiPSCs (iPS(Foreskin)-1) cocultured with (b) overgrown OP9. (c) After 8 days of coculture differentiated blood-forming hPSC colonies with radial sac-like structures are formed. (d) and (e) Typical flow cytometric analysis of differentiated iPS(Foreskin)-1 cells collected after 8 days of OP9 cocluture shows that hiPSC-derived CD34+ cells consist of CD31+CD43− endothelial cells, CD43+ hematopoietic cells, and CD31−CD43− mesenchymal cells. Three major subsets of hematopoietic progenitors could be defined within CD43+ population: CD235a/41a+ erythro-megakaryocytic progenitors, and CD235a/CD41a−CD45− and CD235a/CD41a−CD45+ multipotent progenitors. For the analysis, live (7-AAD−) and human (TRA-1-85+) cells are gated. (f) When differentiated hiPSCs are collected on day 8 of OP9 coculture by enzymatic digestion and cultured in nonadherent conditions in the presence of GM-CSF, cells spontaneously form large aggregates with hematopoietic cells released from these aggregates into suspension. (g) Typical flow cytometric analysis of cells cultured with GM-CSF for 2 days shows a marked expansion of the CD45+ population of hematopoietic progenitors within the CD43+ population. (h) After Percoll separation and (i) magnetic sorting a population of CD45+ progenitors with greater than 90% purity can be obtained. (j) Isolated CD45+ progenitors do not express major lineage markers, but retain expression of CD34, CD90, and CD177 markers characteristic of primitive hematopoietic cells. Scale bars=300µm.
Although we use OP9 coculture to generate lin−CD34+CD43+CD45+ cells from hPSCs, these multipotent progenitors could be potentially obtained by other methods, such as embryoid body or 2D cultures with cytokines, and used for production of mature myelomonocytic cells. However it is important to remember that hPSC differentiation is a continuous process in which multipotent progenitors arise within a specific time frame which is highly influenced by differentiation conditions. This time frame should be determined by careful kinetic analysis of hematopoietic differentiation by flow cytometry and CFC assay. For example, in OP9 coculture almost all CD45+ cells generated at day 8 or 9 of differentiation are lin−CD34+. However, on day 10–12 of culture, most of the CD45+ cells acquire the expression of CD14 and CD15 myelomonocytic lineage markers. This is indicative of their advanced maturation status and associated with decrease in multipotential CFC capability18. Although slight differences in the kinetics of hematopoiesis between different hESC and hiPSC lines have been reported in the embryoid body system25, we found very similar time-frame of hematopoietic development from H1 hESC and hiPSC lines in coculture with OP918. Based on these data, we concluded that the kinetics of hematopoietic differentiation depend mostly on differentiation conditions rather than on intrinsic properties of hPSC lines.
Differentiation of myeloid progenitors
By applying different cytokine combinations, lin−CD34+CD43+CD45+ cells can be differentiated into almost all types of mature myelomonocytic cells (Fig.2). The optimal cytokine combination is GM-CSF, IL-4, and TNF-α for the generation of DCs; GM-CSF, TGF-β1, and TNF-α for LCs; M-CSF and IL-1β for macrophages; G-CSF for neutrophils; and IL-3 and IL-5 for eosinophils. To maximize osteoclast production, we first expand osteoclast progenitors with GM-CSF and Vitamin D3 in pHEMA-coated flasks, then induce their maturation in cultures supplemented with GM-CSF, Vitamin D3, and RANKL. For efficient differentiation into neutrophils and eosinophils, lin−CD34+CD43+CD45+ cells should be cultured on OP9 feeders, because granulocytic differentiation in feeder-free conditions is less effective.
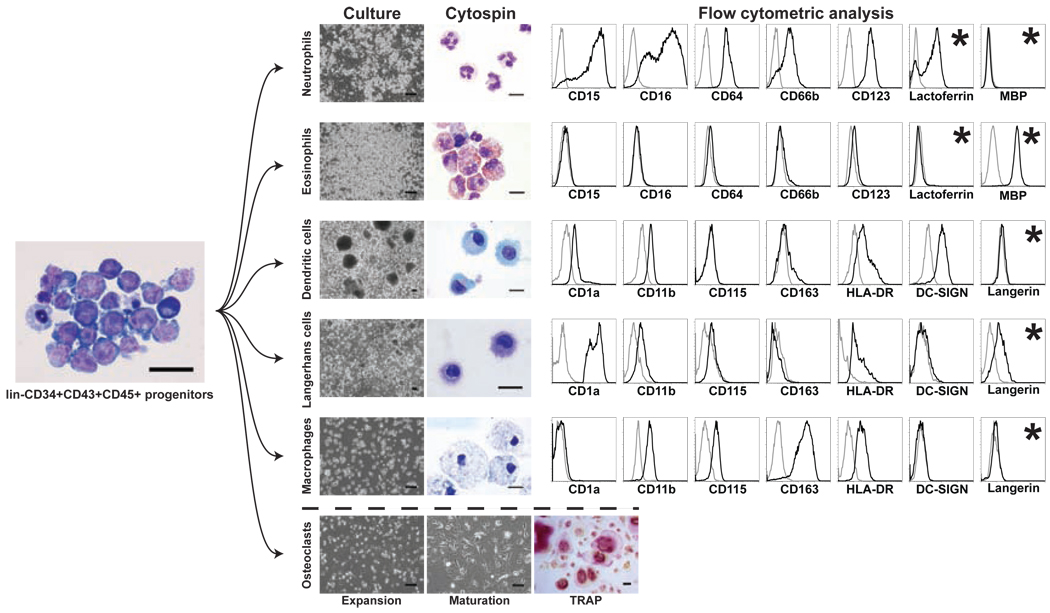
Figure 2.
Directed differentiation of lin−CD34+CD43+CD45+ progenitors into mature myelomonocytic cells. Left-hand side: hPSC-derived lin−CD34+CD43+CD45+ progenitors are shown as cytospin after magnetic sorting (scale bar=30µm). Right-hand images: Cultures (scale bars=300µm), cytospins (scale bars=10µm) and flow cytometric analyses show the morphologic and phenotypic characteristics of mature myelomonocytic cells. * represents intracellular staining for flow cytometry. Bellow dashed line: Osteoclast precursors were expanded without RANKL for 4–5 days (scale bars=300µm). Multi-nucleated mature osteoclasts are TRAP+ in maturation condition. Scale bars=300µm.
Characterizing differentiated cells
The identity of differentiated cells can be established by the analysis of cell morphology via cytospins and phenotype by flow cytometry20,28 (Fig.2). The mature neutrophils in differentiation cultures are identified by the expression of neutrophil marker CD66b and intracellular lactoferrin. The hPSC-derived eosinophils lack CD66b and lactoferrin expression, but express major basic protein (MBP) and eosinophilic peroxidase intracellularly. In addition, hPSC derived eosinophils do not express the CD15, CD16, CD64 and CD123 molecules that are typically detected on neutrophils. Macrophages generated from hPSCs are distinguished from other types of myelomonocytic cells by the CD115+CD163+HLA-DRlowCD1a− phenotype. Distinction between DCs and LCs can be made by analysis of DC-SIGN, CD11b, and intracellular Langerin expression. The typical phenotype of DCs is CD1a+DC-SIGN+CD11b+Langerin−, while LCs have CD1ahighDC-SIGN−CD11blowLangerin+ phenotype. Cytochemical staining for tartrate-resistant acid phosphatase and multinucleation can be used to identify osteoclasts in cultures. It is important to note that both granulocytic and monocytic cells generated in vitro express the CD14 marker that is commonly used to identify monocytes in peripheral blood. Therefore, the analysis of CD14 expression has limited utility for the assessment of monocytic versus granulocytic cell production from hPSCs. It has to be emphasized that in addition to phenotype, differentiated cells should be assayed using a functional test such as migration, reactive oxygen species production or antigen presentation29,30,31 ensure their proper functionality.
Time frame of differentiation
Hematopoietic differentiation in hPSC/OP9 coculture system proceeds very rapidly. The first hematopoietic progenitors with colony-forming potential can be already detected on day 5 of differentiation. Lin−CD34+CD43+CD45+ cells with robust myeloid potential arise on day 6 of differentiation and their number reaches peak over the following 2–3 days19,20. Short treatment (48 hours) with GM-CSF is sufficient to expand significantly myeloid progenitors. Differentiation of myeloid progenitors into mature myeloid cells requires an additional 5–19 days of culture with cytokines, depending on the cell type.
Controls
The differentiation efficiency in OP9 coculture and phenotype of differentiated cells are evaluated by multiparameter flow cytometry. Because non-specific binding depends on Ig isotype, it is important to use appropriate fluorochrome-labeld Ig isotypes as a negative controls. Accurate compensation is critical for multicolor flow cytometric analysis. To set up compensation, human specific antibodies to CD29 or TRA-1-85 surface markers abundantly expressed on differentiated hPSCs can be used.28 To evaluate efficiency of purification and recovery of myeloid progenitors by MACS, both positive and negative fractions should be evaluated by flow cytometry for the presence of CD45+CD235a− cells. Successful purification results in more than 94% of CD45+ cells in positive fraction and less than 5% of CD45dim cells and no CD45bright cells in negative fraction
MATERIALS
REAGENTS
hESC WA01 and WA09 (National stem cell bank, Madison, WI)
Lentivirally reprogrammed iPSCs iPS (Foreskin)-1 (WiscBank, WiCell, Madison, WI)
Transgene-free iPSCs DF-19-9-7T and 4-3-7T (WiscBank, WiCell, Madison, WI)
OP9 mouse bone marrow stromal cell line (ATCC or from Dr. Nakano Osaka University, Osaka, Japan)
Mouse Embryonic Fibroblasts (MEFs, WiCell Research Institute, Madison, WI)
Dulbecco’s modified eagle medium (DMEM), powder (GIBCO-Invitrogen, Carlsbad, CA, USA, Cat. no. 12100-046)
DMEM/nutrient mixture F-12, powder (GIBCO-Invitrogen, Cat. no. 12400-024)
α-MEM basal medium, powder (GIBCO-Invitrogen, Cat. no. 12000-022)
Iscove’s modified Dulbecco’s medium (IMDM), powder (GIBCO-Invitrogen, Cat. no. 12200-036)
KNOCKOUT SR (KO), serum replacement for ES cell (GIBCO-Invitrogen, Cat. no. 10828-023)
CRITICAL: Each lot should be tested for its suitability for hPSC culture
Serum-free medium, Stemline Hematopoietic stem cell expansion medium (SIGMA-ALDRICH, St. Louis, MO, USA, Cat. no. S-0189)
Sodium azide, NaN3 (Fisher Scientific, Cat. no. BP922-500)
Ascorbic acid (SIGMA-ALDRICH, Cat. no. A4544)
L-glutamine (Cellgro, Cat. no. 61-030-RM)
MEM non-essential amino acid, 100× solution (GIBCO-Invitrogen, Cat. no. 11140)
2-mercaptoethanol (SIGMA-ALDRICH, Cat. no. M7522)
Sodium Bicarbonate (Fisher Scientific, Cat. no. S233-500)
Poly (2-hydroxyethyl methacrylate) (p-HEMA) (SIGMA-ALDRICH, Cat. no. P3932)
NaOH (Fisher Scientific, Cat. no. S318)
CAUTION: NaOH is corrosive. Wear rubber gloves and protective eye goggles.
Percoll, solution (SIGMA-ALDRICH, Cat. no. P-1644)
PBS powdered, without calcium and magnesium (GIBCO-Invitrogen, Cat. no. 21600-044)
PBS, 10× (GIBCO-Invitrogen, Cat. no. 70011-044)
Gelatin from porcine skin, Type A (SIGMA-ALDRICH, Cat. no. G-1890)
EDTA 0.5M, pH 8.0 (GIBCO-Invitrogen, Cat. no. 15575-038)
MTG, monothioglycerol (SIGMA-ALDRICH, Cat. no. M-6145)
Fetal bovine serum defined (Hyclone, Logan, UT, USA, Cat. no. SH30070.03)
CRITICAL: FBS used in this protocol is defined FBS. Use FBS directly without the heat inactivation step for OP9 culture, hematopoietic differentiation, expansion of myeloid progenitors, and the generation of mature myelomonocytic cells from human pluripotent stem cells. Heat inactivation does not benefit culture but usually results in a higher adipogenic effect on OP9.
CRITICAL: We have found that different lots of HyClone defined FBS provide relatively stable hematopoietic differentiation in hPSC/OP9 coculture and support efficient OP9 growth without significant adipogenesis. Results from other suppliers are more variable.
Trypsin 0.05%/EDTA 0.5mM (HyClone, Ct. no. SH30236.02)
Cell dissociation buffer, enzyme free, PBS-based (GIBCO-Invitrogen, Cat. no. 13151-014)
Collagenase Type IV (GIBCO-Invitrogen, Cat. no. 17104-019)
Ex-Cyte (Celliance-Millipore, Billerica, MA, USA, Cat. no. 81-129-1)
Neupogen as a human G-CSF (Amgen, Thousand Oaks, CA, USA)
Leukine as a human GM-CSF (Berlex, Richmond, CA, USA)
Human M-CSF (Peprotech, Rocky Hill, NJ, USA, Cat. no. 300-25)
Human FGF-basic (Peprotech, Cat. no. 100-18B)
Human Flt3-Ligand (Peprotech, Cat. no. 300-19)
Human IL-1β (Peprotech, Cat. no. 200-01B)
Human IL-3 (Peprotech, Cat. no. 200-03)
Human IL-4 (Peprotech, Cat. no. 200-04)
Human IL-5 (Peprotech, Cat. no. 200-05)
Human sRANKL (Peprotech, Cat no. 300-01)
Human TGF-β1 (Peprotech, Cat. no. 100-21)
Human TNF-α (Peprotech, Cat no. 300-01A)
1α, 25-Dihydroxyvitamin D3, Biologically active form of vitamin D3 (SIGMA-ALDRICH, Cat. no. D-1530)
Ethanol (EtOH; SIGMA-Aldrich, Cat. no. E7023)
CAUTION: Ethanol is flammable. Avoid exposure to ignition.
Trypan blue solution, 0.4% (SIGMA-ALDRICH, Cat. no. T8154)
Protocol Wright stain (Fisher Scientific, Pittsburgh, PA, USA, Cat. no. 23-264980)
Protocol Phosphate buffer solution for Wright stain, pH 6.4 (Fisher Scientific, Cat. no. 23-262234)
Cytoseal 60 mounting medium (Richard-Allan Scientific-ThermoFisher, Cat. no. 8310-4)
Immersion oil (SIGMA-ALDRICH, Cat. no. 56822)
7-AAD; 7-Aminoactinomycin D (SIGMA-ALDRICH, Cat. no. A9400)
Bovine serum albumin Fraction V (Fisher Scientific, Cat. no. BP1605-100)
Antibodies and magnetic sorting reagents: See Tables 1 and 2.
Table 1.
Antibodies used for magnetic-based cell sorting in this study
| Name | Fluorochrome | Clone | Company | Cat. no. |
|---|---|---|---|---|
| Anti-human CD41a | PE | HIP8 | BD Biosciences | 555467 |
| Anti-human CD45 | FITC | HI30 | BD Biosciences | 555482 |
| Anti-human CD235a | PE | GA-R2 (HIR2) | BD Biosciences | 555570 |
| Anti-Mouse CD29 | PE | HM beta 1-1 | Serotec | MCA2298PE |
| Anti-FITC microbeads | None | None | Miltenyi Biotec | 120-000-293 |
| Anti-PE microbeads | None | None | Miltenyi Biotec | 120-000-294 |
BD Biosciences (San Jose, CA, USA; http://www.bdbiosciences.com), Miltenyi Biotec (Begisch Gladbach, Germany; http://www.miltenyibiotec.com), Serotec (Oxford, UK; http://www.abdserotec.com/).
Table 2.
Antibodies used to analyze differentiation of hematopoietic progenitors and myeloid lineages from human pluripotent stem cells
| Name | Fluorov chrome | Clone | Company | Cat. no. |
|---|---|---|---|---|
| Anti-human CD1a | PE | VIT6b | CALTAG-Invitrogen | MHCD1a04 |
| Anti-human CD2 | FITC | RPA-2.10 | BD Biosciences | 555326 |
| Anti-human CD3 | FITC | SK7 | BD Biosciences | 349201 |
| Anti-human CD7 | PE | M-T701 | BD Biosciences | 555361 |
| Anti-human CD10 | PE | HI10a | BD Biosciences | 555375 |
| Anti-human CD11b | FITC | VIM12 | CALTAG-Invitrogen | CD11b01 |
| Anti-human CD13 | PE | TüK1 | CALTAG-Invitrogen | MHCD1304 |
| Anti-human CD14 | FITC | M5E2 | BD Biosciences | 555397 |
| Anti-human CD15 | FITC | VIMC6 | Miltenyi Biotec | 130-081-101 |
| Anti-human CD16 | PE | 3G8 | CALTAG-Invitrogen | MHCD1604 |
| Anti-human CD19 | PE | HIB19 | BD Biosciences | 555413 |
| Anti-human CD34 | APC | 581 | BD Biosciences | 555824 |
| Anti-human CD41a | PE | HIP8 | BD Biosciences | 555467 |
| Anti-human CD43 | FITC | 1G10 | BD Biosciences | 555475 |
| Anti-human CD45 | APC | HI30 | BD Biosciences | 555485 |
| Anti-human CD64 | FITC | 10.1 | CALTAG-Invitrogen | CD6401 |
| Anti-human CD66b | FITC | G10F5 | BD Biosciences | 555724 |
| Anti-human CD90 (Thy-1) | APC | 5E10 | BD Sciences | 559869 |
| Anti-human CD115 | PE | 61708 | R&D Systems | FAB329P |
| Anti-human CD117 | APC | YB5.B8 | BD Biosciences | 550412 |
| Anti-human CD123 | FITC | AC145 | Miltenyi Biotec | 130-090-897 |
| Anti-human CD163 | PE | 215927 | R&D Systems | FAB1607P |
| Anti-human CD235a | PE | GA-R2(HIR2) | BD Biosciences | 555570 |
| DC-SIGN | FITC | DCN46 | BD Biosciences | 551264 |
| HLA-DR | PE | Tü36 | CALTAG-Invitrogen | MHLDR04 |
| Lactoferrin* | PE | 3C5 | CALTAG-Invitrogen | GIC206 |
| Langerin* | PE | 343828 | R&D Systems | FAB2088P |
| Major basic protein (MBP)* | None | AHE-2 | BD Biosciences | 550843 |
| Myeloperoxidase (MPO)* | FITC | H-43-5 | CALTAG-Invirogen | GIC205 |
| Anti-human TRA-1-85 | APC | TRA-1-85 | R&D Systems | FAB3195A |
represent intracellular staining.
BD Biosciences (San Jose, CA, USA; http://www.bdbiosciences.com), CALTAG-Invitrogen (Carlsbad, CA, USA; http://www.invitrogen.com), R&D Systems (Minneapolis, MN, USA; http://www.rndsystems.com), Miltenyi Biotec(Begisch Gladbach, Germany; http://www.miltenyibiotec.com), Biolegend(San Diego, CA, USA; http://www.biolegend.com).
Equipment
Cell strainer, 40µm (BD Bioscience, San Jose, CA, USA, Cat. no.352340)
Cell strainer, 70µm (BD Bioscience, Cat. no.352350)
MACS separation columns, LD (Miltenyi Biotec, Begisch Gladbach, Germany, Cat. no. 130-042-901)
MACS separation columns, LS (Miltenyi Biotec, Cat. no. 130-042-401)
Midi MACS separation unit (Miltenyi Biotec, Cat. no. 130-042-302)
MACS Multistand (Miltenyi Biotec, Cat. no. 130-042-303)
MACSmix Tube Rotator (Miltenyi Biotec, Cat. no. 130-090-753)
Pre-separation filters with 30µm nylon mesh (Miltenyi Biotec, Cat. no. 130-041-407)
Nalgene Disposable bottle top filter, Polyethersulfone membrane with 0.2µm pore size (Fisher Scientific, Cat. no. 595-4520)
T25 Tissue culture flask canted neck with 0.2µm vented plug seal cap, 50ml, Nonpyrogenic polystyrene (BD Bioscience, Cat. no. 353108)
T75 Tissue culture flask canted neck with 0.2µm vented plug seal cap, 250ml, Nonpyrogenic polystyrene (BD Bioscience, Cat. no. 353136)
Tissue culture dishes, polystyrene 100×20 mm (BD Bioscience, Cat. no. 353003)
Tissue culture 6well plate, Polystyrene flat bottom (BD Bioscience, Cat no. 353046)
0.5ml microcentrifuge tube, autoclavable (Fisher Scientific, Cat. no. 05-408-120)
5ml Polystyrene round-bottom tube, 12×75mm, non-sterile (BD Bioscience, Cat. no. 352008)
5ml Polystyrene round-bottom tube with 35µm cell strainer cap, 12×75mm (BD Bioscience, Cat. no. 352235)
15ml Polypropylene Conical tubes (BD Bioscience, Cat. no. 352097)
50ml Polypropylene Conical tubes (BD Bioscience, Cat. no. 352098)
50ml Vacuum filtration system with 0.22µm pore size membrane (Millipore, Cat. no. SCGP00525)
Serological pipet, 1ml Nonpyrogenic (Fisher Scientific Cat. No. 13-678-11B)
Serological pipet, 5ml Nonpyrogenic (BD Bioscience. Cat. no. 357543)
Serological pipet, 10ml Nonpyrogenic (BD Bioscience. Cat. no. 357551)
Borosilicate glass pipets, 5ml (Corning, Lowell, MA, USA, Cat. no.7077-5N)
Sterling Nitrile-xtra powder-free exam gloves (Kimberly-Clark, Neenah, WI, USA, Cat. no. 53139)
9″ Pasteur pipets, Flint glass (Fisher Scientific, Cat. no. 13-678-6B)
Hemocytometer, Reichert Bright-Line counting chamber (Fisher Scientific, Cat. no. 02-671-5)
Water bath (Fisher Scientific)
Tabletop Centrifuge (Beckman, Brea, CA, USA)
Microcentrifuge (Eppendorf, Hamburg, Germany, Model 5418)
Sterile biosafety cabinet (The Baker Company, Sanford, Maine, USA)
37°C/5% CO2 incubator (Thermo Scientific, Asheville, NC, USA)
Inverted microscope (Olympus, Center Valley, PA, USA)
Object marker, Cell dotter for inverted microscope (Nikon, Melville, NY, USA, Cat. no. MBW10020)
Balance (Denver Instrument, Bohemia, NY, USA)
Milli-Q Water purification system (Millipore, Billerica, MA, USA)
Pipet-Aid, Filler/Dispensers (Drummond, Broomall, PA, USA)
Liquid waste disposal system for aspiration
Flow cytometer FACSCalibur (Becton Dickenson, Franklin Lakes, NJ, USA)
Cytofuge 2 cytocentrifuge system (StatSpin, Westwood, MA, USA)
Cytofuge filter & concentrator (Fisher Scientific, Cat. No. 04-981-3)
1.5 ml microcentrifuge tube, autoclavable (Fisher Scientific, Cat. No. 05-408-129)
Microscope slide glass, Superfrost (Fisher Scientific, Cat. no. 12-550-14)
Cover glass, 18×18mm (Fisher Scientific, Cat. no. 12-548-A)
Nutating mixer, rotator (Fisher Scientific, Cat. no. 22-363-152)
REAGENTS SETUP
Collagenase solution (1mg/ml)
Add 50 mg of collagenase to 50 ml of DMEM/F-12 basal medium, and sterilize the solution by filtration using a 0.22 µm membrane filter. Keep solution at 2–8°C and use for up to one week.
0.1% Gelatin solution (w/v)
Add 500 mg of gelatin to 500 ml of endotoxin-free reagent grade distilled water. Solubilize and sterilize by autoclaving for 20 min at 121°C. Store the solution at 4°C for up to 6 months. Keep sterile.
Magnetic cells sorting (MACS) buffer
MACS buffer contains 5% FBS (v/v) and 2 mM EDTA in PBS (Ca2+ and Mg2+ free). For 500 ml, add 25 ml of FBS and 2 ml of 0.5M EDTA (pH 8.0) into Ca2+ and Mg2+ free-PBS. Sterilize MACS buffer by filtration using a 0.22 µm membrane filter and keep at 2–8°C for up to 6 months. Optional: After filtration, close lid of filter unit and keep MACS buffer under vacuum for about 10–15min for degassing.
Flow cytometry buffer
Flow cytometry buffer contains 2% FBS (v/v), 0.05% sodium azide (NaN3, w/v) and 2 mM EDTA in PBS (Ca2+ and Mg2+ free). For 500 ml, add 10 ml of FBS, 0.25 g of NaN3 and 2 ml of 0.5 M EDTA (pH 8.0) into Ca2+ and Mg2+ free-PBS. Filtrate the buffer using a 0.22 µm membrane filter and store at 2–8°C for up to 6 months.
10% pHEMA coating solution (w/v)
Add 4 g of pHEMA to 40 ml of 95% ethanol containing 10 mM NaOH. Dissolve completely by continuously rotating at room temperature or 37°C overnight. Store at room temperature until needed for use.
CAUTION: It is very hard to dissolve pHEMA crystals completely when they aggregate. Thus, shake the solution immediately after addition of pHEMA to prevent precipitation.
5× Percoll solution
Add 5 ml of 10× PBS (Ca2+ and Mg2+ free) to 45 ml of Percoll solution (90% Percoll and 10% 10× PBS). Store at 4°C for 6 months. For 1× Percoll working solution, dilute 10 ml of 5× Percoll solution in 40 ml of 1× PBS (1/5 dilution). Use fresh.
100× (100 mM) L-glutamine/2-mercaptoethanol solution
Add 146 mg of L-glutamine and 7µl of 2-mercaptoethanol to 10 ml of PBS (Ca2+ and Mg2+ free). Sterilize the solution by filtration using a 0.22µm membrane filter and store up to 2 weeks at 2–8°C.
1000× (100 mM) MTG solution
Add 87 µl of MTG to 10 ml of endotoxin-free reagent grade distilled water. Mix well and divide into 500 µl aliquots. Store up to 6 months at −20°C.
CAUTION: MTG has high viscosity, thus pipet slowly to dispense MTG accurately.
1000× (50 mg/ml) ascorbic acid solution
Add 500 mg of ascorbic acid to 10 ml of endotoxin-free reagent grade distilled water. Dissolve completely, divide into 500 µl aliquots, and store up to 6 months at −20°C.
1mM 1α,25-Dihydroxyvitamin D3 stock solution
Dissolve 10 µg of 1α,25-Dihydroxyvitamin D3 in 24 µl of 95% ethanol (Final concentration is 0.42 µg/µl in 95% EtOH). Store the stock solution at −20°C for up to 6 months.
50× 7-AAD solution
Dissolve 1 mg of 7-AAD in 50µl of absolute methanol, then add 950 µl of 1× PBS. Final concentration is 1 mg/ml. Store the solution in an amber glass bottle or tube at 4°C protected from light. Solution can be stored for at least up to 6 months. For working solution, the stock solution (1 mg/ml) is diluted with flow cytometry buffer to 20 µg/ml concentration. Use 10 µl for staining.
0.1% BSA/PBS solution
Dissolve 25mg of Bovine serum albumin Fraction V in 25ml of PBS (Ca2+ and Mg2+ free). Sterilize the solution by filtration using a 0.22µm membrane filter and store for up to 6 months at 2–8°C.
Reconstitution of cytokines
Centrifuge vials at maximum speed for 1 min to precipitate lyophilized pellet prior to opening vials. Reconstitute cytokines according to the product information provided by manufacturer. Dilute with 0.1% BSA/PBS solution for working concentration and store at −80°C until needed for use. See Table 3 for details.
Table 3.
Reconstitution of cytokines
| Cytokines | Product state | Reconstitution | Working dilution | Storage |
|---|---|---|---|---|
| Neupogen as a human G-CSF | Concentrated aqueous solution | - | 100 µg/ml in 0.1% BSA/PBS solution | Aliquot 100 µl into autoclaved 0.5 ml tubes and store at −80°C until needed for use. |
| Leukine as a human GM-CSF | Concentrated aqueous solution | - | 100 µg/ml in 0.1% BSA/PBS solution | Aliquot 100 µl into autoclaved 0.5 ml tubes and store at −80°C until needed for use. |
| Human FGF-basic | Lyophilized pellet | 1 mg/ml with 5mM Tris (pH7.6) | 100 µg/ml in 0.1% BSA/PBS solution | Aliquot 100 µl into autoclaved 0.5 ml tubes and store at −80°C until needed for use. |
| Human Flt3-Ligand | Lyophilized pellet | 1 mg/ml with distilled water | 100 µg/ml in 0.1% BSA/PBS solution | Aliquot 100 µl into autoclaved 0.5 ml tubes and store at −80°C until needed for use. |
| Human IL-1β | Lyophilized pellet | 100 µg /ml with distilled water | 10 µg/ml in 0.1% BSA/PBS solution | Aliquot 100 µl into autoclaved 0.5 ml tubes and store at −80°C until needed for use. |
| Human IL-3 | Lyophilized pellet | 100 µg /ml with distilled water | 10 µg/ml in 0.1% BSA/PBS solution | Aliquot 100 µl into autoclaved 0.5 ml tubes and store at −80°C until needed for use. |
| Human IL-4 | Lyophilized pellet | 1mg/ml with distilled water | 100 µg/ml in 0.1% BSA/PBS solution | Aliquot 100 µl into autoclaved 0.5 ml tubes and store at −80°C until needed for use. |
| Human IL-5 | Lyophilized pellet | 100 µg /ml with distilled water | 10 µg/ml in 0.1% BSA/PBS solution | Aliquot 100 µl into autoclaved 0.5 ml tubes and store at −80°C until needed for use. |
| Human M-CSF | Lyophilized pellet | 100 µg /ml with distilled water | 10 µg/ml in 0.1% BSA/PBS solution | Aliquot 100 µl into autoclaved 0.5 ml tubes and store at −80°C until needed for use. |
| Human sRANKL | Lyophilized pellet | 100 µg /ml with distilled water | 10 µg/ml in 0.1% BSA/PBS solution | Aliquot 100 µl into autoclaved 0.5 ml tubes and store at −80°C until needed for use. |
| Human TGF-β1 | Lyophilized pellet | 50 µg/ml with 10 mM citric acid solution (pH 3.0) | 5 µg/ml in 0.1% BSA/PBS solution | Aliquot 100 µl into autoclaved 0.5 ml tubes and store at −80°C until needed for use. |
| Human TNF-α | Lyophilized pellet | 1mg/ml with distilled water | 50 µg/ml in 0.1% BSA/PBS solution | Aliquot 100 µl into autoclaved 0.5 ml tubes and store at −80°C until needed for use. |
Vials of cytokine products in lyophilized pellet state should be centrifuged at maximum speed for 1 minutes with microcentrifuge to precipitate pellet prior to opening vials. Distilled water and all buffers should be sterilized by filtration with 0.22 µm membrane filter.
CRITICAL: Perform entire procedure in a sterile biosafety cabinet. Read the product information sheet carefully before preparation of working aliquots of all cytokines.
Medium composition
All basal medium, α-MEM, DMEM, DMEM/F-12, and IMDM should be prepared freshly from powder according to the manufacturer’s instructions, sterilized by filtration using a 0.22µm membrane filter and stored for up to 2 months at 2–8°C.
MEF growth medium (500 ml)
| Composition | Volume | Final concentration |
|---|---|---|
| DMEM | 445 ml | |
| FBS | 50 ml | 10% |
| 100× NEAA solution (10 mM) | 5 ml | 100 µM |
Sterilize the medium by filtration using a 0.22 µm membrane filter and store for up to 3 weeks at 2–8°C
hESC culture medium (250 ml)
| Composition | Volume | Final concentration |
|---|---|---|
| DMEM/F-12 | 195 ml | 78% |
| KO serum replacement | 50 ml | 20% |
| 100× NEAA solution (10 mM) | 2.5 ml | 100 µM |
| L-glutamine/2-mercaptoethanol solution (100 mM) | 2.5 ml | 1 mM |
| Basic FGF (100 µg/ml) for hESCs culture | 10 µl | 4 ng/ml |
Sterilize the medium by filtration using a 0.22 µm membrane filter and store for up to 2 weeks at 2–8°C.
hiPSC culture medium (250 ml)
| Composition | Volume | Final concentration |
|---|---|---|
| DMEM/F-12 | 195 ml | 78% |
| KO serum replacement | 50 ml | 20% |
| 100× NEAA solution (10 mM) | 2.5 ml | 100 µM |
| L-glutamine/2-mercaptoethanol solution (100 mM) | 2.5 ml | 1 mM |
| Basic FGF (100 µg/ml) | 25 µl | 10 ng/ml |
Sterilize the medium by filtration using a 0.22 µm membrane filter and store for up to 2 weeks at 2–8°C. CRITICAL: Note that the basic FGF concentration for hiPSC maintenance should be 2.5 times higher than for hESCs to lessen the effect of variations in MEF quality on culture of hiPSCs.
Mouse OP9 bone marrow stromal cells culture medium (250 ml)
| Composition | Volume | Final concentration |
|---|---|---|
| α-MEM | 200 ml | 80% |
| FBS | 50 ml | 20% |
Sterilize the medium by filtration using a 0.22 µm membrane filter and store for up to 2 weeks at 2–8°C.
Differentiation (hPSC/OP9 coculture) medium (500 ml)
| Composition | Volume | Final concentration |
|---|---|---|
| α-MEM | 450 ml | 90% |
| FBS | 50 ml | 10% |
| 1000× MTG solution (100 mM) | 500 µl | 100 µM |
Sterilize the medium by filtration using a 0.22 µm membrane filter and store for up to 2 weeks at 2–8°C. Optional: Ascorbic acid solution (50 mg/ml) can be added to achieve 50 µg/ml final concentration.
Multipotent myeloid progenitor expansion medium
| Composition | Volume | Final concentration |
|---|---|---|
| Differentiation (hPSC/OP9 coculture) medium | - | |
| GM-CSF (100 µg/ml) | 1/500 dilution | 200 ng/ml |
Volume of the medium will vary depending on input cell number. In this protocol, we use 1 ml of medium for ~1×106 cocultured cells. The complete medium is prepared freshly and GM-CSF is added right before use.
Neutrophil differentiation medium
| Composition | Volume | Final concentration |
|---|---|---|
| IMDM with 20% FBS medium | - | |
| G-CSF (100 µg/ml) | 1/1000 dilution | 100 ng/ml |
Volume of the medium will vary depending on input cell number. In this protocol, we use 1ml of medium for 2×104 cells. The complete medium is prepared freshly and all cytokines and supplements are added right before use.
Eosinophil differentiation medium
| Composition | Volume | Final concentration |
|---|---|---|
| IMDM with 20% FBS medium | - | |
| IL-3 (10 µg/ml) | 1/1000 dilution | 10 ng/ml |
| IL-5 (10 µg/ml) | 1/2000 dilution | 5ng/ml |
Volume of the medium will vary depending on input cell number. In this protocol, we use 1ml of medium for 2×104 cells. The complete medium is prepared freshly and all cytokines and supplements are added right before use.
DC differentiation medium
| Composition | Volume | Final concentration |
|---|---|---|
| Serum free medium (StemLine, SIGMA-Aldrich) | - | |
| GM-CSF (100 µg/ml) | 1/5000 dilution | 20 ng/ml |
| IL-4 (100 µg/ml) | 1/5000 dilution | 20 ng/ml |
| TNF-α (50 µg/ml) | 1/20000 dilution | 2.5 ng/ml |
| Ex-Cyte | 1/500 dilution | 2 µl/ml |
Volume of the medium will vary depending on input cell number. In this protocol, we use 1 ml of medium for 105 cells. The complete medium is prepared freshly and all cytokines and supplements are added right before use.
LC differentiation medium
| Composition | Volume | Final concentration |
|---|---|---|
| Serum free medium (StemLine, SIGMA-Aldrich) | - | |
| GM-CSF (100µg/ml) | 1/5000 dilution | 20ng/ml |
| TGF-β1 (5µg/ml) | 1/2000 dilution | 2.5ng/ml |
| TNF-α (50µg/ml) | 1/50000 dilution | 1ng/ml |
| Ex-Cyte | 1/500 dilution | 2µl/ml |
Volume of the medium will vary depending on input cell number. In this protocol, we use 1ml of medium for 105 cells. The complete medium is prepared freshly and all cytokines and supplements are added right before use.
IMDM medium containing 10% FBS
| Composition | Volume | Final concentration |
|---|---|---|
| IMDM | 225ml | 90% |
| FBS | 25ml | 10% |
Sterilize the medium by filtration with 0.22 µm membrane filter and keep sterile at 2–8°C for up to 2 weeks.
Macrophage differentiation medium
| Composition | Volume | Final concentration |
|---|---|---|
| IMDM with 10% FBS medium | - | |
| M-CSF (10 µg/ml) | 1/500 dilution | 20 ng/ml |
| IL-1β (10 µg/ml) | 1/1000 dilution | 10 ng/ml |
Volume of the medium will vary depending on input cell number. In this protocol, we use 1ml of medium for 105 cells. The complete medium is prepared freshly and all cytokines and supplements are added right before use.
Osteoclast progenitor expansion medium
| Composition | Volume | Final concentration |
|---|---|---|
| Differentiation (hPSC/OP9 coculture) medium | - | |
| GM-CSF (100 µg/ml) | 1/2000 dilution | 50 ng/ml |
| 1α, 25-Dihydroxyvitamin D3 (1mM) | 1/5000 dilution | 200nM |
Volume of the medium will vary depending on input cell number. In this protocol, we use 1 ml of medium for 2×104 cells. The complete medium is prepared freshly and all cytokines and supplements are added right before use.
Osteoclast maturation medium
| Composition | Volume | Final concentration |
|---|---|---|
| Differentiation (hPSC/OP9 coculture) medium | - | |
| GM-CSF (100 µg/ml) | 1/2000 dilution | 50 ng/ml |
| 1α, 25-Dihydroxyvitamin D3 (1 mM) | 1/5000 dilution | 200 nM |
| RANKL (10 µg/ml) | 1/1000 dilution | 10 ng/ml |
Volume of the medium will vary depending on input cell number. In this protocol, we use 1ml of medium for 5×104 cells. The complete medium is prepared freshly and all cytokines and supplements are added right before use.
IMDM medium containing 20% FBS
| Composition | Volume | Final concentration |
|---|---|---|
| α-MEM | 200 ml | 80% |
| FBS | 50 ml | 20% |
Sterilize the medium by filtration using a 0.22µm membrane filter and keep sterile at 2–8°C for up to 2 weeks.
Diluted Buffer for Wright Stain Procedure
Mix 30 ml of Protocol phosphate buffer pH 6.4 with 100 ml of deionized water. Store at room temperature for up to 6 months.
EQUIPMENT SETUP
Gelatin-coated 10 cm culture dish and 6 well tissue-culture plate
Add 7–8 ml of autoclaved gelatin solution to a 10 cm culture dish or 2 ml to each well of a 6 well tissue-culture plate. Allow the gelatin solution to cover the entire plastic surface and incubate for at least 3 hrs at 37°C in an incubator. Dishes and 6-well plates containing gelatin solution can be stored for up to several days at 37°C in CO2 incubator. Do not allow the wells to dry. Before use, aspirate the gelatin solution from the dish.
pHEMA-coated culture flask
Add 5 ml of 10% pHEMA/ethanol solution to a T75 tissue culture flask or 2 ml of the solution to a T25 tissue culture flask. Rotate the flask gently to allow pHEMA solution to cover the entire surface of the flask. Make sure that the flask is completely covered with pHEMA solution. Tip the flask to remove excess pHEMA solution and save to reuse.
After coating, dry the flask overnight in a sterile biosafety cabinet, close the cap and store under sterile conditions at room temperature until needed for use.
CRITICAL Treatment with pHEMA should be done quickly to avoid irregular coating due to rapid ethanol evaporation.
PROCEDUREs
MEFs preparation for Human ES/iPSC culture TIMING 24 hours
-
1
Prepare MEFs according to WiCell protocol (http://www.wicell.org/index.php?option=com_content&task=category§ionid=7&id=246&Itemid=248).
-
2
Inactivate MEFs with gamma irradiation at 8,000 rad
-
3
Resuspend MEFs at 2×105 cells/ml in prewarmed MEF growth.
-
4
Add 2 mL/well of prewarmed MEF growth medium and then dispense MEF suspension on gelatin- coated 6-well plate (1ml/well).
CRITICAL STEP Distribute MEFs evenly with a back/forth and right/left movement twice. Irregular distribution of MEFs may cause death and unwanted differentiation of hPSC colonies during culture.
-
5
Incubate MEF plates in a CO2 incubator at 37°C for at least 24 hrs before adding hPSCs. CRITICAL STEP MEFs should be used for hPSC passage within one week. Before plating hPSCs, aspirate MEF medium, add 2ml of PBS, swirl once and aspirate PBS. Add 2 ml of prewarmed hPSC medium and place plate into CO2 incubator at 37°C. Now MEF feeders are ready for hPSC plating (step 15).
hES/iPSC culture TIMING 7 days
-
6
Aspirate hPSC growth medium from one well of the 6-well plate of hESCs or iPSCs.
CRITICAL STEP: Note that cells will need to be split every 6–7 days.
-
7
Wash cells with 2 ml/well of PBS (Ca2+ and Mg2+ free) stored at room temperature.
-
8
Add 1 ml/well of collagenase IV solution (1 mg/ml) and incubate at 37°C in a CO2 incubator until the edges of the hPSC colonies begin to curl (approximately 7–10 minutes).
-
9
Add 1 ml of hESC or hiPSC growth medium and break up the colonies into small cell aggregates by gently pipetting.
CRITICAL STEP: After collagenase treatment, hPSC colonies are loosely attached and can be collected by gentle pipetting. Do not use excessive mechanical force or scraping which can provoke spontaneous differentiation.
-
10
Transfer cells to a 15 ml conical tube.
-
11
Centrifuge at 200×g at room temperature for 3–5 min.
-
12
Aspirate the medium gently without disturbing the pellet.
-
13
Resuspend cells in 3 ml of hESC or hiPSC growth medium, and wash cells by repeating steps 11 and 12.
-
14
Resuspend cell pellet in 3 ml of hESC or hiPSC growth medium.
-
15
Plate 0.5 ml/well of cell suspension onto MEF-grown 6-well plate from Step 5.
TROUBLESHOOTING
-
16
Feed hPSCs daily by replacing the old medium with 3ml of prewarmed hESC or hiPSC medium.
-
17
Passage undifferentiated hES/iPSCs (Fig. 1a) weekly at 1.2–1.5×106 cells/well density on MEFs.
CRITICAL STEP: If spontaneous differentiation of hESCs or hiPSCs occurs, differentiated hPSC colonies should be eliminated during the maintenance; observe hPSCs every day before changing medium. Mark differentiated colonies with an objective marker under the inverted microscope and aspirate marked areas using a glass Pasteur pipette while feeding hPSC with fresh medium.
CRITICAL STEP: Alternatively, hPSCs can be maintained under feeder-free conditions32. In OP9 coculture system, we did not observe significant differences in the efficiency of hematopoietic differentiation of hPSCs maintained on MEFs or in feeder-free cultures.
Culture of mouse OP9 cells TIMING 8–12 days
-
18
Aspirate OP9 growth medium and wash cells twice with 10 ml of PBS.
CRITICAL STEP: Note that cells will need to be split every 4 days.
-
19
Add 5 ml of trypsin/EDTA(0.05%/0.5 mM) solution and incubate for 5 min at 37°C in a CO2 incubator.
CRITICAL STEP: OP9 feeders consist of heterogeneous cell populations which include cells with at least adipogenic and osteogenic potential. To maintain a proper balance of cells following passage, OP9 feeders should be digested and detached completely by trypsin treatment. Inadequate washing of cells with PBS or using an old trypsin may results in partial detachment and enrichment in adipogenic cells.
-
20
Add 5 ml of OP9 growth medium and collect cells by pipetting.
-
21
Transfer cell suspension into a 15 ml conical tube and centrifuge for 5 min at 300×g at room temperature.
-
22
Aspirate supernatant and resuspend cells in 1 ml of OP9 growth medium.
-
23
Add 100 µl of cell suspension to 10 ml of OP9 growth medium and plate cells onto 10 cm gelatin-coated culture dishes.
CRITICAL STEP: It is essential to culture OP9 on gelatin-coated plates to prevent spontaneous adipogenesis.
TROUBLESHOOTING
-
24
When cultures are confluent, split 1 dish for maintenance. This should occur after approximately 4 days of growth.
-
25
To prepare overgrown OP9 for coculture with hPSCs, change half of the medium after 4 days of culture on gelatin-coated plates and incubate for an additional 4–8 days to achieve a dense OP9 monolayer.
CRITICAL STEP: OP9 should be split every 4 days for maintenance/expansion. OP9 used for hPSC differentiation should be fed with fresh media at confluence (day 4) and incubated for an additional 4–8 days to form a dense monolayer embedded in extracellular matrix.
Hematopoietic differentiation on OP9 TIMING 9 days
-
26
Remove overgrown OP9 dishes prepared for coculture from the CO2 incubator.
-
27
Aspirate OP9 growth medium.
-
28
Add 10 ml of differentiation medium and keep at 37°C in a CO2 incubator.
-
29
From one well of a 6-well hPSC plate from Step 17, aspirate hES/hiPSC growth medium. Add 1 ml of collagenase IV solution (1 mg/ml), and incubate cells for 10 min at 37°C.
-
30
Add 1 ml/well of differentiation medium directly to the well and break up colonies into small cell aggregates by gentle pipetting. Transfer cells into a 15 ml conical tube.
CRITICAL STEP: hPSCs for differentiation studies should be prepared as small aggregates. Single hPSCs will not survive on OP9.
-
31
Centrifuge cells at 200×g for 3–5 min at room temperature.
-
32
Aspirate the medium gently without disturbing the pellet.
-
33
Resuspend the cell pellet with 1mL of differentiation medium.
-
34
Add 1mL of hES/hiPSC suspension to 1 OP9 dish prepared in steps 26–28.
CRITICAL STEP: Efficiency of hematopoietic differentiation is significantly affected by the density of hPSCs plated on OP9. In our experience, the optimal plating density of hPSCs is 1.0–1.5 × 106 cells per 10 cm dish of OP9. To estimate the number of cells in a suspension consisting of small hPSC aggregates, one well of a 6-well plate can be used to prepare a single cell suspension by treatment with trypsin for cell counting. Alternatively, aliquots of hPSC aggregates can be collected, treated with trypsin, and counted.
-
35
Distribute cells evenly with a back/forth and right/left movement twice.
-
36
The following day (day 1), aspirate all of the media to waste and replace with 20 ml of prewarmed differentiation medium.
-
37
On day 4, change half of the medium.
-
38
On day 6, change half of the medium
-
39
To collect cells on day 9, aspirate the supernatant and add 5 ml of prewarmed collagenase solution (1mg/ml) to each dish of hPSC/OP9 coculture and incubate for 30 minutes at 37°C in a CO2 incubator.
-
40
Remove the collagenase solution and keep it on ice in a 15 ml conical tube for subsequent collection of trypsin digested cells (cell collection tube).
CRITICAL STEP: hPSC/OP9 coculture produces collagen-rich matrix and pretreatment with collagenase is essential to achieve efficient digestion of cells with trypsin. Because some cultures might form an excessive amount of extracellular matrix, the time of treatment with collagenase can be extended up to 40–50 minutes to achieve complete dissociation and maximize cell recovery.
-
41
Add 5 ml of prewarmed Trypsin/EDTA solution (0.05%/0.5mM) to the dish from Step 39 and incubate for 15–20 minutes at 37°C CO2 incubator.
-
42
Add 2 ml/dish of MACS buffer, suspend coculture cells by pipetting and transfer to the collection tube from Step 40.
-
43
Add an additional 5 ml/dish of MACS buffer to the coculture dish and collect the remaining cells into the collection tube.
-
44
Centrifuge cell suspension at 300×g for 5min at room temperature.
-
45
Wash cells once by adding 5 ml of MACS buffer to cell pellet followed by pipetting and centrifugation at 300×g for 5 min at room temperature.
-
46
Cells are ready to use in further applications such as flow cytometry, CFC assay, and further differentiation.
CRITICAL STEP: The success of subsequent steps in this differentiation protocol largely depends on effective induction of hematopoietic differentiation and lin-CD34+CD43+CD45+ progenitors in coculture with OP9. Therefore analysis of CD43 expression and simultaneous detection of CD235a/CD41a+ and CD45+ cells within CD43+ population can be performed to confirm myeloid commitment28 (see Fig.1).
CRITICAL STEP: Because we observed a decline in the hematopoietic differentiation capacity of hESCs after passage 50, we do not recommend the use of hESC lines beyond this passage. OP9 cells should not be used beyond passage 60, because of significant decrease in hematopoiesis-inductive potential.
TROUBLESHOOTING
Short-term expansion of multipotent myeloid progenitors TIMING 2 days
-
47
Wash out pHEMA-coated flasks with 20 ml (T75 flask) PBS.
-
48
Resuspend differentiated hPSCs (from Step 46) in multipotent myeloid progenitor expansion medium at a concentration of ~ 1×106 cells/ml.
CRITICAL STEP: Note that typically, 1.5–2×107 cells are recovered from one 10 cm dish of hPSC/OP9 coculture. We usually culture cells collected from 2 dishes in one T75 flask.
: GM-CSF is a single key factor required for expansion of hPSC-derived myeloid progenitors. The addition of SCF and/or FLT3L to expansion cultures has little effect on the growth of myeloid precursors, but significantly increases the proportion of CD235a+ erythroid cells.
-
49
Incubate 2 days at 37°C in a CO2 incubator.
CRITICAL STEP: Differentiated hPSCs cultured in non-adherent conditions spontaneously reaggregate and form large floating cellular conglomerates with myeloid progenitors proliferating in suspension as single cells.
Purification of multipotent myeloid progenitors TIMING 3 hrs
-
50
Collect myeloid cultures (from Step 49) and filter through a 70 µm cell strainer into a 50 ml tube to remove cell aggregates.
-
51
Pellet the cells by centrifugation at 250×g for 5 min at room temperature
-
52
Resuspend cells with 5 ml of MACS buffer in a 15 ml tube.
-
53
Underlay cell suspension with 1.0–1.5 ml of Percoll solution. Place a 1 ml plastic serological pipet filled with Percoll solution into the tube with cell suspension, so the pipet tip touches the bottom of the tube. Very carefully dispense Percoll solution to underlay cell suspension avoiding mixing.
-
54
Centrifuge tube at 300×g for 10–15 min at room temperature.
-
55
Aspirate supernatant and interface containing dead cells and debris.
-
56
Resuspend cells with 5 ml of MACS buffer and centrifuge at 250×g for 5 min at room temperature. Take an aliquot of the resuspended cells before centrifugation to count the number of isolated cells.
-
57
Aspirate supernatant and add 0.2 ml of MACS buffer to cell pellet.
-
58
Add 1 µl of anti-human CD235a-PE, and 5 µl of anti-human CD41a-PE antibodies per 106 cells.
CRITICAL STEP: Usually cells collected after Percoll separation are free of residual OP9 cells. If significant contamination of human cells with OP9 cells occurs, 10 µl of anti-mouse CD29-PE Ab can optionally be added to the cell pellet to deplete the mouse cells. The presence of contaminating OP9 cells in suspension can be evaluated by flow cytometry using mouse-specific CD29 antibodies.28
-
59
Set up tube on the MACS mixer and incubate at the lowest rotation speed at 4°C for 15–20 minutes.
-
60
Wash cells with ice-cold MACS buffer by adding 5 ml of MACS buffer to cell pellet followed by pipetting and centrifugation at 300×g for 5 min at 4°C, resuspend in 0.4 ml of MACS buffer, and add 10 µl of anti-PE magnetic beads.
-
61
Repeat step 59.
-
62
Wash cells with ice-cold MACS buffer as described in step 60 and resuspend in 1 ml of MACS buffer.
-
63
Filter cells through a 30 µm pre-separation filter.
CRITICAL STEP: It is important to filter cells before magnetic separation to remove large cell aggregates which may block the magnetic column.
-
64
Assemble the MACS-LD separation column according to the manufacturer’s instructions.
-
65
Wash column with 2 ml of MACS buffer.
-
66
Apply the cell suspension from Step 63 to the LD column allowing cells to pass completely through the column into 15 ml collection tube.
-
67
Wash column with 2 ml of MACS buffer and collect in same collection tube.
-
68
Recap and remove collection tube with unlabeled cells (CD235a-CD41a- human cells).
-
69
Centrifuge cells at 300×g for 5 min at 4°C.
-
70
Aspirate supernatant and add 80 µl of MACS buffer and 20 µl of anti-human CD45-FITC Ab.
-
71
Place the tube on the MACS mixer and incubate at the lowest rotation speed at 4°C for 15–20 minutes.
-
72
Wash cells with ice-cold MACS buffer as described in step 60, resuspend in 80 µl of MACS buffer, and add 20 µl of anti-FITC magnetic beads.
-
73
Repeat step 69.
-
74
Wash cells with ice-cold MACS buffer as described in step 60 and resuspend in 1ml of MACS buffer.
-
75
Filter cells through a 30 µm pre-separation filter.
-
76
Assemble MACS-LS separation unit according to the manufacturer’s instructions.
-
77
Rinse column with 2 ml of MACS buffer.
-
78
To purify CD235a/CD41a−CD45+ multipotent myeloid progenitors, apply the cell suspension from Step 75 to the LS column allowing cells to pass completely through the column into collection tube.
-
79
Wash column with 2 ml of MACS buffer and collect in same collection tube then discard.
-
80
Remove the column from the magnet and place in an empty 15 ml tube.
-
81
Wash out CD45+ cells with 5 ml MACS buffer using the plunger supplied with column.
-
82
Centrifuge cells at 300×g for 5 min at 4°C.
-
83
Resuspend cells in 0.2 ml of MACS buffer and keep on ice. Cells are ready to use for further differentiation.
TROUBLESHOOTING
Differentiation of hPSC-derived myelomonocytic cells
-
84
Differentiate hPSC-derived lin−CD34+CD43+CD45+ progenitors using option A for neutrophils, option B for eosinophils, option C for macrophages, option D for DCs, option E for LCs, and option F for osteoclasts. Protocol for cytospin preparation and staining of differentiated cells is provided as option G.
(A) Neutrophil differentiation TIMING 8–10 days
Prepare 6-well plates with semiconfluent OP9 monolayer.
-
Add 5×104 purified CD45+ cells in 2.5 ml of neutrophil differentiation medium per well of the 6-well plate.
CRITICAL STEP: To obtain a pure population of neutrophils, it is essential to limit cytokine addition to G-CSF alone. Although IL-3, IL-6, and GM-CSF have been used to increase the output of neutrophils from somatic CD34+ cells, we found that addition of these cytokines to the neutrophil differentiation cultures resulted in production of a mixture of eosinophils and neutrophils, with eosinophils often predominating.
On day 3, add 2.5 ml of neutrophil differentiation medium.
On day 6, change half of the neutrophil differentiation medium.
-
On day 8–10, collect differentiated cells from OP9 by gentle pipetting.
CRITICAL STEP: The presence of mature neutrophils in cultures can be quickly evaluated using Wright-stained cytospins (see protocol G below). The mature neutrophils have segmented nucleus (usually 2–5 segments joined by thin filaments) and pale pink cytoplasm (see Fig. 2). Because the viability and functionality of neutrophils are decreased with the extension of culture time, the optimal time for cell collection should be determined using vital dye staining and functional analysis of collected cells.
(B) Eosinophil differentiation TIMING 12–14 days
Prepare 6-well plates with semiconfluent OP9 monolayer.
-
Add 5×104 purified CD45+ cells in 2.5 ml of eosinophil differentiation medium per well of the 6-well plate.
CRITICAL STEP: IL5 alone is sufficient to differentiate hPSC-derived myeloid progenitors into eosinophils, however the addition of IL3 significantly increases the total cell output.
On day 3, add 2.5ml of eosinophil differentiation medium.
Change half of the eosinophil differentiation medium on days 6 and 9.
-
On day 12–14, collect differentiated cells by gentle pipetting.
CRITICAL STEP: The presence of mature eosinophils in cultures can be quickly evaluated using Wright-stained cytospins (see protocol G below). The mature eosinophils have classic bilobed nucleus and abundant eosinophilic cytoplasm filled by numerous coarse, orange-red granules of uniform size (see Fig. 2). In contrast, immature myeloid cells have round nucleus and less abundant basophilic or amphophilc cytoplasm. The optimal collection time for eosinophils should be determined based on predominance of viable mature cells in cultures.
(C) Macrophage differentiation TIMING 5–7 days
Wash a pHEMA-coated T25 tissue culture flask with 10 ml of PBS.
-
Add 1–5×105 purified CD45+ cells in 5mL of macrophage differentiation medium to the T25 flask.
CRITICAL STEP: M-CSF and IL-1β are two critical cytokines for the differentiation of myeloid precursors into macrophages. We do not recommend using GM-CSF in macrophage differentiation cultures, because cells continue to proliferate and do not mature into macrophages in presence of GM-CSF.
At day 3, add an additional 5ml of macrophage differentiation medium.
-
At day 5–7, collect differentiated cells.
CRITICAL STEP: The presence of mature macrophages in cultures can be quickly evaluated using Wright-stained cytospins (see protocol G below).The macrophages have round nucleus and abundant foamy cytoplasm (see Fig. 2). Because the viability and functionality of macrophages are decreased with the extension of culture time, the optimal time for cell collection should be determined using vital dye staining and functional analysis of collected cells.
(D) Dendritic cell differentiation TIMING 7 days
Wash a pHEMA-coated T25 tissue culture flask with 10 ml of PBS.
-
Add 5×105 isolated CD45+ cells in 5ml of DC differentiation medium without TNF-α to the T25 flask.
CRITICAL STEP: It is essential to avoid adding TNF-α during the first day of culture. We noted a significant decrease in cell viability if TNF-α was added to the freshly isolated cells.
On day 2–3, add 5ml of DC differentiation medium with TNF-α (2.5 ng/ml).
-
On day 7, collect differentiated cells.
CRITICAL STEP: On Wright-stained cytospins, cells of dendritic lineage can be recognized by the presence of cytoplasmic veils and dendritic projections (see Fig.2). However, flow cytometric analysis20,28 is essential to identify myeloid DCs which have CD1a+HLA-DR+DC-SIGN+CD11b+Langerin− phenotype.
(E) Langerhans cell differentiation TIMING 7 days
Wash a pHEMA-coated T25 tissue culture flask with 10 ml of PBS.
-
Add 5×105 cells in 5 ml LC differentiation medium without TNF-α and TGF-β1 to the T25 flask.
CRITICAL STEP: It is essential to avoid adding TNF-α and TGF-β1 during the first day of culture. We noted a significant decrease in cell viability if these cytokines were added to the freshly isolated cells.
On day 2–3, add 5ml of complete LC differentiation medium with TNF-α and TGF-β1 (2.5 ng/ml).
-
On day 7, collect differentiated cells.
CRITICAL STEP: On Wright-stained cytospins, cells of dendritic lineage can be recognized by the presence of cytoplasmic veils and dendritic projections (see Fig.2). However, flow cytometric analysis20,28 is essential to identify LCs which have CD1ahighHLA-DR+DC-SIGN−CD11blowLangerin+ phenotype.
(F) Osteoclast differentiation TIMING 16–19 days
Expansion step for osteoclast precursors. Wash a pHEMA-coated T25 tissue culture flask with 10 ml of PBS.
Add 5×104 to 1×105 cells in 5–10 ml of osteoclast progenitor expansion medium to the T25 flask.
Change half of the medium on day 2–3.
On day 4–5, collect cells; during the expansion step, osteoclast precursors adhere to the surface of the pHEMA coated-culture flask. Therefore, treat with 3 ml PBS (Ca2+ and Mg2+ free) or enzyme free cell dissociation buffer for 5–10min and collect cells by gentle pipetting.
Maturation step for osteoclasts. Plate 1–2×105 cells/well of a 6-well plate in 2.5 ml of osteoclast maturation medium.
-
Change half of the osteoclast maturation media every 3 days until the osteoclasts mature (12–14 days of culture).
CRITICAL STEP: Large, adhesive and multinucleated cells are markers for mature osteoclasts. However, immature myeloid cells (small round and non-adherent cells) keep proliferating due to the presence of GM-CSF and these proliferating cells interfere with maturation of the osteoclasts. Therefore, eliminate non-adherent cells by aspiration during regular medium changes. We found that the addition of M-CSF had no effect on development of osteoclasts in our differentiation system. Moreover, the addition of M-CSF to osteoclast cultures shifted differentiation hiPSC-derived myeloid progenitors toward macrophages.
(G) Cytospin prepartion and Wright staining
Use 2.5–3×104 cells per slide for Cytospin preparation and Wright staining
Cytospin preparation. Collect cells in 1.5 ml microcentrifuge tube and centrifuge at 400×g for 4 min.
Wash cells once by adding ice cold 0.5 ml of flow cytometry buffer followed centrifugation at 400×g for 4 minutes. Discard supernate and resuspend cells with 250µl of flow flow cytometry buffer.
During centrifugation, label glass slide and assemble with Cytofuse filter concentrator unit according to manufacturer’s instruction.
Transfer cell suspension into sample chamber of assembled glass slide-Cytofuse filter concentrator unit.
Spin samples at 700 rpm (27 ×g) for 4 min at room temperature.
Dry slide completely at room temperature.
Fix slide with absolute methanol for 30–60 seconds at room temperature.
Wright staining. Cover cytospin area with 300µl of Protocol Wright Stain and allow to stain for 3 minutes at room temperature. Make sure that stain covers the entire area with cells to ensure uniform staining of cells on cytospin.
Add 450 µl of pH 6.4 Diluted Buffer to the Wright stain-covered slide.
Mix the stain and buffer together by gently rocking slide for 1 minute and incubate the mixture for additional 4 minutes at room temperature.
Rinse the slide with deionized water and dry completely in the air.
Drop 2–3 droplets of Cytoseal 60 mounting medium on the stained cell area and cover with cover glass.
Observe under the microscope.
TIMING
See Fig. 3.
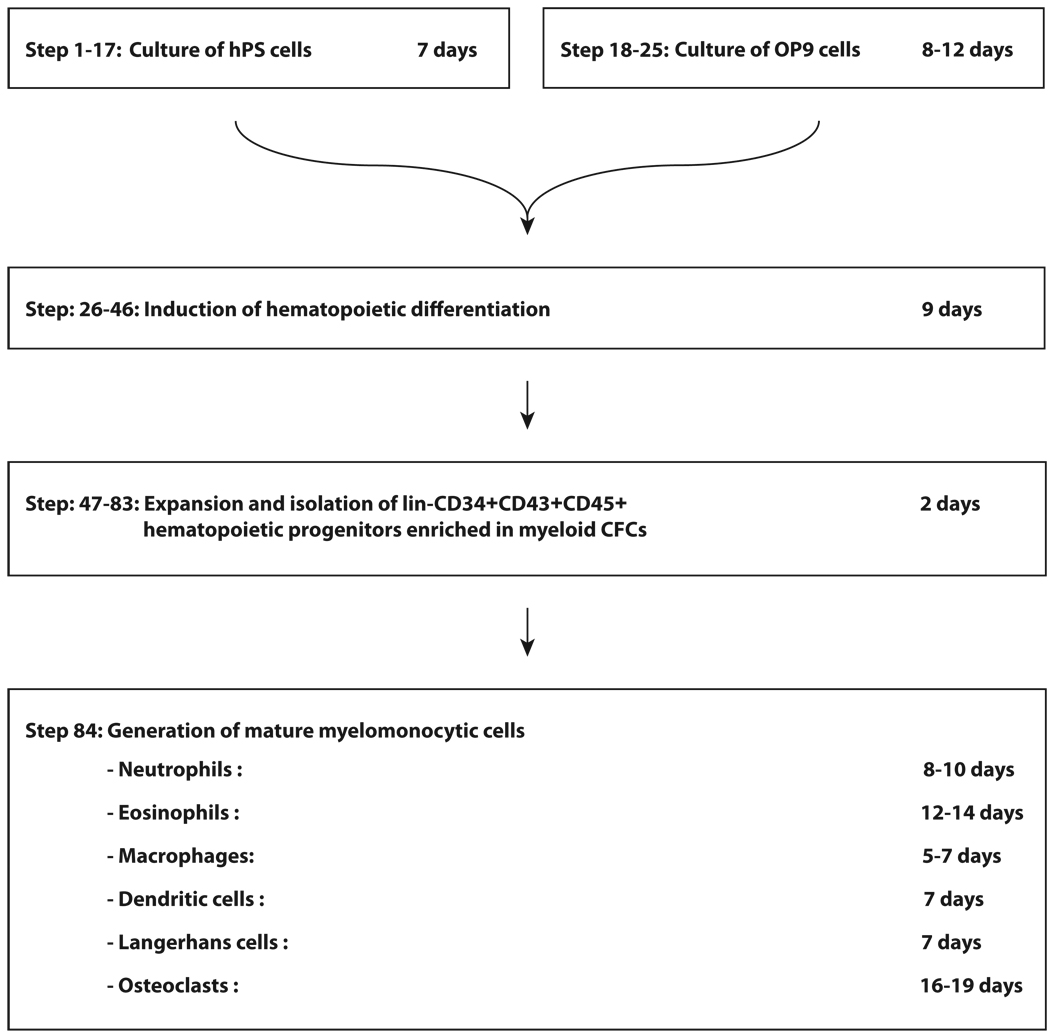
Figure 3.
Timing for directed differentiation of mature myelomonocytic cells from human pluripotent stem cells. To induce hematopoietic differentiation, hES/iPSCs are collected from MEF cocultures and transferred onto overgrown OP9 cells. After 9 days of coculture with OP9, cells should be collected and treated with enzymes to prepare single cell suspensions containing lin−CD34+CD43+CD45+ hematopoietic progenitors. To expand these progenitors, single cells should be cultured in non-adherent conditions (pHEMA-coated flasks) in the presence of GM-CSF for 2 days. Expanded lin−CD34+CD43+CD45+ progenitors enriched in myeloid CFCs should be isolated by magnetic sorting. Through cell culture with a particular cytokine combination, myeloid progenitors can be differentiated into mature myeloid cells of the desired type. Depending on the cell type, 5 to 19 days of culture are required to obtain a mature cell population.
TROUBLEHSOOTING
Troubleshooting advice can be found in Table 3.
ANTICIPATED RESULTS
Successful hematopoietic differentiation of hPSCs through coculture with OP9 yields colonies with radial sac-like structures (Fig. 1c). In cocultures with efficient differentiation, few undifferentiated colonies can be also observed (Fig. 4). The predominance of undifferentiated colonies is only seen in hPSC/OP9 cocultures that fail to generate good hematopoiesis. The efficiency of hematopoietic differentiation significantly varies among different hPSC lines18. Using H1 hESC, iPS(foreskin)-1 or iPSCs DF-19-9-7T, we can usually obtain differentiated cells containing 9–17% of CD43+ hematopoietic progenitors. CD235a+CD41a+/− erythromegakaryocytic progenitors comprise the majority of CD43+ cells (60–70%), while multipotent CD235a/CD41a−CD45+ cells comprise only 10–20% of the total number of CD43+ cells or 1–2% of the total number of hPSC-derived cells (Fig. 1e). After expansion with GM-CSF, the proportion of CD45+ cells within the population of hematopoietic progenitors significantly increases with up to 40–80% of cells being CD45+ (Fig. 1i). The total yield of myeloid progenitors after expansion depends primarily on the efficiency of the hematopoietic differentiation, measured as the absolute number of cells generated in OP9 coculture. We can typically obtain 1×106 lin−CD34+CD43+CD45+ cells from one 10 cm dish of H1/OP9 coculture after GM-CSF expansion. The yield of differentiated cells from the same starting material should be expected as follows: neutrophils 60 × 106, eosinophils up to 250 × 106, DCs 2 × 106, LCs 0.5 × 106, macropahges 1 × 106, and osteoclasts 6 × 106. The yield of macrophages and DCs can be increased by extending the GM-CSF expansion step to 8 days. However, prolonged expansion is associated with the decrease of granulocytic potential, and there is a significantly lower output of neutrophils and eosinophils from day 8 expanded CD45+ cells20.
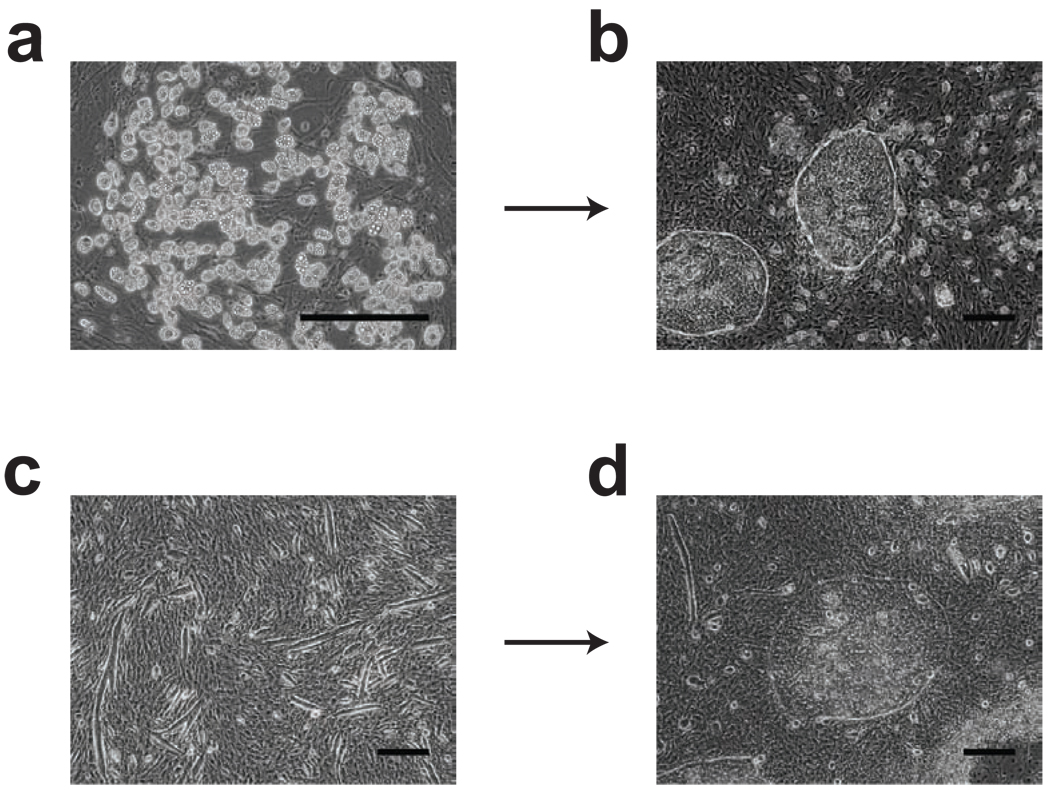
Figure 4.
Demonstration of morphological changes in OP9 associated with a loss of hematopoiesis-inductive properties. (a) Spontaneous adipogenesis in overgrown OP9 cells. (b) After 8 days of hPSC/adipogenic OP9 coculture, the majority of hPSC colonies retain undifferentiated morphology. (c) Heterogeneous overgrown OP9 cultures with spindle cells forming long cords. (d) After 8 days of hPSC/heterogeneous OP9 coculture, the majority of hPSC colonies retain undifferentiated morphology. Scale bars=300µm.
Table 4.
Troubleshooting
| Step | Trouble | Reason | Trouble shootings |
|---|---|---|---|
| Step 15 | Spontaneous hPSC differentiation, loss of pluripotency | Feeder-layer cells of low quality or inadequate density | - MEF lots have to be pretested for their ability to support hPSC growth. Make sure to use a semiconfluent MEF monolayer |
| - Mark differentiated colonies with objective markers under a microscope and eliminate differentiated hPSC colonies by aspiration of the marked area with glass Pasteur pipettes during medium change. | |||
| Step 23 | High spontaneous adipogenesis in OP9 cultures or loss of homogeneity of OP9 with formation of cord-like structures (Fig.4) | - Adipogenic serum | - Select FBS lots with minimal adipogenic effect. |
| - Incorrect split ratio | - Adjust the split ratio with a new FBS lot to achieve confluence by day 4 of differentiation. | ||
| - Incomplete enzymatic dissociation of OP9 cells during passage | - Make sure OP9 cells are completely detached after trypsinization. | ||
| Step 46 | Low efficiency of hematopoietic differentiation | - hPCSs spontaneously differentiate in maintenance culture | - See troubleshooting for step 15 |
| - Massive adipogenesis in OP9 cells | - See troubleshooting for step 23 | ||
| - Non-optimal plating density of hPSCs | - Optimize plating density using an initial range of 0.5×106 cells per 10 cm dish with a 0.5×106 interval. | ||
| - Low density of OP9 monolayer used to initiate differentiation of hPSCs | - Make sure to use overgrown OP9 cells before plate hPSCs. OP9 cells should be cultured for additional 4–6 days after forming a confluent monolayer. | ||
| - Irregular feeding | - Strictly adhere to the feeding schedule described in this protocol. | ||
| Step 79 | Number of isolated CD45+ cells is low | - Low efficiency of hematopoietic differentiation | - See troubleshooting for step 46. |
| - Loss of cells during magnetic separation | - Avoid bubble formation in column during separation procedure by using degassed MACS separation buffer. | ||
| - Remove cell aggregates by filtration before applying cell suspension to the column. | |||
| - Make sure that the primary antibodies have been appropriately titrated. |
ACKNOWLEDGMENTS
This study is supported by funds from the National Institute of Heart, Lung and Blood Institute (R01 HL081962, R21 HL085223) and NIH grant P51RR000167 to the National Primate Research Center, University of Wisconsin Madison. We thank Dr. Toru Nakano for providing OP9 cells.
Footnotes
AUTHOR CONTRIBUTION
K-D.C. designed and performed experiments, analyzed data and wrote the paper. M.V. design and performed experiments. I.S. supervised project, designed experiments, analyzed data, and wrote the paper.
CONFLICT OF INTERESTS
IS owns stock and is a scientific founder of Cellular Dynamics International.
Contributor Information
Kyung-Dal Choi, Email: kyungdalchoi@wisc.edu.
Maxim Vodyanik, Email: mvodyanik@hotmail.com.
References
- 1.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. Epub 2006 Aug 2010. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. Epub 2007 Nov 1920. [DOI] [PubMed] [Google Scholar]
- 5.Chadwick K, et al. Cytokines and BMP-4 promote hematopoietic differentiation of human embryonic stem cells. Blood. 2003;102:906–915. doi: 10.1182/blood-2003-03-0832. [DOI] [PubMed] [Google Scholar]
- 6.Ng ES, Davis RP, Azzola L, Stanley EG, Elefanty AG. Forced aggregation of defined numbers of human embryonic stem cells into embryoid bodies fosters robust, reproducible hematopoietic differentiation. Blood. 2005;106:1601–1603. doi: 10.1182/blood-2005-03-0987. Epub 2005 May 1624. [DOI] [PubMed] [Google Scholar]
- 7.Zambidis ET, Peault B, Park TS, Bunz F, Civin CI. Hematopoietic differentiation of human embryonic stem cells progresses through sequential hematoendothelial, primitive, and definitive stages resembling human yolk sac development. Blood. 2005;106:860–870. doi: 10.1182/blood-2004-11-4522. Epub 2005 Apr 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaufman DS, Hanson ET, Lewis RL, Auerbach R, Thomson JA. Hematopoietic colony-forming cells derived from human embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10716–10721. doi: 10.1073/pnas.191362598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiu C, et al. Differentiation of human embryonic stem cells into hematopoietic cells by coculture with human fetal liver cells recapitulates the globin switch that occurs early in development. Exp Hematol. 2005;33:1450–1458. doi: 10.1016/j.exphem.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 10.Ledran MH, et al. Efficient hematopoietic differentiation of human embryonic stem cells on stromal cells derived from hematopoietic niches. Cell Stem Cell. 2008;3:85–98. doi: 10.1016/j.stem.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 11.Vodyanik MA, Bork JA, Thomson JA, Slukvin II. Human embryonic stem cell-derived CD34+ cells: efficient production in the coculture with OP9 stromal cells and analysis of lymphohematopoietic potential. Blood. 2005;105:617–626. doi: 10.1182/blood-2004-04-1649. [DOI] [PubMed] [Google Scholar]
- 12.Wang ZZ, et al. Endothelial cells derived from human embryonic stem cells form durable blood vessels in vivo. Nat Biotechnol. 2007;25:317–318. doi: 10.1038/nbt1287. doi:nbt1287 [pii] 10.1038/nbt1287. [DOI] [PubMed] [Google Scholar]
- 13.Timmermans F, et al. Generation of T cells from human embryonic stem cell-derived hematopoietic zones. J Immunol. 2009;182:6879–6888. doi: 10.4049/jimmunol.0803670. doi:182/11/6879 [pii] 10.4049/jimmunol.0803670. [DOI] [PubMed] [Google Scholar]
- 14.Galic Z, et al. T lineage differentiation from human embryonic stem cells. Proc Natl Acad Sci U S A. 2006;103:11742–11747. doi: 10.1073/pnas.0604244103. Epub 12006 Jul 11714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takayama N, et al. Generation of functional platelets from human embryonic stem cells in vitro via ES-sacs, VEGF-promoted structures that concentrate hematopoietic progenitors. Blood. 2008;111:5298–5306. doi: 10.1182/blood-2007-10-117622. doi:blood-2007-10-117622 [pii] 10.1182/blood-2007-10-117622. [DOI] [PubMed] [Google Scholar]
- 16.Gaur M, et al. Megakaryocytes derived from human embryonic stem cells: a genetically tractable system to study megakaryocytopoiesis and integrin function. J Thromb Haemost. 2006;4:436–442. doi: 10.1111/j.1538-7836.2006.01744.x. [DOI] [PubMed] [Google Scholar]
- 17.Martin CH, Woll PS, Ni Z, Zuniga-Pflucker JC, Kaufman DS. Differences in lymphocyte developmental potential between human embryonic stem cell and umbilical cord blood-derived hematopoietic progenitor cells. Blood. 2008;112:2730–2737. doi: 10.1182/blood-2008-01-133801. doi:blood-2008-01-133801 [pii] 10.1182/blood-2008-01-133801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi K, et al. Hematopoietic and endothelial differentiation of human induced pluripotent stem cells. Stem Cells. 2009;27:559–567. doi: 10.1634/stemcells.2008-0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vodyanik MA, Thomson JA, Slukvin II. Leukosialin (CD43) defines hematopoietic progenitors in human embryonic stem cell differentiation cultures. Blood. 2006;108:2095–2105. doi: 10.1182/blood-2006-02-003327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi KD, Vodyanik MA, Slukvin II. Generation of mature human myelomonocytic cells through expansion and differentiation of pluripotent stem cell-derived lin-CD34+CD43+CD45+ progenitors. J Clin Invest. 2009;119:2818–2829. doi: 10.1172/JCI38591. doi:38591 [pii] 10.1172/JCI38591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vodyanik MA, Slukvin II. Directed differentiation of human embryonic stem cells to dendritic cells. Methods Mol Biol. 2007;407:275–293. doi: 10.1007/978-1-59745-536-7_19. [DOI] [PubMed] [Google Scholar]
- 22.Karlsson KR, et al. Homogeneous monocytes and macrophages from human embryonic stem cells following coculture-free differentiation in M-CSF and IL-3. Exp Hematol. 2008;36:1167–1175. doi: 10.1016/j.exphem.2008.04.009. Epub 2008 Jun 1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su Z, Frye C, Bae KM, Kelley V, Vieweg J. Differentiation of human embryonic stem cells into immunostimulatory dendritic cells under feeder-free culture conditions. Clin Cancer Res. 2008;14:6207–6217. doi: 10.1158/1078-0432.CCR-08-0309. [DOI] [PubMed] [Google Scholar]
- 24.Senju S, et al. Genetically manipulated human embryonic stem cell-derived dendritic cells with immune regulatory function. Stem Cells. 2007;25:2720–2729. doi: 10.1634/stemcells.2007-0321. Epub 2007 Aug 2729. [DOI] [PubMed] [Google Scholar]
- 25.Grigoriadis AE, et al. Directed differentiation of hematopoietic precursors and functional osteoclasts from human ES and iPS cells. Blood. 2010;115:2769–2776. doi: 10.1182/blood-2009-07-234690. doi:blood-2009-07-234690 [pii] 10.1182/blood-2009-07-234690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yokoyama Y, et al. Derivation of functional mature neutrophils from human embryonic stem cells. Blood. 2009;113:6584–6592. doi: 10.1182/blood-2008-06-160838. doi:blood-2008-06-160838 [pii] 10.1182/blood-2008-06-160838. [DOI] [PubMed] [Google Scholar]
- 27.Saeki K, et al. A Feeder-Free and Efficient Production of Functional Neutrophils from Human Embryonic Stem Cells. Stem Cells. 2009;27:59–67. doi: 10.1634/stemcells.2007-0980. [DOI] [PubMed] [Google Scholar]
- 28.Vodyanik MA, Slukvin II. Hematoendothelial differentiation of human embryonic stem cells. Curr Protoc Cell Biol. 2007;Chapter doi: 10.1002/0471143030.cb2306s36. Unit 23.26. [DOI] [PubMed] [Google Scholar]
- 29.Immunologic Studies in Humans. Curr Protoc Immunol. 2010;Chapter doi: 10.1002/0471142735.im0700s91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Innate Immunity. Curr Protoc Immunol. 2010;Chapter [Google Scholar]
- 31.Antigen processing and presentation. Curr Protoc Immunol. 2010;Chapter [Google Scholar]
- 32.Ludwig TE, et al. Feeder-independent culture of human embryonic stem cells. Nat Methods. 2006;3:637–646. doi: 10.1038/nmeth902. [DOI] [PubMed] [Google Scholar]