Summary
To reconstruct a minimum complement of notochord genes evolutionarily conserved across chordates, we scanned the Ciona intestinalis genome using the sequences of 182 genes reported to be expressed in the notochord of different vertebrates and identified 139 candidate notochord genes. For 66 of these Ciona genes expression data were already available, hence we analyzed the expression of the remaining 73 genes and found notochord expression for 20. The predicted products of the newly identified notochord genes range from the transcription factors Ci-XBPa and Ci-miER1 to extracellular matrix proteins. We examined the expression of the newly identified notochord genes in embryos ectopically expressing Ciona Brachyury (Ci-Bra) and in embryos expressing a repressor form of this transcription factor in the notochord, and we found that while a subset of the genes examined are clearly responsive to Ci-Bra, other genes are not affected by alterations in its levels. We provide a first description of notochord genes that are not evidently influenced by the ectopic expression of Ci-Bra and we propose alternative regulatory mechanisms that might control their transcription.
Keywords: Ciona, ascidian, notochord, chordate, evolution, Brachyury
INTRODUCTION
The notochord is one of the distinctive features of the Chordate phylum. Chordate embryos rely on the notochord as a source of structural support and patterning signals. In invertebrate chordates, the notochord is composed of a small number of cells (less than one hundred) and its function appears to be predominantly structural (Jiang and Smith, 2007; Satoh, 1994). In vertebrate embryos, the notochord lays out a blueprint for the vertebral column and provides stability and key developmental cues to the developing neural tube, cardiac field, and early endoderm (Cleaver and Krieg, 2001; Fekany et al., 1999; Placzek et al., 1993; Stemple, 2005).
Relatively little is known about the evolutionary origins of the notochord and about the molecular steps responsible for its increasing sophistication in the vertebrate lineage. The transcription factor Brachyury appears to be a major component of an evolutionarily conserved kernel in the notochord gene regulatory network, being expressed in the notochord of all the chordates analyzed so far, and being required for its proper formation (Di Gregorio et al., 2002; Schulte-Merker et al., 1994; Wilson et al., 1995). However, the current knowledge of additional evolutionary landmarks that are required for notochord formation across chordates is still fragmentary.
The ascidian Ciona intestinalis is a member of the tunicate clade possessing a primitive yet functional notochord. Ascidians are among the simplest experimentally amenable animals to have a notochord (Passamaneck and Di Gregorio, 2005; Satoh, 1994). Because ascidians have not undergone the genome duplications that vertebrates have, their gene networks are simpler and easier to decipher (Imai et al., 2006), even though they have likely experienced lineage-specific gene losses after diverging from the lineage leading to vertebrates in the Pre-Cambrian, roughly 600 million years ago (Hughes and Friedman, 2005; Swalla and Smith, 2008). Since Ciona embryos are translucent and contain 40 easily identifiable notochord cells, it is easy to visualize the effect of mutations and other perturbations on notochord development (e.g., Nakatani et al., 1999).
The basic structure of vertebrate and ascidian notochords is remarkably similar. In zebrafish, on which most research on the vertebrate notochord has focused, the notochord consists of a single row of vacuolated cells surrounded by a basement membrane (Scott and Stemple, 2005). The basement membrane, which has been shown to be required for notochordal integrity (Coutinho et al., 2004; Parsons et al., 2002), is composed of an inner basal lamina, a medial layer of collagen lattice, and an outer layer of loosely organized matrix (Scott and Stemple, 2005). In other vertebrates, such as chicks and mice, the basement membrane has a comparable ultrastructure, although it surrounds a notochord composed of more than a single row of cells (Camon et al., 1990). Ciona notochord cells are also arranged in a single row and are enveloped by a perinotochordal sheath composed of a basal lamina and a layer of circumferentially arranged extracellular matrix filaments (Cloney, 1990; Miyamoto and Crowther, 1985).
As a first step toward the reconstruction of the essential complement of evolutionarily conserved genes that are sufficient to form a functional notochord, and to form an understanding of the notochord genes that represent vertebrate innovations, we analyzed the expression of Ciona orthologs of previously described vertebrate notochord genes by whole-mount in situ hybridization (WMISH).
Through the analysis of the expression patterns of 73 Ciona intestinalis putative orthologs of vertebrate notochord genes, we identified 17 Ciona genes expressed in the ascidian notochord and/or its precursor blastomeres and three genes expressed in a nearly ubiquitous fashion during the developmental stages which encompass notochord formation. The predicted products of these genes are transcription factors (XBPa and miER1), an RNA-binding protein (Quaking), a signaling molecule (SWiP1), extracellular matrix proteins (Laminins α1, α4, and β1; Cadherin 8, Entactin, Fibronectin, Thrombospondin 3), enzymes (Carbonic anhydrase III, Furin, Lysyl oxydases 1, and 4), a cytoskeletal component (Cofilin), and an axon-guidance factor (Semaphorin 3A). We also found that 11 of the genes analyzed are expressed in a variety of embryonic tissues that do not include the notochord. Lastly, expression of 42 genes was not detected at any of the premetamorphic stages examined.
To investigate the hierarchical relationships between the newly identified notochord genes and the notochord-specific transcription factor Ciona Brachyury (Ci-Bra), we performed WMISH on embryos ectopically expressing Ci-Bra in neural and endodermal cells (Takahashi et al., 1999). We also analyzed the expression of these genes in embryos expressing in their notochord a repressor form of Ci-Bra, and we found that a significant fraction of these genes does not respond to alterations in the levels, function, and expression territory of Ci-Bra.
RESULTS
In Silico Identification of Putative Ciona intestinalis Orthologs of Vertebrate Notochord Genes
Reports of genes expressed even transiently in the vertebrate notochord (hereinafter: vertebrate notochord genes) were identified mainly through extensive searches of the public biomedical literature database of the U.S. National Library of Medicine (http://www.nlm.nih.gov/). As of the time of the preparation of this manuscript, there were at least 182 genes reported to be expressed in the notochord of a variety of vertebrate species, including Homo, Mus, Gallus, Danio, etc. The protein sequences encoded by these genes were retrieved and used to perform BLAST searches (Altschul et al., 1990) against the C. intestinalis genome (http://genome.jgi-psf.org/Cioin2/Cioin2.home.html).
For 66 of the Ciona genes of interest, expression patterns had been previously published or were available from the Ghost database (http://ghost.zool.kyoto-u.ac.jp/indexr1.html; Fujiwara et al., 2002; Imai et al., 2004; Kusakabe et al., 2002; Satou et al., 2001); in particular, 20 of these 66 genes had been previously described as being expressed in the Ciona notochord (Table S1, and references therein; see also Supplemental References), while 46 had been previously reported to be expressed in tissues other than the notochord (Table S2, and references therein). One of the genes we analyzed, cadherin 8, represents a putative ortholog of a Ciona savignyi gene previously examined, cadherinII, which is expressed at early developmental stages in notochord precursors, among other tissues (Imai, 2003).
Forty-three of the 182 vertebrate notochord genes did not have clear Ciona counterparts and were excluded from further study (Table S3). Finally, for 73 putative Ciona orthologs of vertebrate notochord genes no expression data were available, and it is on these genes that the present study is focused.
Identification of Novel Ciona Notochord Genes
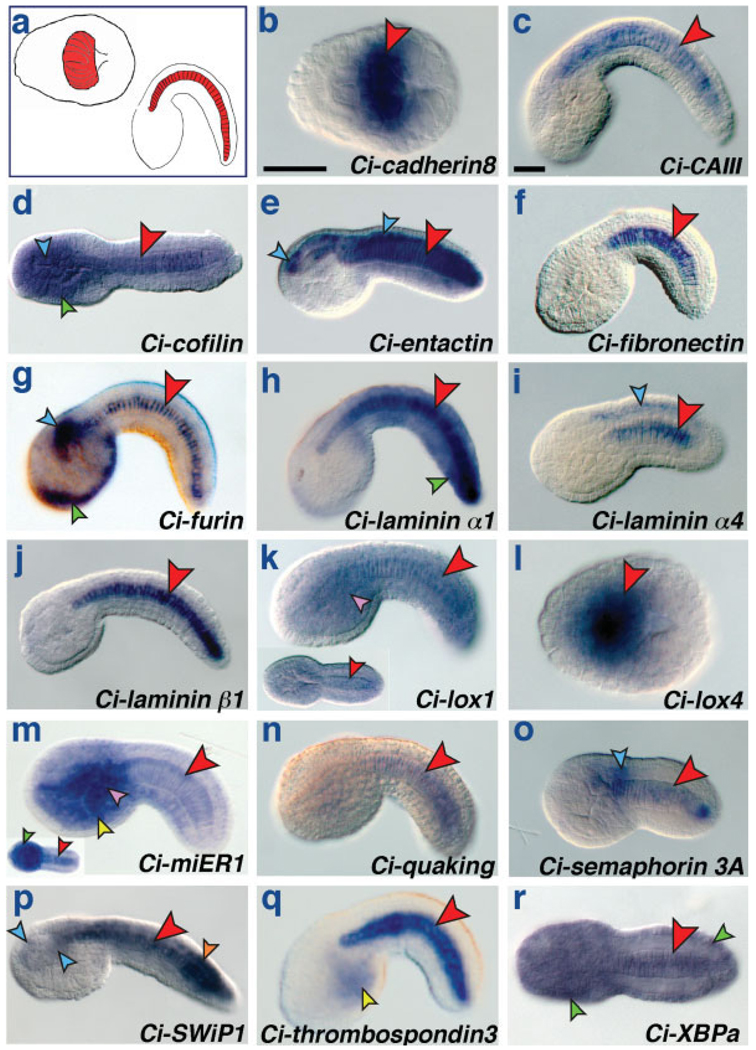
We analyzed the expression patterns of 73 putative C. intestinalis orthologs of vertebrate notochord genes by WMISH on C. intestinalis embryos fixed at stages ranging from 32-cell to late tailbud. 54 of the 73 genes were covered by available ESTs (Satou et al., 2002), and cDNAs for the remaining 19 genes were cloned by RT-PCR from total RNAs extracted from C. intestinalis embryos at different developmental stages (Table S4). Our results show that 17 genes are expressed in the Ciona notochord at various stages of embryonic development (Fig. 1a). The predicted products of these genes cover a wide range of probable functions, and include 7 putative extracellular matrix components, Cadherin 8 (Fig. 1b), Entactin (Fig. 1e), Fibronectin (Fig. 1f), Laminins α1 (Fig. 1h), α4 (Fig. 1i) and β1 (Fig. 1j), and Thrombospondin 3 (Fig. 1q), the enzymes Carbonic anhydrase III, Furin and Lysyl oxidases Lox1 and Lox4 (Fig. 1c,g,k,l), the cytoplasmic proteins Cofilin and SWiP1 (Fig. 1d,p), the transcription factors miER1 and XBPa (Fig. 1m,r), the RNA-binding protein Quaking (Fig. 1n), and the axon guidance factor Semaphorin 3A (Fig. 1o).
FIG. 1.
Novel notochord genes in Ciona intestinalis. (a) schematics of gastrula (left; vegetal view) and late tailbud (right; lateral view) embryos; anterior is to the left. The notochord and its precursors are highlighted in red. (b–r) Whole-mount C. intestinalis embryos hybridized in situ with digoxygenin-labeled antisense RNA probes for putative orthologs of vertebrate notochord genes. The genes are identified at the bottom right of each panel. Arrowheads highlight expression in various embryonic tissues and are color-coded as follows: red: notochord; blue: CNS; green: epidermis; purple: mesenchyme; yellow: endoderm; orange: muscle. Embryos in panels b and l are at the gastrula stage; embryos in all other panels are at various stages of tailbud development. Dorsal, up; anterior, left. Panels d and r and insets in k and m are dorsal views, with anterior to the left. Scale bars: 40 µm.
The majority of genes found to have expression in the notochord were also expressed in additional tissues, most notably the CNS (entactin, furin, laminin α4, semaphorin 3A, and SWiP1; blue arrowheads in Fig. 1e,g,i,o,p), epidermis (cofilin, furin, laminin α1, miER1, and XBPa; green arrowheads in Fig. 1d,g,h,m,r), mesenchyme (lox1 and miER1; purple arrowheads in Fig. 1k,m), muscle (SWiP1; orange arrowhead in Fig. 1p), and endoderm (miER1 and thrombospondin 3; yellow arrowheads in Fig. 1m,q).
Expression time-courses for stages ranging from 110-cell to mid/late tailbud are shown in Figure S1 and additional WMISH images are available online (http://cornell-celldevbiology.org/digregorio/research.html; hereinafter indicated as additional images online, AIO).
Of the 17 notochord genes, only three, carbonic anhydrase III, fibronectin, and laminin β1 were exclusively expressed in notochord cells at all stages analyzed (Fig. 1c,f j; Fig. S1; AIO); SWiP1 and thrombospondin 3 were notochord-specific only during early developmental stages, and expanded to additional expression domains at the tailbud stages (epidermis and trunk endoderm, respectively; Fig. 1p,q; Fig. S1; AIO). quaking expression was detectable only in muscle cells during early development and, interestingly, switched to notochord cells by the end of neurulation (Fig. 1n; AIO).
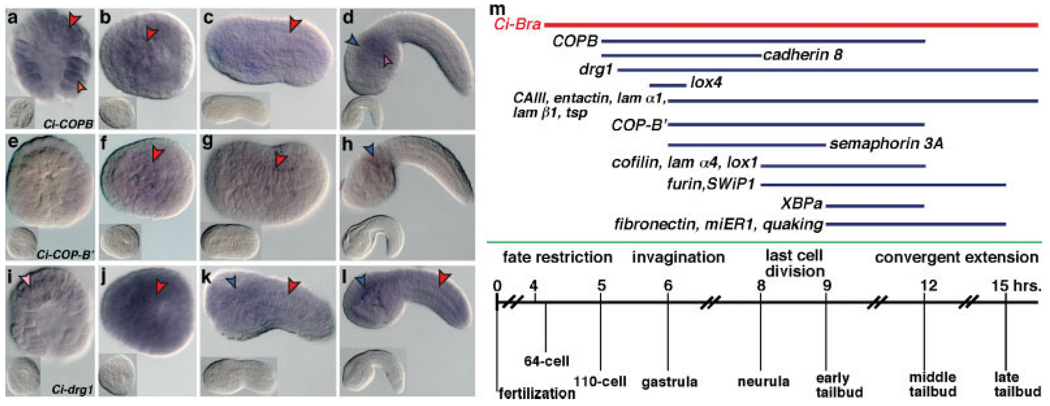
Three genes appeared to be expressed in all the embryonic tissues discernible by WMISH, including the notochord. These genes encode two subunits of the coatomer vesicular coat complex, COP β (COPB) and COP β′ (COPB′) and Drg1, a GTP-binding protein (see Fig. 2). COPB expression was detected at high levels in the majority of the blastomeres of the 110-cell embryo, including notochord and muscle precursors (Fig. 2a, red and orange arrowheads, respectively). Expression remained nearly ubiquitous throughout gastrulation and neurulation (Fig. 2b,c) but appeared somewhat more intense in cells of the CNS and mesenchyme from the mid-tailbud stage onward (Fig. 2d, blue and purple arrowheads, respectively). COPB′ expression was seen beginning at gastrulation and encompassed the notochord and its precursors during gastrulation and neurulation (Fig. 2f,g, red arrowheads). By the mid-tailbud stage, the levels of COPB′ transcripts decreased significantly in the majority of tissues, including the notochord, and were detected only in a subset of cells, possibly belonging to the CNS (Fig. 2h, blue arrowhead). drg1 was faintly expressed in the notochord precursors at the 110-cell stage (Fig. 2i, pink arrowhead); expression peaked at gastrulation, when this gene appeared expressed ubiquitously at high levels (Fig. 2j), and diminished at the tailbud stages, when the hybridization signal became more localized to the ventral region of the sensory vesicle and to the notochord (Fig. 2k,l). These diffuse patterns were validated by carrying out WMISH with sense probes under the same experimental conditions (insets in Fig. 2a–l).
FIG. 2.
Diffusely expressed genes and temporal expression windows of notochord genes. (a–l) Whole-mount C. intestinalis embryos hybridized in situ with digoxygenin-labeled antisense RNA probes showing a diffused signal. Stages: 110-cell (a,e,i), gastrula (b,f,j), early tailbud (c,g,k), and late tailbud (d,h,l). (a–d) Ci-COPB. (e–h) Ci-COP-B′. (i–l) Ci-drg1. Insets show embryos hybridized with the appropriate sense probes. Red arrowheads indicate the notochord territory, a pink arrowhead indicates faint staining in notochord precursors; blue arrowheads: CNS; purple arrowhead: mesenchyme; an orange arrowhead in a indicates a muscle precursor. m: temporal expression windows of the Ciona notochord genes identified in this study. The Ciona Brachyury (Ci-Bra) expression time-course is shown at the top as a reference (red bar; from Corbo et al., 1997). The blue bars indicate the time span during which different genes are expressed. The diagram at the bottom summarizes the developmental stages during which gene expression is observed and reports the main steps in notochord formation. The numbers on the scale indicate the number of hours necessary to reach the indicated developmental stages when embryos are incubated at 18°C (after Whittaker, 1977).
In Figure 2m, the windows of notochord expression of these 20 genes are superimposed to a time-course diagram summarizing the developmental milestones in notochord formation. Compared with Ci-Bra (red horizontal bar in Fig. 2m), which is first detected at the 64-cell stage, the time of notochord fate restriction (Corbo et al., 1997), the genes with the earliest onset are cadherin 8 and COPB, which are both detected in notochord precursors starting at the 110-cell stage, while the genes that are activated last in notochord cells are fibronectin, miER1, quaking, and XBPa, which begin being expressed in notochord cells at the early tailbud stage (blue bars in Fig. 2m; red bars in Fig. S1). lox4, miER1, and XBPa are only transiently expressed in the notochord (Fig. 2m), but continue to be detected in other tissues after their notochord transcription has faded.
Expression Patterns of Genes Expressed in Tissues Other Than the Notochord
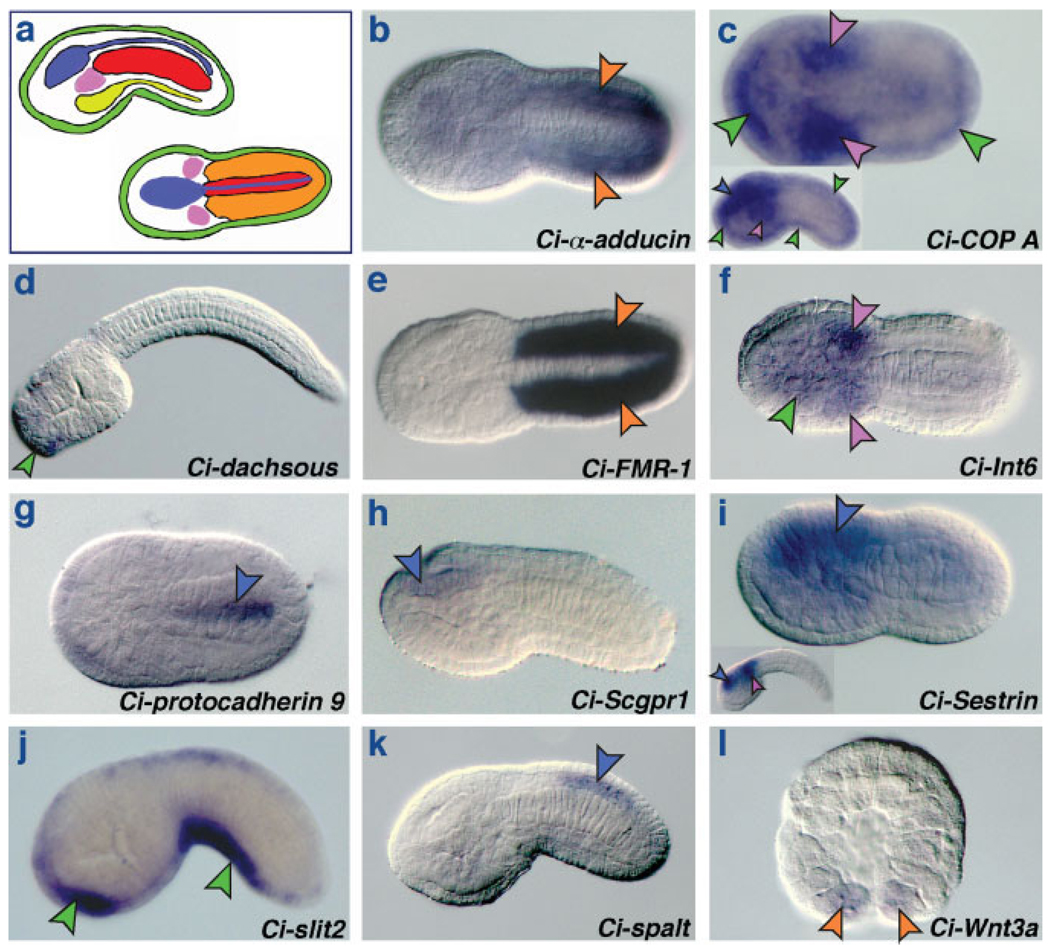
Eleven Ciona genes that were tested based on their sequence similarities with vertebrate notochord genes did not show notochord expression, but yielded specific expression patterns in tissues other than the notochord, including muscle, CNS, mesenchyme, and epidermis (color-coded in Fig. 3a). Representative embryos hybridized in situ with antisense probes specific for these genes are shown in Figure 3b–l. In particular, three genes are expressed in muscle and/or muscle precursors of developing Ciona embryos (orange arrowheads in Fig. 3b,e,l; AIO). α-adducin, encoding a putative cytoskeletal component, is expressed in muscle precursors from the 32-cell stage onward, and remains expressed in both primary and secondary muscle lineages throughout embryonic development (Fig. 3b and AIO). FMR-1, encoding the fragile-X mental retardation protein, is expressed in both primary and secondary muscle starting from the early tailbud stage (Fig. 3e and AIO) and Wnt3a, which encodes a signaling molecule, is transiently expressed in precursors of both secondary muscle and trunk ventral cells (heart progenitors) at the 110-cell stage and at gastrulation, but it is not detected afterward (Fig. 3l and AIO).
FIG. 3.
Ciona genes expressed in tissues other than the notochord. (a) Schematics of mid-tailbud stage ascidian embryos from a lateral view (top left) and a dorsal view (bottom right); anterior is to the left. Tissues are color-coded as follows: red: notochord; blue: CNS; green: epidermis; yellow: endoderm; purple: mesenchyme; orange: muscle. In panel l, the precursors of muscle and trunk ventral cells are indicated by an orange arrowhead for simplicity. (b–l) Whole-mount C. intestinalis embryos hybridized in situ with antisense RNA probes for putative orthologs of vertebrate notochord genes. The genes are identified at the bottom right of each panel. The embryo in panel d is at the late tailbud stage, the embryo in panel l is at the 110-cell stage. All other embryos are at the early or mid-tailbud stage.
Five genes are expressed in the developing nervous system (blue arrowheads in Fig. 3). COP A, a coatomer subunit, is expressed in the sensory vesicle from the early tailbud stage onward (Fig. 3c and AIO). Protocadherin 9, a cadherin-type cell adhesion protein, is expressed in the neural precursors at gastrula stage and is then restricted to the neural tube at early tailbud stages before expression tapers off at mid-tailbud (Fig. 3g and AIO). Scgpr1, (Spinal cord G-protein receptor 1), an endothelin-type receptor, is expressed specifically in the sensory vesicle from the early tailbud stage onward (Fig. 3h and AIO), unlike its vertebrate counterpart which is also detected in notochord, neural tube and spinal cord (Odani et al., 2007). Sestrin, a protein of unknown function, is expressed ubiquitously at the gastrula stage, but from neurula forward its expression is refined to the sensory vesicle (Fig. 3i and AIO). Spalt, a zinc-finger transcription factor, is expressed in the presumptive sensory vesicle at the neurula stage before its expression territory is confined to a segment of the posterior neural tube of the early tailbud embryo; this expression is also seen in subsequent stages (Fig. 3k and AIO).
Three genes, COP A (Fig. 3c), Int6, an ortholog of the p48 oncogene (Fig. 3f), and Sestrin (Fig. 3i) are expressed in trunk mesenchyme cells and their precursors (purple arrowheads in Fig. 3).
Finally, four genes are expressed in the epidermis: COP A, dachsous, Int6, and slit2 (Fig. 3c,d,f,j). Two of these genes, dachsous, encoding an atypical cadherin (Fig. 3d) and slit2, encoding an axon guidance factor (Fig. 3j), are both expressed in the epidermis near the tip of the trunk, in the presumptive adhesive organ territory; however, slit2 is expressed in a wider territory that encompasses also the ventral tail epidermis, where signal is detected at considerably high levels from neurulation on (Fig. 3j and AIO). COP A is expressed in both trunk and tail epidermis (green arrowheads in Fig. 3c), while expression of Int6 seems to be present only in cells of the dorsal trunk epidermis (Fig. 3f and AIO).
For the remaining 42 Ciona counterparts of vertebrate notochord genes, we could not detect expression at any of the stages analyzed. However, for 10 of these genes the EST counts indicated that their mRNAs were predominantly found at postmetamorphosis stages, and 11 genes were poorly represented in the EST counts overall (Satou et al., 2002; http://ghost.zool.kyoto-u.ac.jp/indexr1.html), suggesting that a large fraction of the undetectable genes might be expressed after metamorphosis and/or transcribed at levels too low to be revealed under the experimental conditions employed (see Methods).
Hierarchical Relationships of Ciona intestinalis Notochord Genes With Brachyury
At least 44 genes expressed in the developing Ciona notochord have been shown to be controlled, directly or indirectly, by Ci-Bra (Di Gregorio and Levine, 1999; Hotta et al., 2000, 2008; Oda-Ishii and Di Gregorio, 2007; Takahashi et al., 1999). Ci-Bra mRNA becomes detectable at the 64-cell stage, at which point the notochord precursors become fate-restricted (Corbo et al., 1997). When the function of Ci-Bra is perturbed, an organized notochord fails to form, notochord cells fail to intercalate and differentiate and, as a consequence, the tail fails to extend (Di Gregorio et al., 2002). These previous observations, together with the consideration that all the notochord genes identified in this study are expressed after Ci-Bra (Fig. 2m) suggest that, potentially, all the newly identified genes could be controlled by this transcription factor. To assess the relationship between the notochord genes described here and Ci-Bra, we performed a round of WMISH on embryos that ectopically expressed Ci-Bra in CNS and endoderm and on embryos that expressed a repressor form of this transcription factor in the notochord. For these experiments, we utilized the same probes used for the WMISH shown in Figure 1.
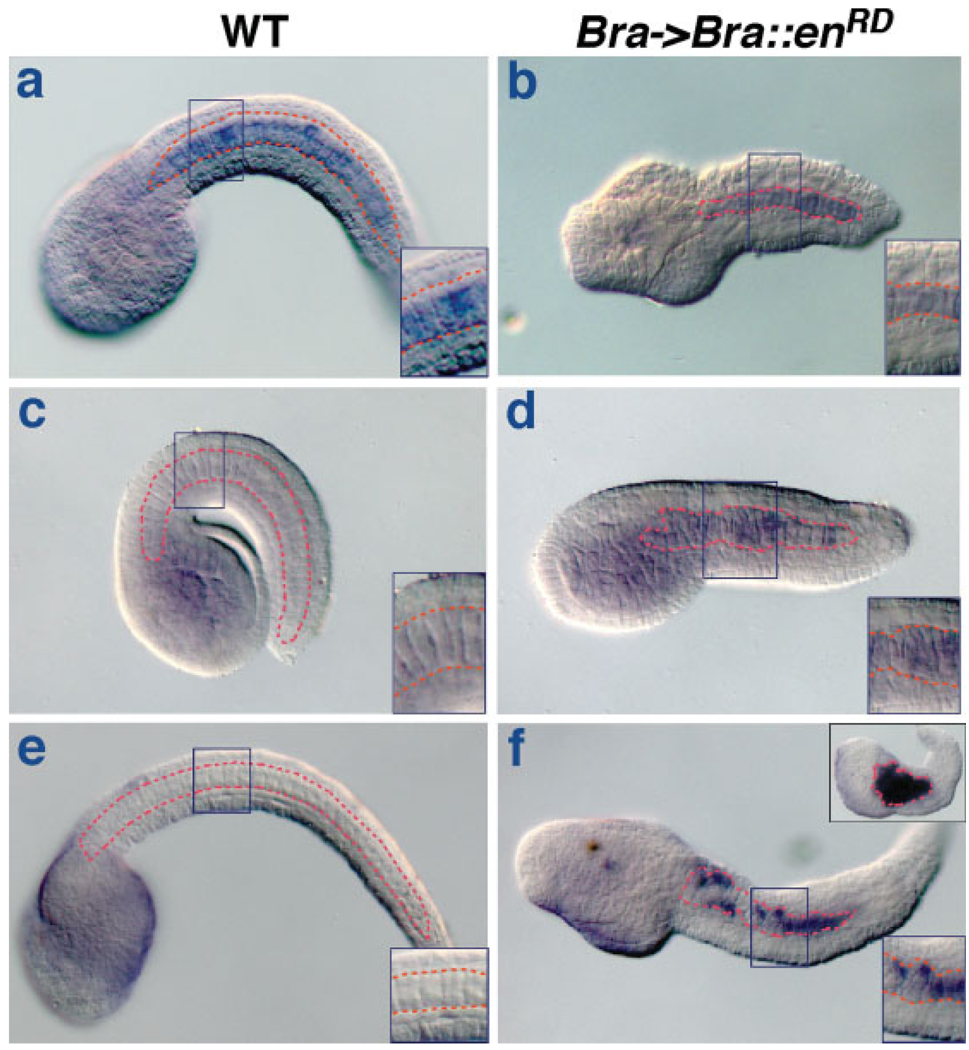
The plasmid used to ectopically express Ci-Bra was Ci-fkh->Ci-Bra (abbreviated as fkh->Bra), which drives expression of Ci-Bra in CNS, endoderm and notochord by means of the Ci-fkh promoter region (Di Gregorio et al., 2001; Takahashi et al., 1999). Embryos electroporated with fkh->Bra are easily identifiable since they display a characteristic phenotype, consisting of a short tail bearing a large number of cells in its ventro-medial region (Takahashi et al., 1999). To create a repressor form of Ci-Bra, we fused part of the Ci-Bra coding sequence to the Drosophila engrailed repression domain (enRD) and cloned the resulting sequence downstream of the Ci-Bra cis-regulatory region; the resulting plasmid, Bra->Bra::enRD, when used for transient transgenesis, is expected to repress transcription of Ci-Bra targets in the notochord (Jaynes et al., 1991). Consistent with this hypothesis, embryos electroporated with Bra->Bra::enRD display a characteristic “short-tail” phenotype, whereby the notochord is abnormal and, as a consequence, the tail is considerably shortened compared with that of control embryos (Fig. 4a,c,e; compare to b,d,f). In particular, in mid-tailbud embryos deriving from zygotes electroporated with the Bra->Bra::enRD plasmid, the notochord territory (surrounded by a dashed red line in Fig. 4) is disorganized, as individual notochord cells fail to acquire the “stack-of-coins” arrangement characteristic of this developmental stage (insets in panels a–d).
FIG. 4.
Effect of a repressor form of Ci-Bra on notochord development. (a,b) Whole-mount mid-tailbud stage embryos hybridized in situ with an antisense Ci-Bra probe. (c–f) Whole-mount mid-tailbud embryos hybridized with a transgene-specific probe against the Drosophila engrailed repression domain. (a–d) Mid-tailbud stage embryos. (e,f) Late-tailbud stage embryos. (a,c,e) Wildtype embryos. (b,d,f) Embryos electroporated at the one-cell stage with Bra->Bra::enRD. The inset on the top right of panel f depicts an embryo displaying high levels of transgene incorporation and a particularly severe phenotype. The notochord is outlined in red in all panels. Blue rectangles highlight the sites of the higher magnification views of the notochord shown in the insets on the bottom right of each panel. Embryos in each row derive from a single clutch and were incubated, fixed, and hybridized in parallel and stained for the same amount of time.
Similarly, at the late tailbud stage, the notochord cells of Bra->Bra::enRD embryos appear to have failed to complete intercalation (Fig. 4f; compare with 4e) and not to have started elongating along the anterior–posterior axis (compare insets in Fig. 4e,f). These morphological abnormalities are evident in Bra->Bra::enRD embryos hybridized in situ with a Ci-Bra probe (Fig. 4b) as well as in Bra->Bra::enRD embryos hybridized in situ with a transgene-specific probe encompassing only the engrailed repression domain (Fig. 4d,f), which was employed to assess the efficiency of incorporation of the Bra->Bra::enRD plasmid after electroporation (Table S5). We noticed that the severity of the phenotype observed was directly proportional to the amount of cells expressing the repressor form of Ci-Bra (Fig. 4f, inset on the top right side); however, embryos showing clear incorporation of the Bra->Bra::enRD transgene only in a small number of notochord cells also displayed tail malformations (Fig. 4f, Table S5 and data not shown). The short-tail phenotype seen in embryos electroporated with the Bra->Bra::enRD plasmid is clearly distinct from the aberrant morphology that can result from electroporation damage or random developmental malformations, as revealed by comparison with control embryos electroporated with a developmentally neutral reporter construct (data not shown).
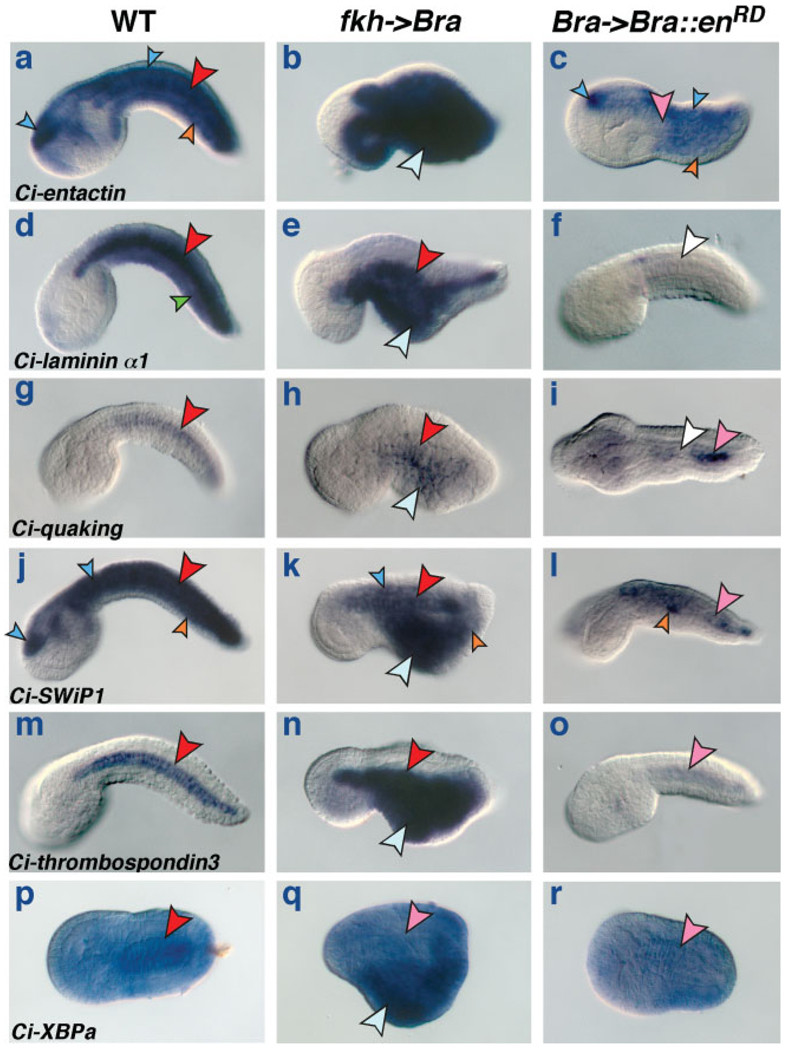
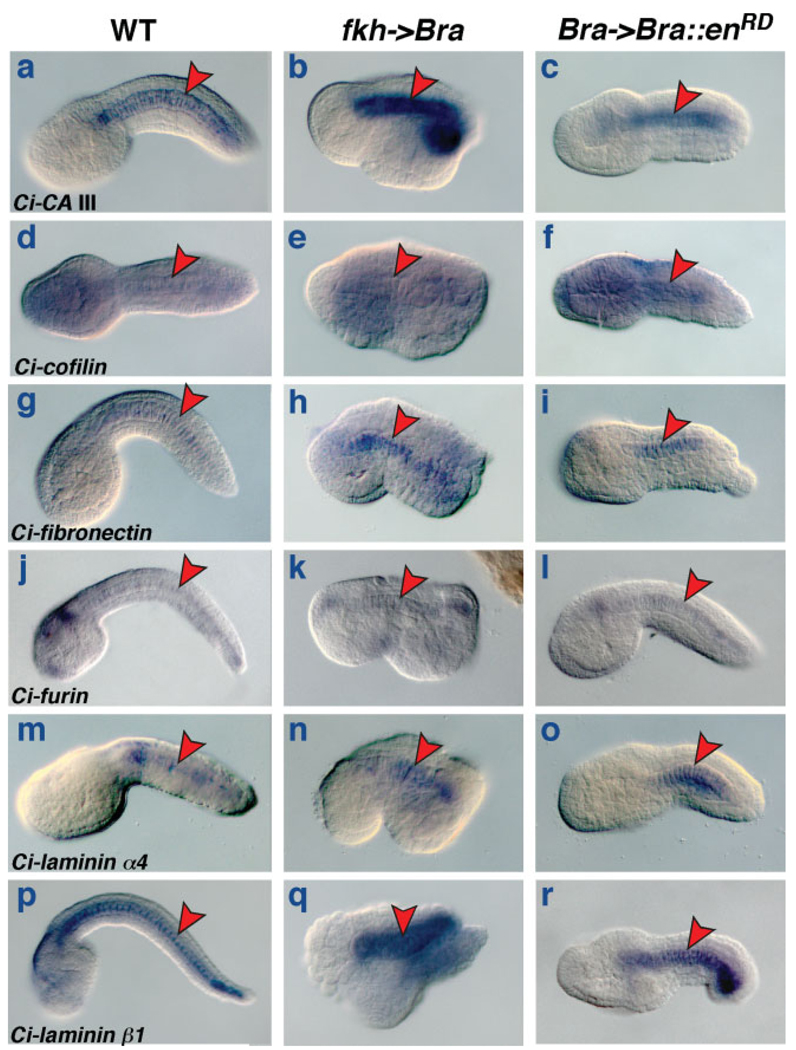
It has been previously shown that notochord genes with widely different expression patterns, such as Ci-ERM, Ci-trop, and Ci-multidom, respond to the ectopic expression of Ci-Bra induced by the fkh->Bra plasmid (Di Gregorio and Levine, 1999; Oda-Ishii and Di Gregorio, 2007; Takahashi et al., 1999), which is also sufficient, in transient transgenic assays, to ectopically activate notochord cis-regulatory modules, such as the Ci-trop notochord enhancer (Di Gregorio and Levine, 1999). By carrying out WMISH in parallel on control embryos, embryos electroporated at the 1-cell stage with the fkh->Bra plasmid and embryos electroporated at the 1-cell stage with the Bra->Bra::enRD plasmid, we found that six of the 17 newly identified notochord genes are responsive to changes in the levels of Ci-Bra (see Fig. 5), while six genes do not visibly respond to these perturbations (see Fig. 6). This analysis did not yield clear results in the case of cadherin 8, lox1, lox4, miER1, and semaphorin 3A (data not shown).
FIG. 5.
Evolutionarily conserved notochord genes responsive to Brachyury. (a–r) whole-mount C. intestinalis embryos hybridized in situ with antisense RNA probes for newly identified notochord genes (see Fig. 1). In each row, the gene analyzed is identified on the bottom left of the first panel. Embryos in each row derive from a single clutch and were incubated, fixed and hybridized in parallel and stained for the same amount of time. (a,d,g,j,m) Wild-type mid-tailbud embryos. (p) Wild-type early tailbud embryo. (b,e,h,k,n,q) Embryos electroporated at the 1-cell stage with fkh->Bra. (c,f,i,l,o,r) Embryos electroporated at the 1-cell stage with Bra->Bra::enRD. Pink and white arrowheads indicate weak or no notochord staining, respectively. Light blue arrowheads indicate neural and endodermal cells ectopically expressing Ci-Bra. Other arrowheads are color-coded as in previous figures.
FIG. 6.
Evolutionarily conserved notochord genes that do not respond to the ectopic expression of Brachyury. (a–r) Whole-mount C. intestinalis embryos hybridized in situ with antisense RNA probes for newly identified notochord genes (see Fig. 1). In each row, the gene analyzed is identified on the bottom left of the first panel. Embryos in each row derive from a single clutch and were incubated, fixed and hybridized in parallel and stained for the same amount of time. (a,d,g,j,m) Wild-type mid-tailbud embryos. (p) Wild-type late tailbud embryo. (b,e,h,k,n,q) Embryos electroporated at the 1-cell stage with fkh->Bra. (c,f,i,l,o,r) Embryos electroporated at the 1-cell stage with Bra->Bra::enRD. Red arrowheads indicate the notochord.
Entactin (Fig. 5a–c), laminin α1 (Fig. 5d–f), quaking (Fig. 5g–i), SWiP1 (Fig. 5j–l), thrombospondin 3 (Fig. 5m–o), and XBPa (Fig. 5p–r) are clearly ectopically expressed in CNS and endoderm cells of the fkh->Bra embryos, which are displaced to the ventro-medial region of the tail by the ectopic expression of Ci-Bra (light blue arrowheads in Fig. 5b,e,h,k,n,q). Likewise, these genes appear down-regulated in embryos transfected with the Bra->Bra::enRD plasmid (Fig. 5c,f,i,l,o,r). In this latter case, the effects of Bra->Bra::enRD on gene expression in notochord cells appear more variable, that is, expression seems to be more severely down-regulated in a subset of notochord cells (white and pink arrowheads in Fig. 5c,f,i,l,o,r), likely due to mosaic incorporation of the Bra->Bra::enRD plasmid (see Fig. 4 and Table S5).
In contrast, carbonic anhydrase III, cofilin, fibronectin, furin, laminin α4, laminin β1 (Fig. 6a,d,g,j,m,p,s) are still expressed in the notochord of fkh->Bra embryos but are not expressed ectopically in any other tissue (Fig. 6b,e,h,k,n,q). Likewise, the expression levels of these genes in Bra->Bra::enRD embryos seemed undiminished overall, even in notochord cells that appeared to have been efficiently transfected with the Bra->Bra::enRD plasmid (Fig. 6c,f,i,l,o,r).
DISCUSSION
Evolutionary Conservation of Vertebrate Notochord Genes in Ciona intestinalis
The work presented in this study, combined with previously published results from other laboratories, indicates that of the 182 vertebrate notochord genes targeted by this analysis, 40 have evolutionarily conserved notochord expression (20 are presented in this study, Figs. 1 and 2) and 57 are expressed in tissues other than the notochord (11 presented in this study, Fig. 3). For 42 genes we could not detect expression at any of the stages analyzed, which ranged from 64-cell to late tailbud; the larval stage provided technical challenges and unclear results and was therefore disregarded. The remaining 43 genes did not have clear orthologs in the Ciona intestinalis genome (Tables S1–S3).
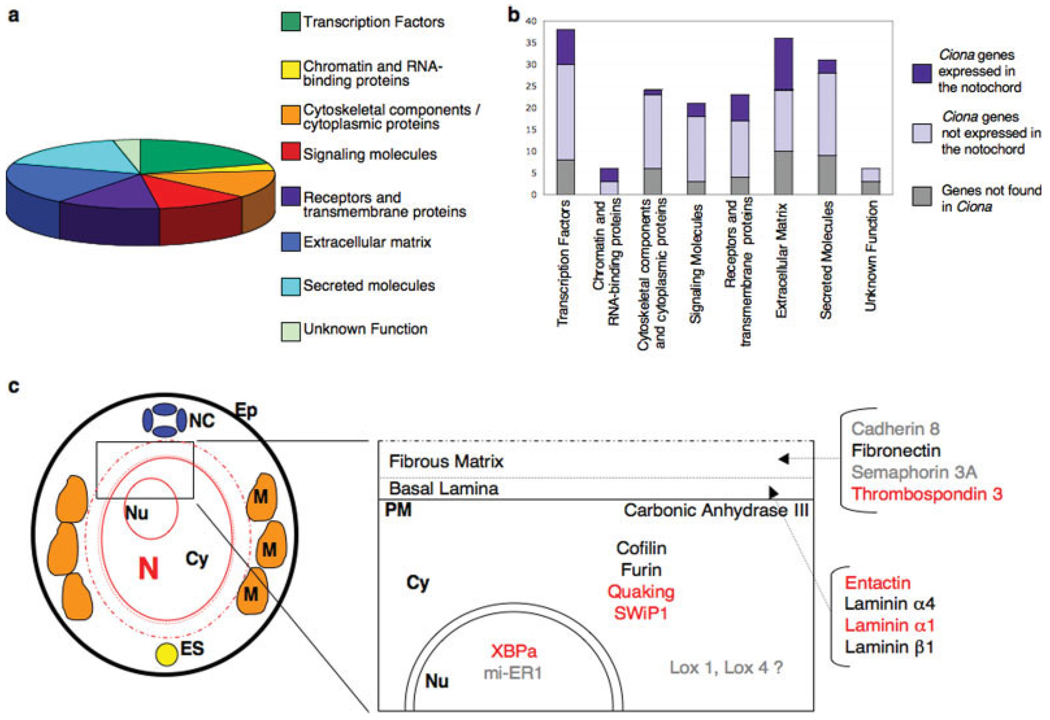
These results are summarized in Figure 7. The two largest fractions of the 182 vertebrate notochord genes encode transcription factors and extracellular matrix components (Fig. 7a). Interestingly, even though the majority of the genes in both categories are present in the Ciona genome (graph in Fig. 7b; dark blue and light blue sections of the bars; compare with the grey sections), the fraction of the extracellular matrix components expressed in the Ciona notochord is considerably higher than the fraction of transcription factors. These data suggest that the evolutionary conservation of structural components is higher than the conservation seen in putative transcription factors. Nevertheless, the Ciona notochord does express several putative transcription factors (Imai et al., 2004, 2006; Miwata et al., 2006), some of which might be still uncharacterized in vertebrates or simply represent lineage-specific acquisitions. Perhaps, the most representative example of transcription factors that are “missing” from either the Ciona genome or the Ciona notochord is provided by the Hox genes. Ciona intestinalis possesses an incomplete Hox cluster (Di Gregorio et al., 1995; Spagnuolo et al., 2003) and none of its Hox genes is expressed in the notochord (Ikuta et al., 2004). However, studies on the tunicate larvacean Oikopleura dioica show that five of the nine Hox genes found in its genome are expressed in the notochord (Seo et al., 2004), suggesting that Hox gene expression in the notochord might represent an ancestral condition rather than a trait acquired ex novo multiple times during chordate evolution.
FIG. 7.
Notochord genes in vertebrates and in Ciona. (a) pie graph showing the distribution of vertebrate notochord genes based on their putative function. Putative functions were ascertained from the literature (Tables S1–S3). Percentages are as follows: transcription factors: 20.2%; chromatin and RNA-binding proteins: 3.2%; cytoskeletal components/cytoplasmic proteins: 13.1%; signaling molecules: 11.4%; receptors and transmembrane proteins: 12.5%; extracellular matrix: 19.6%; secreted molecules: 16.8%; unknown function: 3.2%. (b) Bar graph showing the conservation of vertebrate notochord genes in Ciona. The y-axis reports the number of genes in each category. This graph includes the data presented in this study combined with previously published results; see Supplemental Tables and References for details. (c) Left panel: schematic showing a cross-section of the tail of a Ciona embryo at the mid-tailbud stage. Tissues are color-coded as described previously. A rectangle indicates the area which is enlarged to the right. Right panel: predicted subcellular localization of the newly identified Ciona notochord gene products. Products of genes responsive to the ectopic expression of Ci-Bra are highlighted in red, products of non-responsive genes are in black and genes not tested or uncertain are in grey. Abbreviations: Ep: epidermis; NC: nerve cord; M: muscle cell; ES: endodermal strand; N: notochord cell; Nu: nucleus; Cy: cytoplasm; PM: plasma membrane.
Nevertheless, the evolutionary conservation identified for the genes described in this study indicates that the minimum complement of evolutionarily conserved notochord genes found in Ciona can provide useful information on the essential repertoire of genes that are, potentially, necessary and/or sufficient to form a functional notochord, without the elaboration seen in vertebrates. Examples of effectors that likely underlie the increasing sophistication undergone by the notochord during chordate evolution are provided by genes not present in the Ciona genome, such as, for example, cdmp-1 which encode a cartilage-derived morphogenetic protein able to stimulate expression of osteogenic markers (Yeh et al., 2005), or chondromodulin 1 and the related genes encoding cartilage-derived growth factors (Suzuki, 1996).
Additionally, the analysis of genes that are not expressed in the Ciona notochord but are specifically expressed in other embryonic tissues (see Fig. 3) allows us to tentatively reconstruct their ancestral function and might help in pinpointing the evolutionary timing of their co-option to the notochord. This is possibly the case for the transcription factor gene spalt, which in Drosophila is a homeotic gene expressed in the embryonic CNS and required for its organization (Cantera et al., 2002). spalt is expressed in the developing nervous system in Ciona (see Fig. 3) but in mouse has acquired additional functions in mesodermal derivatives, including the notochord (Ott et al., 1996).
Lastly, our finding of the evolutionarily conserved expression of semaphorin 3A in the Ciona notochord, together with the earlier identification of a netrin gene expressed in notochord cells (Hotta et al., 1999, 2000), argues in favor of an active patterning role of the ascidian notochord in CNS development.
Potential Functions of the Newly Identified Ciona Notochord Genes
Figure 7c provides a tentative map of the putative intra- and extracellular localization of the products of the notochord genes identified in this study. In this diagram, extracellular matrix molecules such as the Laminins and Entactin are predicted as components of the basal lamina, while Cadherins and Thrombospondin 3 are possibly part of the outer fibrous matrix of the notochordal sheath, which supplies the notochord with the rigidity it needs to ensure support to the developing embryo. The axon guidance factor Semaphorin 3A likely provides directional cues to the developing neurons. Interestingly, recent work from the sister ascidian species Ciona savignyi demonstrates that one of the laminin genes, highly related to the laminin α1 gene analyzed here, encodes a laminin localized externally to the notochord; chongmague mutants, lacking the function of this laminin, show severe defects in notochord morphogenesis (Veeman et al., 2008).
The evolutionary conservation in the expression of laminin genes in the notochord is of particular interest, since it has been shown that certain laminins are essential for the structural integrity of the zebrafish notochord (Parsons et al., 2002). Laminin trimers form a meshwork that becomes the perinotochordal basement membrane, which envelopes the notochord, helping it to maintain a rigid structure. Individual mutations in laminin β1, γ1, or compound mutations of α1 and α4, which are redundant, abolish formation of the perinotochordal basement membrane (Parsons et al., 2002). Additionally, zebrafish mutants for coatomer complex subunits, such as COP β or COP β′, display disrupted Golgi apparati and lack the outer layers of the perinotochordal basement membrane (Coutinho et al., 2004).
The conservation of genes encoding coatomer proteins (COP) and lysyl oxidases (lox) reinforces the molecular similarities between ascidian and vertebrate notochords. Lysyl oxidases are copper-binding enzymes that are implicated in collagen and elastin cross-linking in vertebrates (Hornstra et al., 2003). Work carried out in zebrafish has demonstrated a necessary role for two specific lysyl oxidase genes, loxl1, and loxl5b, in notochord development. Simultaneous morpholino knockdown of both loxl1 and loxl5b results in a distorted, wavy notochord, a phenotype which can be rescued by microinjection of either loxl1 or loxl5b mRNA (Gansner et al., 2007). It can be therefore hypothesized that the cross-linking function of these enzymes is crucial for the integrity of the extracellular matrix that composes the notochordal sheath. We tentatively assigned a cytoplasmic localization to the Ciona enzymes (question mark in Fig. 7c), based on reports from vertebrates (Guo et al., 2007), although lysyl oxidases have also been demonstrated to be secreted extracellularly (Jansen and Csiszar, 2007).
Transcriptional Regulation of Evolutionarily Conserved Notochord Genes
In Ciona intestinalis, ectopic expression of Ci-Bra induces up-regulation of at least 44 notochord genes (Hotta et al., 2008; Takahashi et al., 1999). The observation that all the notochord genes identified in this study are expressed after the onset of Ci-Bra transcription (see Fig. 2) prompted us to analyze the effects of the ectopic expression of Ci-Bra and of its conversion to a putative transcriptional repressor (Ci-Bra-engrailedRD) on the expression of these genes. When expressed in developing notochord cells, the Ci-Bra-engrailedRD fusion protein caused a specific and distinctive phenotype, characterized by impaired notochord formation and a resulting short, improperly developed tail (see Fig. 4). In particular, the number of distinguishable notochord cells appeared reduced in embryos transfected with Bra >Bra::enRD as compared with wild-type embryos.
We hypothesize that this might be because of a block in cell division attributable, in turn, to the repression of Ci-Bra targets associated with mitosis and cell division, such as Ci-cdc45, a cell-cycle modulator, and Ci-PCNA, an essential component of the DNA replication machinery (Hotta et al., 2000, 2008). Surprisingly, even tailbud embryos expressing the Ci-Bra-engrailedRD fusion at detectable levels only in a small number of notochord cells displayed a phenotype (Table S5); one possible explanation for this phenomenon is that several Ci-Bra targets that might be repressed by Ci-Bra-engrailedRD are involved in cell-cell signaling pathways that are crucial for notochord morphogenesis. One of these genes is likely to be prickle, which has been shown to be required for correct intercalation of notochord cells in both Ciona intestinalis and Ciona savignyi (Hotta et al., 2007; Jiang et al., 2005). In addition, recent work has shown that also other components of the Wnt/PCP signaling pathway, such as Ci-cdc42, Ci-zipper, Ci-wnt-a, and Ci-wnt9/14/15 are expressed in the Ciona notochord and are controlled by Ci-Bra (Hotta et al., 2008). Finally, the incomplete intercalation and the morphological aberrations observed in the notochord of embryos expressing Ci-Bra-engrailedRD are consistent with the phenotypes described in individual morpholino-mediated knockdowns of the Ci-Bra targets Ci-ACL, Ci-Noto3, and Ci-pellino (Hotta et al., 2007).
Of the notochord genes identified here, six appeared to be ectopically expressed as a consequence of the ectopic expression of Ci-Bra driven by the Ci-fkh promoter in CNS and endoderm (see Fig. 5). These genes encode the extracellular matrix proteins Entactin, Laminin α1, and Thrombospondin, the signaling molecule SWiP1, the transcription factor XBPa, and, remarkably, the RNA-binding protein Quaking, which in Xenopus is required for the accumulation of mRNAs required for notochord development, including the Xenopus Brachyury mRNA (Zorn and Krieg, 1997).
Although their response to the expression in notochord cells of a Ci-Bra-engrailedRD fusion was not as clear as the response seen in the previous case, likely as a result of mosaic incorporation of the plasmid in notochord precursors, these genes appeared down-regulated in the short tails of the embryos electroporated with the Bra>Bra::enRD construct. These results suggest that these genes might be regulated by Ci-Bra either directly or via transcriptional relays and/or cofactors that are not specifically localized to the notochord. A relay mechanism and a required interaction of Ci-Bra with cofactors characterized by narrower expression windows might also explain the differences that we observed in the onset of transcription of the notochord genes analyzed here, as well as the differences that were previously observed in the onset of Ci-Bra targets (Hotta et al., 2000; Oda-Ishii and Di Gregorio, 2007).
Conversely, the expression of six other genes did not seem affected by these perturbations (see Fig. 6). These genes encode extracellular matrix proteins, such as laminin subunits and Fibronectin, a cytoskeletal component, Cofilin, and the enzymes Carbonic anhydrase III and Furin. The finding that these genes do not respond to the ectopic expression of Ci-Bra in neural and endodermal cells still leaves open the possibility that their expression in the notochord might be controlled by an obligate cooperation between Ci-Bra and a notochord-specific cofactor. This might indeed be the case for one such gene, carbonic anhydrase III, which appears to be slightly up-regulated in the notochord of fkh->Bra embryos, possibly in response to the increase in the levels of Ci-Bra caused by the transgene (Fig. 6b), even though this gene does not appear to be down-regulated in Bra>Bra::enRD embryos (Fig. 6c), suggesting that Ci-Bra is not required for its expression to occur. In the case of the remaining five genes, the observation that the notochord expression is not evidently affected by either the over-expression of Ci-Bra in the notochord induced by the Ci-fkh promoter or by the expression of the Ci-Bra-engrailedRD fusion protein indicates that their expression relies on a Ci-Bra-independent mechanism. The discovery of a group of notochord genes insensitive to gross alterations in the levels and territories of Ci-Bra expression is relevant since it might also partially explain the observation that neural and endodermal precursors adopt a “notochord-like” phenotype in response to the ectopic expression of Ci-Bra, but fail to form a fully organized ectopic notochord.
In conclusion, these results are a first hint at the existence of Brachyury-independent shunts in the gene regulatory circuitry intrinsic to the Ciona notochord.
METHODS
Animals and Electroporations
Adult Ciona intestinalis were purchased from Marine Research and Educational Products (M-REP; Carlsbad, CA) and kept at 18°C in recirculating artificial sea water. Culturing and electroporations were carried out as previously described (Oda-Ishii and Di Gregorio, 2007). Embryos to be used for whole-mount in situ hybridization (WMISH) were fixed in 4% paraformaldehyde in 0.1 M MOPS (pH 7.5), 0.5 M NaCl at 4°C overnight, then put through washes containing increasing concentrations of ethanol, ending with 70% ethanol/ddH20, and stored at −20°C.
Plasmid Construction
The Bra>Bra::enRD construct was generated by amplifying the Ci-Bra coding sequence from the published Ci-Bra cDNA clone (Corbo et al., 1997) and ligating it into the Ci-Bra->eGFP plasmid (Corbo et al., 1997) as a NotI/BlpI fragment. An 897-bp fragment of Drosophila engrailed encoding the repression domain (enRD) was amplified with the primers:
Dm-en-F1: 5′-AATGGCCCTGGAGGATCGCTGC-3′ and
Dm-en-R1: 5′-AGGGATCCCAGAGCAGATTTCTC-3′
and subsequently inserted in-frame into the Ci-Bra coding sequence at a BstZ17I site near its 3′ end, to generate the Bra::enRD fusion.
Probe Preparation
The complete list of the EST clones used in this study is provided in Table S4. The Ci-Bra probe was prepared from EST clone 13h10 (Satou et al., 2001). The Drosophila enRD probe was prepared as follows: the enRD fragment was PCR-amplified from the Bra>Bra::enRD construct using the primers Dm-en-F1 and Dm-en-R1 and Hi-Fi Taq polymerase (Invitrogen, Carlsbad, CA), then ligated into pGEM-T (Promega, Madison, WI).
cDNAs not represented in the EST collection were amplified by RT-PCR, essentially as previously described (Oda-Ishii and Di Gregorio, 2007). Plasmid DNA was purified using the QIAprep Spin Miniprep kit (Qiagen, Valencia, CA), linearized using appropriate restriction enzymes (New England Biolabs, Ipswich, MA), and purified by standard phenol-choloroform extraction followed by ethanol precipitation. One microgram of each purified plasmid DNA was used as a template for in vitro transcription in the presence of 11-Digoxigenin-UTP (Roche, Indianapolis, IN). Probes were purified by lithium chloride precipitation, then resuspended in: 50% formamide, 5× SSC, 100 µg/ml yeast tRNA, 50 µg/ml Heparin, and 0.1% Tween-20, and stored at −20°C.
Whole-Mount In Situ Hybridization
WMISH experiments were performed essentially as previously described (Oda-Ishii and Di Gregorio, 2007), using a hybridization temperature of 42°C. After the detection reactions were satisfactorily completed (~4–48 h, depending on the probes), embryos were washed six times in 100% ethanol, rinsed briefly in xylenes, and mounted in Permount (Sigma, St. Louis, MO).
Supplementary Material
ACKNOWLEDGMENTS
The authors are indebted to Dr. Nori Satoh (Kyoto University) for providing the Ciona intestinalis cDNA collection. We thank Dr. Yutaka Nibu and members of the Di Gregorio and Nibu labs for discussion and comments on the manuscript. We thank Dr. José Xavier-Neto for valuable editorial comments. J.E.K. was supported in part by a Jacques Cohenca predoctoral fellowship from the Weill Cornell Graduate School of Medical Sciences. A.D.G. is an Irma T. Hirschl Scholar. This manuscript is respectfully dedicated to Prof. Nick Cozzarelli.
Contract grant sponsor: NIH/NICHD, Contract grant number: R01HD050704; Contract grant sponsor: March of Dimes Birth Defects Foundation, Contract grant number: 5-FY03-153.
Abbreviations
- bp
base pair(s)
- kb
kilobase(s), or 1000 base pairs
- PCR
polymerase chain reaction
- CNS
central nervous system
- WT
wild-type
- eGFP
enhanced green fluorescent protein
Footnotes
Additional Supporting Information may be found in the online version of this article.
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Camón J, Degollada E, Verdú J. Ultrastructural aspects of the production of extracellular matrix components by the chick embryonic notochord in vitro. Acta Anat (Basel) 1990;137:114–123. doi: 10.1159/000146869. [DOI] [PubMed] [Google Scholar]
- Cantera R, Lüer K, Rusten TE, Barrio R, Kafatos FC, Technau GM. Mutations in spalt cause a severe but reversible neurodegenerative phenotype in the embryonic central nervous system of Drosophila melanogaster. Development. 2002;129:5577–5586. doi: 10.1242/dev.00158. [DOI] [PubMed] [Google Scholar]
- Cleaver O, Krieg PA. Notochord patterning of the endoderm. Dev Biol. 2001;234:1–12. doi: 10.1006/dbio.2001.0214. [DOI] [PubMed] [Google Scholar]
- Cloney RA. Urochordata: Ascidiacea. In: Adiyodi KG, Adiyodi RG, editors. Reproductive Biology of Invertebrates. New Delhi: Oxford and IBH; 1990. pp. 391–451. [Google Scholar]
- Corbo JC, Levine M, Zeller RW. Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian Ciona intestinalis. Development. 1997;124:589–602. doi: 10.1242/dev.124.3.589. [DOI] [PubMed] [Google Scholar]
- Coutinho P, Parsons MJ, Thomas KA, Hirst EM, Saúde L, Campos I, Williams PH, Stemple DL. Differential requirements for COPI transport during vertebrate early development. Dev Cell. 2004;7:547–558. doi: 10.1016/j.devcel.2004.07.020. [DOI] [PubMed] [Google Scholar]
- Di Gregorio A, Corbo JC, Levine M. The regulation of forkhead/HNF-3β expression in the Ciona embryo. Dev Biol. 2001;229:31–43. doi: 10.1006/dbio.2000.9964. [DOI] [PubMed] [Google Scholar]
- Di Gregorio A, Harland RM, Levine M, Casey ES. Tail morphogenesis in the ascidian, Ciona intestinalis, requires cooperation between notochord and muscle. Dev Biol. 2002;244:385–395. doi: 10.1006/dbio.2002.0582. [DOI] [PubMed] [Google Scholar]
- Di Gregorio A, Levine M. Regulation of Ci-tropomyosin-like, a Brachyury target gene in the ascidian, Ciona intestinalis. Development. 1999;126:5599–5609. doi: 10.1242/dev.126.24.5599. [DOI] [PubMed] [Google Scholar]
- Di Gregorio A, Spagnuolo A, Ristoratore F, Pischetola M, Aniello F, Branno M, Cariello L, Di Lauro R. Cloning of ascidian homeobox genes provides evidence for a primordial chordate cluster. Gene. 1995;156:253–257. doi: 10.1016/0378-1119(95)00035-5. [DOI] [PubMed] [Google Scholar]
- Fekany K, Yamanaka Y, Leung T, Sirotkin HI, Topczewski J, Gates MA, Hibi M, Renucci A, Stemple D, Radbill A, Schier AF, Driever W, Hirano T, Talbot WS, Solnica-Krezel L. The zebrafish bozozok locus encodes Dharma, a homeodomain protein essential for induction of gastrula organizer and dorsoanterior embryonic structures. Development. 1999;126:1427–1438. doi: 10.1242/dev.126.7.1427. [DOI] [PubMed] [Google Scholar]
- Fujiwara S, Maeda Y, Shin-I T, Kohara Y, Takatori N, Satou Y, Satoh N. Gene expression profiles in Ciona intestinalis cleavage-stage embryos. Mech Dev. 2002;112:115–127. doi: 10.1016/s0925-4773(01)00651-7. [DOI] [PubMed] [Google Scholar]
- Gansner JM, Mendelsohn BA, Hultman KA, Johnson SL, Gitlin JD. Essential role of lysyl oxidases in notochord development. Dev Biol. 2007;307:202–213. doi: 10.1016/j.ydbio.2007.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Pischon N, Palamakumbura AH, Trackman PC. Intracellular distribution of the lysyl oxidase propeptide in osteoblastic cells. Am J Physiol Cell Physiol. 2007;292:C2095–C2102. doi: 10.1152/ajpcell.00613.2006. [DOI] [PubMed] [Google Scholar]
- Hornstra IK, Birge S, Starcher B, Bailey AJ, Mecham RP, Shapiro SD. Lysyl oxidase is required for vascular and diaphragmatic development in mice. J Biol Chem. 2003;278:14387–14393. doi: 10.1074/jbc.M210144200. [DOI] [PubMed] [Google Scholar]
- Hotta K, Takahashi H, Asakura T, Saitoh B, Takatori N, Satou Y, Satoh N. Characterization of Brachyury-downstream notochord genes in the Ciona intestinalis embryo. Dev Biol. 2000;224:69–80. doi: 10.1006/dbio.2000.9765. [DOI] [PubMed] [Google Scholar]
- Hotta K, Takahashi H, Erives A, Levine M, Satoh N. Temporal expression patterns of 39 Brachyury-downstream genes associated with notochord formation in the Ciona intestinalis embryo. Dev Growth Differ. 1999;41:657–664. doi: 10.1046/j.1440-169x.1999.00467.x. [DOI] [PubMed] [Google Scholar]
- Hotta K, Takahashi H, Satoh N, Gojobori T. Brachyury-downstream gene sets in a chordate, Ciona intestinalis: Integrating notochord specification, morphogenesis and chordate evolution. Evol Dev. 2008;10:37–51. doi: 10.1111/j.1525-142X.2007.00212.x. [DOI] [PubMed] [Google Scholar]
- Hotta K, Yamada S, Ueno N, Satoh N, Takahashi H. Brachyury-downstream notochord genes and convergent extension in Ciona intestinalis embryos. Dev Growth Differ. 2007;49:373–382. doi: 10.1111/j.1440-169X.2007.00935.x. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Friedman R. Loss of ancestral genes in the genomic evolution of Ciona intestinalis. Evol Dev. 2005;7:196–200. doi: 10.1111/j.1525-142X.2005.05022.x. [DOI] [PubMed] [Google Scholar]
- Ikuta T, Yoshida N, Satoh N, Saiga H. Ciona intestinalis Hox gene cluster: Its dispersed structure and residual colinear expression in development. Proc Natl Acad Sci USA. 2004;101:15118–15123. doi: 10.1073/pnas.0401389101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai KS. Isolation and characterization of β-catenin downstream genes in early embryos of the ascidian Ciona savignyi. Differentiation. 2003;71:346–360. doi: 10.1046/j.1432-0436.2003.7106001.x. [DOI] [PubMed] [Google Scholar]
- Imai KS, Hino K, Yagi K, Satoh N, Satou Y. Gene expression profiles of transcription factors and signaling molecules in the ascidian embryo: Towards a comprehensive understanding of gene networks. Development. 2004;131:4047–4058. doi: 10.1242/dev.01270. [DOI] [PubMed] [Google Scholar]
- Imai KS, Levine M, Satoh N, Satou Y. Regulatory blueprint for a chordate embryo. Science. 2006;312:1183–1187. doi: 10.1126/science.1123404. [DOI] [PubMed] [Google Scholar]
- Jansen MK, Csiszar K. Intracellular localization of the matrix enzyme lysyl oxidase in polarized epithelial cells. Matrix Biol. 2007;26:136–139. doi: 10.1016/j.matbio.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaynes JB, O’Farrell PH. Active repression of transcription by the engrailed homeodomain protein. EMBO J. 1991;10:1427–1433. doi: 10.1002/j.1460-2075.1991.tb07663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Munro EM, Smith WC. Ascidian prickle regulates both mediolateral and anterior-posterior cell polarity of notochord cells. Curr Biol. 2005;15:79–85. doi: 10.1016/j.cub.2004.12.041. [DOI] [PubMed] [Google Scholar]
- Jiang D, Smith WC. Ascidian notochord morphogenesis. Dev Dyn. 2007;236:1748–1757. doi: 10.1002/dvdy.21184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusakabe T, Yoshida R, Kawakami I, Kusakabe R, Mochizuki Y, Yamada L, Shin-i T, Kohara Y, Satoh N, Tsuda M, Satou Y. Gene expression profiles in tadpole larvae of Ciona intestinalis. Dev Biol. 2002;242:188–203. doi: 10.1006/dbio.2002.0538. [DOI] [PubMed] [Google Scholar]
- Miwata K, Chiba T, Horii R, Yamada L, Kubo A, Miyamura D, Satoh N, Satou Y. Systematic analysis of embryonic expression profiles of zinc finger genes in Ciona intestinalis. Dev Biol. 2006;292:546–554. doi: 10.1016/j.ydbio.2006.01.024. [DOI] [PubMed] [Google Scholar]
- Miyamoto DM, Crowther RJ. Formation of the notochord in living ascidian embryos. Embryol Exp Morphol. 1985;86:1–17. [PubMed] [Google Scholar]
- Nakatani Y, Moody R, Smith WC. Mutations affecting tail and notochord development in the ascidian Ciona savignyi. Development. 1999;126:3293–3301. doi: 10.1242/dev.126.15.3293. [DOI] [PubMed] [Google Scholar]
- Oda-Ishii I, Di Gregorio A. Lineage-independent mosaic expression and regulation of the Ciona multidom gene in the ancestral notochord. Dev Dyn. 2007;236:1806–1819. doi: 10.1002/dvdy.21213. [DOI] [PubMed] [Google Scholar]
- Odani N, Pfaff SL, Nakamura H, Funahashi J. Cloning and developmental expression of a chick G-protein-coupled receptor SCGPR1. Gene Expr Patterns. 2007;7:375–380. doi: 10.1016/j.modgep.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Ott T, Kaestner KH, Monaghan AP, Schütz G. The mouse homolog of the region specific homeotic gene spalt of Drosophila is expressed in the developing nervous system and in mesoderm-derived structures. Mech Dev. 1996;56:117–128. doi: 10.1016/0925-4773(96)00516-3. [DOI] [PubMed] [Google Scholar]
- Parsons MJ, Pollard SM, Saúde L, Feldman B, Coutinho P, Hirst EM, Stemple DL. Zebrafish mutants identify an essential role for laminins in notochord formation. Development. 2002;129:3137–3146. doi: 10.1242/dev.129.13.3137. [DOI] [PubMed] [Google Scholar]
- Passamaneck YJ, Di Gregorio A. Ciona intestinalis: Chordate development made simple. Dev Dyn. 2005;233:1–19. doi: 10.1002/dvdy.20300. [DOI] [PubMed] [Google Scholar]
- Placzek M, Jessell TM, Dodd J. Induction of floor plate differentiation by contact-dependent, homeogenetic signals. Development. 1993;117:205–218. doi: 10.1242/dev.117.1.205. [DOI] [PubMed] [Google Scholar]
- Satoh N. Developmental Biology of Ascidians. New York: Cambridge University Press; 1994. [Google Scholar]
- Satou Y, Takatori N, Fujiwara S, Nishikata T, Saiga H, Kusakabe T, Shin-i T, Kohara Y, Satoh N. Ciona intestinalis cDNA projects: Expressed sequence tag analyses and gene expression profiles during embryogenesis. Gene. 2002;287:83–96. doi: 10.1016/s0378-1119(01)00826-5. [DOI] [PubMed] [Google Scholar]
- Satou Y, Takatori N, Yamada L, Mochizuki Y, Hamaguchi M, Ishikawa H, Chiba S, Imai K, Kano S, Murakami SD, Nakayama A, Nishino A, Sasakura Y, Satoh G, Shimotori T, Shin-I T, Shoguchi E, Suzuki MM, Takada N, Utsumi N, Yoshida N, Saiga H, Kohara Y, Satoh N. Gene expression profiles in Ciona intestinalis tailbud embryos. Development. 2001;128:2893–2904. doi: 10.1242/dev.128.15.2893. [DOI] [PubMed] [Google Scholar]
- Schulte-Merker S, van Eeden FJ, Halpern ME, Kimmel CB, Nüsslein-Volhard C. no tail (ntl) is the zebrafish homologue of the mouse T (Brachyury) gene. Development. 1994;120:1009–1015. doi: 10.1242/dev.120.4.1009. [DOI] [PubMed] [Google Scholar]
- Scott A, Stemple DL. Zebrafish notochordal basement membrane: Signaling and structure. Curr Top Dev Biol. 2005;65:229–253. doi: 10.1016/S0070-2153(04)65009-5. [DOI] [PubMed] [Google Scholar]
- Seo HC, Edvardsen RB, Maeland AD, Bjordal M, Jensen MF, Hansen A, Flaat M, Weissenbach J, Lehrach H, Wincker P, Reinhardt R, Chourrout D. Hox cluster disintegration with persistent anteroposterior order of expression in Oikopleura dioica. Nature. 2004;431:67–71. doi: 10.1038/nature02709. [DOI] [PubMed] [Google Scholar]
- Spagnuolo A, Ristoratore F, Di Gregorio A, Aniello F, Branno M, Di Lauro R. Unusual number and genomic organization of Hox genes in the tunicate Ciona intestinalis. Gene. 2003;309:71–79. doi: 10.1016/s0378-1119(03)00488-8. [DOI] [PubMed] [Google Scholar]
- Stemple DL. Structure and function of the notochord: An essential organ for chordate development. Development. 2005;132:2503–2512. doi: 10.1242/dev.01812. [DOI] [PubMed] [Google Scholar]
- Suzuki F. Roles of cartilage matrix proteins, chondromodulin-I and -II, in endochondral bone formation: A review. Connect Tissue Res. 1996;35:303–307. doi: 10.3109/03008209609029204. [DOI] [PubMed] [Google Scholar]
- Swalla BJ, Smith AB. Deciphering deuterostome phylogeny: Molecular, morphological and palaeontological perspectives. Philos Trans R Soc Lond B Biol Sci. 2008;363:1557–1568. doi: 10.1098/rstb.2007.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Hotta K, Erives A, Di Gregorio A, Zeller RW, Levine M, Satoh N. Brachyury downstream notochord differentiation in the ascidian embryo. Genes Dev. 1999;13:1519–1523. doi: 10.1101/gad.13.12.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeman MT, Nakatani Y, Hendrickson C, Ericson V, Lin C, Smith WC. chongmague reveals an essential role for laminin-mediated boundary formation in chordate convergence and extension movements. Development. 2008;135:33–41. doi: 10.1242/dev.010892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker JR. Segregation during cleavage of a factor determining endodermal alkaline phosphatase development in ascidian embryos. J Exp Zool. 1977;202:139–153. doi: 10.1002/jez.1402020202. [DOI] [PubMed] [Google Scholar]
- Wilson V, Manson L, Skarnes WC, Beddington RS. The T gene is necessary for normal mesodermal morphogenetic cell movements during gastrulation. Development. 1995;121:877–886. doi: 10.1242/dev.121.3.877. [DOI] [PubMed] [Google Scholar]
- Yeh LC, Tsai AD, Lee JC. Cartilage-derived morphogenetic proteins induce osteogenic gene expression in the C2C12 mesenchymal cell line. J Cell Biochem. 2005;95:173–188. doi: 10.1002/jcb.20402. [DOI] [PubMed] [Google Scholar]
- Zorn AM, Krieg PA. The KH domain protein encoded by quaking functions as a dimer and is essential for notochord development in Xenopus embryos. Genes Dev. 1997;11:2176–2190. doi: 10.1101/gad.11.17.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.