Abstract
Analysis of mRNA levels in cells that express or lack signal transducers and activators of transcription 1 (Stat1) reveals that Stat1 mediates the constitutive transcription of many genes. Expression of the low molecular mass polypeptide 2 (LMP2), which requires Stat1, has been studied in detail. The overlapping interferon consensus sequence 2/γ-interferon-activated sequence (ICS-2/GAS) elements in the LMP2 promoter bind to interferon regulatory factor 1 (IRF1) and Stat1 and are occupied constitutively in vivo. The point mutant of Stat1, Y701F, which does not form dimers involving SH2–phosphotyrosine interactions, binds to the GAS element and supports LMP2 expression. Unphosphorylated Stat1 binds to IRF1 directly and we conclude that this complex uses the ICS-2/GAS element to mediate constitutive LMP2 transcription in vivo. The promoter of the IRF1 gene, which also contains a GAS site but not an adjacent ICS-2 site, is not activated by Stat1 Y701F. The promoters of other genes whose constitutive expression requires Stat1 may also utilize complexes of unphosphorylated Stat1 with IRF1 or other transcription factors.
Keywords: differential mRNA levels/DNA binding affinity/DNA micro-arrays/Stat1 monomer/unphosphorylated Stat1 dimer
Introduction
The signal transducers and activators of transcription (Stats) mediate many effects of cytokines and peptide growth factors (for a recent review see Stark et al., 1998). Stat1 is a major transcription factor in the interferon (IFN) α/β and IFN-γ signal transduction pathways that lead to the activation of antiviral, antiproliferative and immunomodulatory functions. IFN-γ stimulates gene expression through the phosphorylation of Stat1 by Janus kinases (JAKs) at the cell membrane (Darnell et al., 1994; Ihle, 1995), followed by homodimerization of Stat1, nuclear translocation and binding to IFN-γ-activated sequence (GAS) elements. TTCNNNG/TAA is the GAS consensus (Decker et al., 1997). Homodimerization of Stat1 is mediated by the binding of the phosphorylated Tyr701 of one Stat1 monomer to the Src homology 2 (SH2) domain of another (Shuai et al., 1994).
Among the genes that mediate IFN-γ-induced antiviral, antitumor and immunomodulatory responses are class II transactivator (CIITA) (Muhlethaler-Mottet et al., 1998), intercellular adhesion molecule 1 (ICAM-1) (Walter et al., 1997), the human monokine inducer by IFN-γ 1 (MIG1) chemokine (Guyer et al., 1995), interferon regulatory factor 1 (IRF1) (Li et al., 1996), low molecular mass polypeptide 2 (LMP2), transporter associated with antigen processing 1 (TAP1) (Min et al., 1996; Chatterjee-Kishore et al., 1998) and human p21 (Su et al., 1997). Stat1 is required for the efficient constitutive expression of the ICE, Cpp32 and Ich-1 (caspase 1–3) genes, which are also expressed well in Stat1-deficient U3A cells reconstituted with the Y701F mutant of Stat1 (U3A-701). This result indicates that unphosphorylated Stat1 is necessary for the expression of certain genes, since Stat1 Y701F is incapable of being phosphorylated at residue 701 and therefore of forming classical Stat1 homodimers (Kumar et al., 1997).
We have analyzed differential gene expression in U3A and U3A-701 cells using DNA arrays and have identified additional genes whose expression can be mediated by Stat1 Y701F. Many of these genes have GAS or GAS-like elements in their promoters. However, not all genes regulated by GAS-bound proteins are activated by Stat1 Y701F. For example, constitutive expression of the IRF1 gene, whose IFN-γ-induced expression is regulated by the binding of a phosphorylated Stat1 homodimer to a GAS site, does not depend on Stat1. Constitutive expression of the LMP2 gene, on the other hand, does require Stat1. LMP2 RNA was not detected in U3A cells but is present in almost equal amounts in U3A-701 and 2fTGH cells. Thus, comparison of the requirements for basal expression of the IRF1 and LMP2 genes should be informative in studying how Stat1 regulates constitutive expression.
LMP2 is a subunit of the 20S proteasome, which processes viral and tumor antigens for presentation to CD8+ T cells in the context of major histocompatibility complex (MHC) class I molecules (Lehner et al., 1998). Transcription of LMP2 is regulated by a bidirectional promoter that also controls transcription of the TAP1 gene (Wright et al., 1995). In melanoma cell lines, LMP2 transcription is activated only in the presence of both Stat1 and IRF1 (Chatterjee-Kishore et al., 1998). Stat1-null (Lee et al., 1999) and IRF1-null mice (White et al., 1996) express the LMP2 gene at much lower levels than do wild-type mice. In the present study, we show that unphosphorylated Stat1 and IRF1 bind to the partially overlapping interferon consensus sequence 2 (ICS-2) and GAS sites in the LMP2 promoter to activate transcription.
Results
Stat1 mediates constitutive gene expression
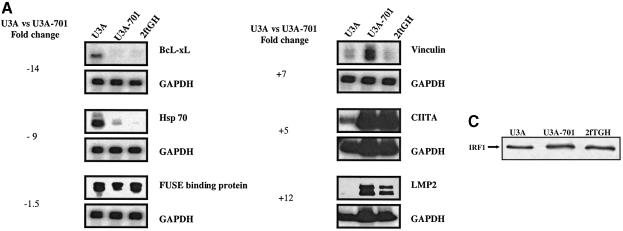
Affymetrix arrays were used to compare the expression of ∼6800 genes in U3A cells (lacking Stat1) and in U3A-701 cells (expressing Stat1 Y701F). Some of the data were confirmed by using northern and S1 nuclease analyses. Table I shows the Affymetrix data for the most differentially expressed genes, according to the sort scores. Unphosphorylated Stat1 can down-regulate as well as up-regulate the constitutive expression of a wide variety of genes (Table I). Many genes down-regulated by unphosphorylated Stat1 encode cell cycle regulatory proteins (e.g. CDC2, cyclin A and cyclin B). It is interesting that the expression of these genes can also be down-regulated by IFN-α and -γ (Iwase et al., 1997; Sibinga et al., 1999). Notable among the genes whose expression is higher in U3A-701 cells are those encoding MHC class I, β2-microglobulin, CIITA and LMP2. The proteins encoded by these genes are all involved in antigen presentation via the MHC class I- or class II-restricted pathways and the promoters of all of these genes contain Stat1-binding sites (Gobin et al., 1997; Solheim, 1999). The expression of some of the genes identified in the DNA array experiment has also been analyzed by other techniques (Figure 1A). Comparison of the fold changes from the Affymetrix data with those obtained from northern or S1 nuclease analyses shows that the Affymetrix data serve at best as a qualitative indicator of relative gene expression.
Table I. Differential gene expression in U3A and U3A-701 cells.
| Probe set | Gene | Fold change | Sort score |
|---|---|---|---|
| U73514 | human alcohol dehydrogenase | –29.4 | –17 |
| M27826 | human endogenous retroviral protease | –10.4 | –14.06 |
| HG2887 | Sry-related Hmg box 12 protein | –58 | –13 |
| U04847 | human Ini 1 | –9.4 | –7.9 |
| M11717 | human Hsp70 | –4.6 | –3.3 |
| X69819 | human ICAM3 | –4.9 | –2.7 |
| X51688 | human cyclin A | –4.3 | –2.4 |
| HG2175 | human myosin | –4.1 | –1.7 |
| X05360 | human CDC2 | –2.9 | –1.1 |
| L23959 | human E2F-related transcription factor DP-1 | –2.5 | –0.8 |
| Z23115 | human Bcl-xL | –1.9 | –0.6 |
| M25753 | human cyclin B | –2.1 | –0.6 |
| M64174 | human JAK1 | –1.4 | –0.1 |
| U69126 | human FUSE-binding protein | –4.6 | –0.1 |
| X13238 | human cytochrome c oxidase Cox VIIc | 3.6 | 4.9 |
| S73591 | human HHCPA78 homolog | 3.4 | 4.5 |
| X04500 | human pro-IL1βa | 3.6 | 3.9 |
| M62402 | human insulin-like growth factor-binding protein 6 | 6.1 | 2.65 |
| HG3597 | human MHC class I | 2.7 | 1.5 |
| J00105 | human β2-microglobulin | 1.7 | 1.4 |
| U28014 | human cysteine protease ICE rel-II | 3 | 1.04 |
| D50310 | cyclin I | 1.9 | 1 |
| M33308 | human vinculin | 7.4 | 1 |
| U18259 | human CIITA | 13.4 | 0.8 |
| X66401 | human LMP2 | 2.3 | 0.7 |
| L08246 | human myeloid cell differentiation protein (MCL1) | 2.2 | 0.4 |
| X15187 | human tra1 (human homolog of gp96) | 1.9 | 0.4 |
| U32114 | human caveolin 2 | 2.7 | 0.4 |
| U23850 | human inositol 1,4,5 triphosphate receptor type 1 | 2.4 | 0.2 |
The expression of 6800 genes was analyzed using Affymetrix expression arrays. Thirty differentially expressed genes are listed according to their sort scores and fold changes in expression. A negative change and sort score indicate decreased expression while a positive change and sort score indicate increased expression. Higher sort scores indicate higher significance of the data set. U3A cells were taken as baseline and U3A-701 as sample. Expression of genes indicated in bold has been analyzed using northern or S1 nuclease assays.
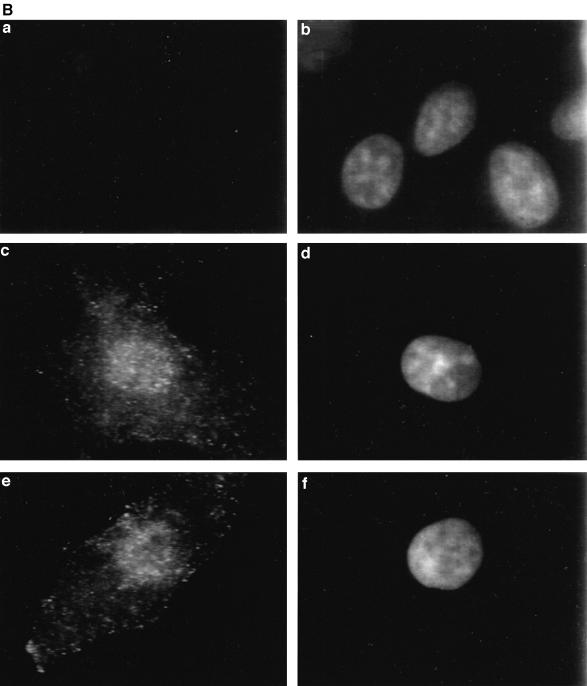
Fig. 1. Stat1 regulates constitutive gene expression. (A) Confirmation of Affymetrix data by northern and S1 nuclease analyses. The expression of some of the differentially expressed genes (in bold in Table I) was analyzed in U3A, U3A-701 and 2fTGH cells. S1 nuclease analysis was used for expression of the LMP2, CIITA and vinculin genes, with GAPDH as control. The sizes of the protected fragments were 677 bp for LMP2, 503 bp for CIITA, 450 bp for GAPDH and 286 bp for vinculin. BcL-xL, Hsp-70 and FUSE-binding protein mRNA expression was analyzed in northern transfers using PCR-derived probes. The transfers were stripped and reprobed for GAPDH expression. Fold changes were estimated by densitometry. In the case of CIITA, the change is probably underestimated due to the large signal in U3A-701 and 2fTGH cells. (B) Unphosphorylated Stat1 translocates to the nucleus. Formaldehyde-fixed U3A, 2fTGH and U3A-701 cells (a, c and e, respectively) were stained with anti-Stat1 and FITC-conjugated goat anti-rabbit antibody and DAPI was used to stain the nuclei (b, d and f, respectively). (C) IRF1 expression in U3A, U3A-701 and 2fTGH cells. Approximately 2 × 106 cells were labeled in vivo using [35S]methionine. Proteins were extracted after 18 h of labeling and counts per minute were determined. Aliquots containing equal numbers of counts were immunoprecipitated with anti-IRF1 and the immunoprecipitates were separated by 12% SDS–PAGE.
Stat1 can be detected in the nucleus of >90% of untreated U3A-701 and 2fTGH cells. Figure 1B shows a representative photomicrograph of U3A, U3A-701 and 2fTGH cells stained with anti-Stat1. Therefore, tyrosine phosphorylation is not a prerequisite for Stat1 to be present in the nucleus.
These experiments provide initial evidence for the fact that unphosphorylated Stat1 may be involved in the constitutive expression of a wide variety of genes, although the means used by Stat1 to regulate the constitutive expression of each of these genes may be different. Analysis of the promoters of several of the genes regulated by Stat1 Y701F revealed GAS or GAS-like sequences. However, not all GAS-containing genes are activated by unphosphorylated Stat1. The promoter of the IRF1 gene has a GAS sequence and IFN-γ-mediated IRF1 expression is driven primarily by phosphorylated Stat1 homodimers, which bind to this element. Constitutive IRF1 expression, however, is similar in U3A, U3A-701 and 2fTGH cells (Figure 1C). On the other hand, constitutive expression of the LMP2 gene, whose IFN-γ-induced expression is driven by an overlapping ICS-2/GAS element (Figure 2), was apparent in U3A-701 and 2fTGH cells but not in Stat1-deficient U3A cells (Figure 1A). The ICS-2/GAS element in the LMP2 promoter binds to IRF1 and Stat1 (Chatterjee-Kishore et al., 1998) and no other consensus Stat1-binding sites are evident in this promoter. Therefore, we analyzed the LMP2 promoter in greater detail in order to determine how unphosphorylated Stat1 mediates constitutive gene expression.
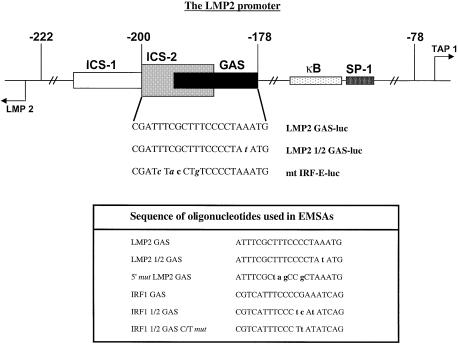
Fig. 2. Functional diagram of the LMP2/TAP1 bi-directional promoter and the sequences of wild-type and mutant constructs representing the ICS-2/GAS region. The table shows the sequences of oligonucleotides used in EMSAs.
The ICS-2/GAS element of the LMP2 promoter is occupied constitutively in vivo
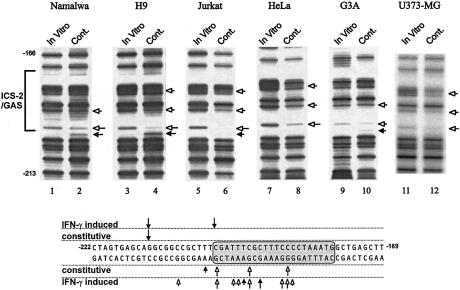
Many cancer cell lines express LMP2 constitutively (Thor Straten et al., 1997; Chatterjee-Kishore et al., 1998). We used in vivo footprinting to analyze the LMP2 ICS-2/GAS for constitutive binding of transcription factors in six cell lines that express LMP2 constitutively (data not shown). All the cell lines also express Stat1 and IRF1. Constitutive tyrosine phosphorylation of Stat1 could not be detected in any of these cell lines (data not shown). Methylated genomic DNA obtained from Namalwa, H9, Jurkat, HeLa, G3A and U373-MG cells was used in ligation-mediated PCR (LM-PCR), using specific primers (White et al., 1996), and the LM-PCR products were separated in denaturing gels. Genomic DNA methylated in vitro was used as a control. Comparison of the footprints from DNA methylated in vitro and in vivo shows that two bases are protected constitutively in vivo within the ICS-2 region and that one base is protected at the 5′ end of the GAS (open arrows, Figure 3). One base is hypermodified within the ICS-2 (filled arrow, Figure 3). It is not possible to learn much about the occupancy of the 3′ end of the GAS by methylation protection because this region is AT rich. However, it is clear from these data that the 3′ end of the ICS-2 and the 5′ end of the GAS element bind to one or more proteins constitutively, even though there is some cell-to-cell variation in the pattern of occupancy. The occupancy in untreated cells is quite different from that observed following IFN-γ treatment (Figure 3, bottom; White et al., 1996). These experiments suggest that the ICS-2/GAS region is occupied in cells that express LMP2 constitutively.
Fig. 3. Constitutive in vivo occupancy of the ICS-2/GAS. Dimethyl sulfate treatment, genomic DNA preparation and LM-PCR for the ICS-2/GAS (IRF-E) region (lower strand) of the LMP2 promoter were described by White et al. (1996). DNA samples treated with DMS in vitro or in vivo were analyzed simultaneously. Open arrows mark the bases protected from methylation and the filled arrows mark methylation enhancements. The sequence of the ICS-2/GAS region is marked with the constitutive and IFN-γ-induced protections and enhancements determined in HeLa cells by White et al. (1996).
In the absence of IFN stimulation, Stat1 binds to the ICS-2/GAS element of the LMP2 gene but not to the IRF1 GAS
The in vivo footprinting experiments do not provide any information about the nature of the factors that bind to the ICS-2/GAS constitutively. Since Stat1 and IRF1 are the only proteins known to bind to the ICS-2/GAS, and since tyrosine phosphorylation of Stat1 could not be detected in any of the cell lines analyzed, we asked whether unphosphorylated Stat1 could also bind to the ICS-2/GAS region in vivo.
We used U3A, U3A-701 and 2fTGH cells in an in vivo chromatin immunoprecipitation (ChIP) assay since the AT-rich nature of the LMP2 GAS element precludes the use of dimethyl sulfate-mediated footprinting to analyze Stat1 binding to this site. U3A cells do not express either Stat1 or LMP2, and transcription of the LMP2 gene cannot be induced in these cells following IFN-γ treatment (data not shown). U3A-701 cells express Stat1 Y701F and LMP2. IFN-γ treatment of these cells does not lead to up-regulation of LMP2 RNA levels (data not shown). 2fTGH cells express wild-type Stat1 and LMP2 constitutively. IFN-γ treatment of these cells leads to higher levels of LMP2 expression (data not shown).
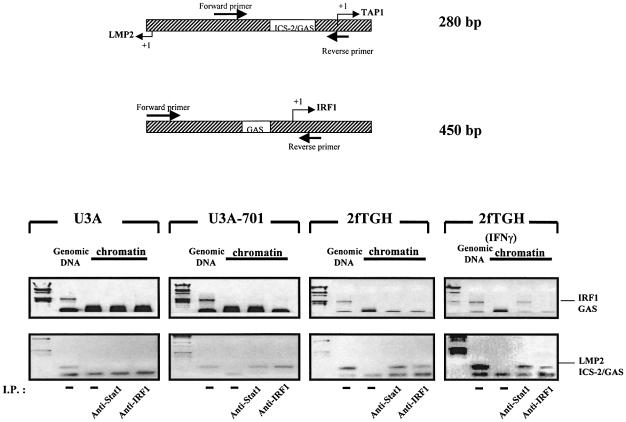
Proteins were cross-linked to DNA in growing U3A, U3A-701 2fTGH and IFN-γ treated 2fTGH cells, using formaldehyde. Chromatin was recovered by isopycnic centrifugation in a cesium chloride gradient and DNA-bound Stat1 and IRF1 were immunoprecipitated, cross-links were reversed and the purified DNA was amplified by PCR, using primers for either the LMP2 ICS-2/GAS or the IRF1 GAS (Figure 4). Anti-Stat1 co-immunoprecipitated the IRF1 GAS only from parental 2fTGH cells treated with IFN-γ. However, the LMP2 ICS-2/GAS was amplified from DNA co-immunoprecipitated with anti-Stat1 in U3A-701, 2fTGH and 2fTGH cells treated with IFN-γ. DNA co-immunoprecipitated with anti-IRF1 did not contain the IRF1 GAS in any cell line, as expected, since there are no IRF1-binding sites within this region of the promoter (Miyamoto et al., 1988). The LMP2 ICS-2/GAS was amplified from DNA samples co-immunoprecipitated with anti-IRF1 from U3A-701, 2fTGH and 2fTGH cells treated with IFN-γ. In U3A cells, very little PCR product corresponding to the LMP2 ICS-2/GAS region (not easily visible in the photograph of the gel) was obtained following immunoprecipitation with anti-IRF1. The poor binding of IRF1 to the ICS-2/GAS region in U3A cells probably indicates that Stat1 bound to the overlapping GAS site helps to recruit IRF1 to the ICS-2 sequence. This experiment shows that the LMP2 ICS-2/GAS is constitutively occupied by unphosphorylated Stat1 and IRF1 in untreated U3A-701 and 2fTGH cells in vivo.
Fig. 4. In vivo cross-linking of unphosphorylated Stat1 and IRF1 on the ICS-2/GAS region of the LMP2 promoter. Proteins were cross-linked to DNA in growing U3A and U3A-701 cells and in untreated and IFN-γ-treated (1000 IU/ml for 1 h) 2fTGH cells. DNA-bound Stat1 and IRF1 were immunoprecipitated. Co-immunoprecipitated DNA from uncross-linked genomic DNA (lanes marked genomic DNA) or in vivo cross-linked chromatin (lanes marked chromatin) was amplified by PCR, using primers (position shown at the top of the figure) for the LMP2 ICS/GAS or IRF1 GAS region. The positions of the 450 and 280 bp PCR products corresponding to the IRF1 GAS and the LMP2 ICS-2/GAS regions are marked.
The complete palindromic GAS element is not required for constitutive activity of the LMP2 promoter
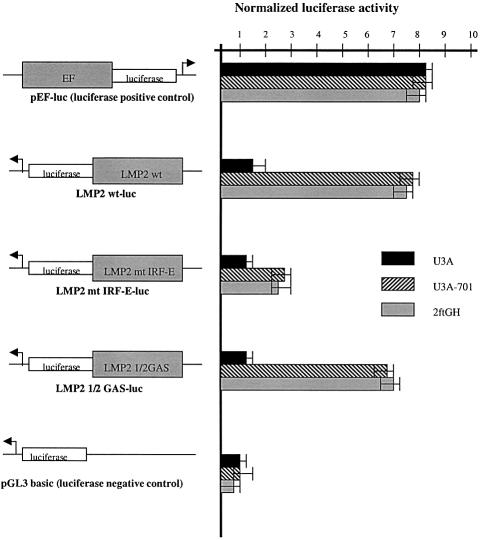
To delineate further the sequence requirements for constitutive transcription factor binding to the ICS-2/GAS element, we used three different promoter constructs linked to luciferase. The LMP2 GAS-luc (wild-type LMP2) and mt IRF-E-luc constructs (Figure 2) have been described previously (White et al., 1996). The mt IRF-E-luc construct has mutations within the ICS-2 subregion and at the 5′ end of the overlapping GAS element. The LMP2 1/2 GAS-luc construct contains a point mutation at the 3′ end of the GAS (Figure 2). U3A, U3A-701 and 2fTGH cells were co-transfected with pBabePURO and the LMP2 GAS-luc, mt IRF-E-luc or LMP2 1/2 GAS-luc constructs, and luciferase activity was assayed in extracts prepared from pools of stable transfectants (Figure 5). All three constructs have little activity in U3A cells. In both U3A-701 and 2fTGH cells, mutation of the four nucleotides that inactivate the ICS-2 and the 5′ region of the GAS (mt IRF-E-luc) reduces constitutive activity to less than half that of the wild-type LMP2 construct (LMP2 GAS-luc). However, mutation of a single base at the 3′ end of the GAS (LMP2 1/2 GAS-luc), which abrogates binding of the IFN-γ-induced Stat1 homodimer (Figure 6A), has little effect on constitutive activity. Thus, in both U3A-701 and 2fTGH cells, the constitutive activity of the LMP2 promoter requires the ICS-2 and the 5′ end of the GAS.
Fig. 5. Activity of wild-type or mutant LMP2 promoter constructs in U3A, U3A-701 and 2fTGH cells. Cells were stably transfected with LMP2 GAS-luc, mt IRF-E-luc, LMP2 1/2GAS-luc, pEF-luc as positive control and pGL3-basic as negative control. Luciferase activity was assayed in duplicate in extracts from pools of stably transfected cells. Luciferase activity units were normalized against total protein levels, assayed spectrophotometrically.
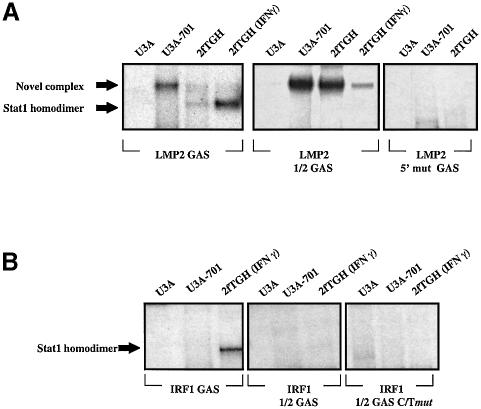
Fig. 6. A novel complex binds to the LMP2 GAS in U3A-701 cells. (A) Binding activities towards the wild-type and mutant LMP2 GAS probes in extracts prepared from U3A, U3A-701 and 2fTGH cells. Extracts from untreated and IFN-γ-treated cells were assayed by EMSA. (B) Binding activities towards the wild-type and mutant IRF1 GAS probes in extracts prepared from U3A, U3A-701 and 2fTGH cells. Extracts from untreated cells were assayed in EMSAs.
The LMP2 GAS element binds to a novel Stat1-containing complex in U3A-701 cells
To correlate the studies on promoter activity and occupancy of the ICS-2/GAS in vivo with in vitro experiments and to analyze the nature of the complex that binds to the ICS-2/GAS element constitutively, we investigated by electrophoretic mobility shift assay (EMSA) the effects of mutations in the ICS-2/GAS region of the LMP2 promoter on transcription factor binding in untreated U3A, U3A-701 and 2fTGH cells and in IFN-γ-treated 2fTGH cells. These data were compared with constitutive and IFN-γ-induced binding to the IRF1 GAS. EMSAs were performed with the following probes: the GAS site and part of the adjoining ICS-2 of the LMP2 promoter (LMP2 GAS), a single base mutation at the 3′ end of the LMP2 GAS (LMP2 1/2 GAS) and the 5′ mut LMP2 GAS (for sequence see Figure 2). The LMP2 GAS varies from the IRF1 GAS (T1T2C3C4C5C6G7A8A9) at position 7 (T instead of G). A novel LMP2 GAS-binding complex, formed with extracts of U3A-701 cells (Figure 6A), can also be detected with extracts of untreated 2fTGH cells, but the amount is less. The novel DNA-bound complex migrates more slowly than the Stat1 homodimer detected in 2fTGH cells treated with IFN-γ, indicating that it is larger. The novel complex was also detected when the LMP2 1/2 GAS was used in EMSAs, whereas binding of the IFN-γ-induced Stat1 homodimer was completely abrogated. A small amount of the novel complex was also detected in extracts of 2fTGH cells treated with IFN-γ. The novel complex was not seen with any extract when the 5′ mut LMP2 GAS was used. Anti-Stat1α blocked formation of the novel complex, confirming that it contains Stat1 (Chatterjee-Kishore et al., 2000). Thus, Stat1 Y701F does form distinct complexes with the LMP2 GAS and LMP2 1/2 GAS elements. However, it does not bind detectably to either the IRF1 GAS or the IRF1 1/2 GAS elements (Figures 2 and 6B). No specific DNA-binding activity towards the IRF1 GAS was seen with extracts of U3A-701 cells even when the IRF1 1/2 GAS C/T mut probe (the sequence of which is very similar to that of the LMP2 1/2 GAS) was used. The results of these experiments are consistent with the data obtained from analysis of LMP2 promoter mutants. Formation of the novel complex containing Stat1 Y701F does not require the 3′ end of the GAS, but mutations in the 5′ end abrogate binding. Thus, this novel complex in U3A-701 cells may account for constitutive occupancy in vivo of the ICS-2/GAS region.
Unphosphorylated Stat1 can bind to DNA as either a dimer or a monomer
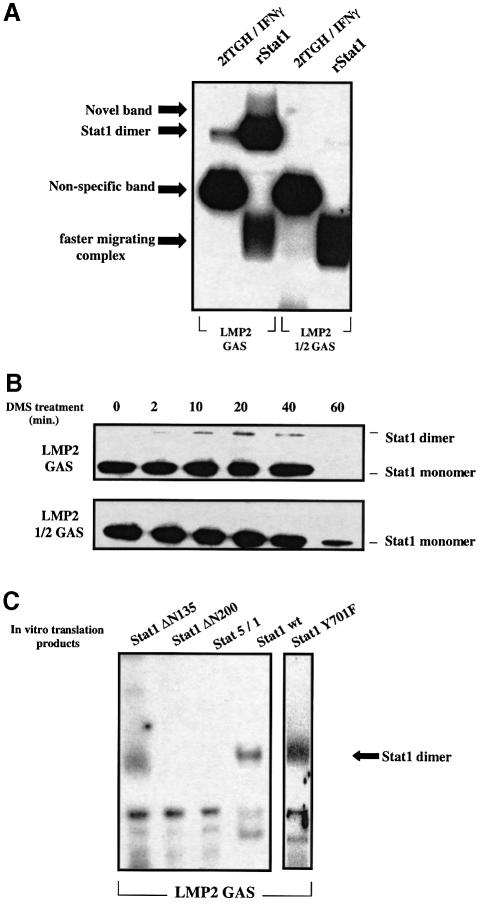
His-tagged Stat1 was purified from Escherichia coli extracts and the tag was removed with thrombin to yield recombinant Stat1 (rStat1). rStat1 was unphosphorylated, as judged by western analysis with anti-phosphotyrosine Stat1 (Y701) and anti-phosphoserine Stat1 (S727) (data not shown). We investigated the binding of rStat1 to the LMP2 GAS, LMP2 1/2 GAS and the LMP2 5′ mut GAS elements. EMSAs (Figure 7A) show that LMP2 GAS bound to rStat1 migrates differently from the complex found in extracts of U3A-701 cells. rStat1 binds to the LMP2 GAS as a dimer, and the complex has a mobility similar to that of the complex formed with phosphorylated Stat1 in extracts of IFN-γ-treated 2fTGH cells. The single base mutation in the LMP2 1/2 GAS abrogates the binding of unphosphorylated Stat1 as a dimer, but allows formation of a complex with faster mobility. This complex was also seen when the LMP2 GAS was used as probe.
Fig. 7. rStat1 can bind DNA as a dimer or monomer, but with low affinity. (A) EMSA analysis of the binding of rStat1 to the LMP2 GAS and LMP2 1/2 GAS probes. Approximately 200 ng of purified rStat1 and 5 µg of extracts from IFN-γ-treated 2fTGH cells were used in EMSAs with the LMP2 GAS probe. A 500 ng aliquot of rStat1 was used in EMSAs with the LMP2 1/2 GAS and 5′ mut LMP2 GAS probes. (B) Time course of DMS-mediated cross-linking of rStat1 on LMP2 GAS or LMP2 1/2 GAS probes. Neutravidin–agarose beads saturated with either the LMP2 GAS or 1/2 GAS probes were allowed to bind to rStat1 (2–5 µg). Proteins were cross-linked while bound to DNA using DMS and analyzed in western transfers using anti-Stat1. (C) The N-terminal region of unphosphorylated Stat1 is required for dimer formation in vitro. Stat1, Stat1 Y701F, Stat1 ΔN135, Stat1 ΔN200 and Stat5/1 were translated in vitro and partially purified on heparin–agarose. The DNA-binding activity of equal amounts of Stat1, Stat1 ΔN135, Stat1 ΔN200, Stat5/1 and Stat1 Y701F to the LMP2 GAS was analyzed in EMSAs.
Cross-linking experiments with dimethylsuberimidate (DMS) confirm that rStat1 is indeed a dimer on the LMP2 GAS element. A time course of cross-linking shows the formation of Stat1 homodimers (Figure 7B, top panel). The intensity of the dimer band is low, probably due to the alteration of protein–DNA contacts accompanying the DMS-mediated cross-linking of Stat1 monomers. We have also observed that DMS treatment for >40 min leads to rapid loss of DNA-bound rStat1 (Figure 7B, top panel, extreme right lane). Cross-linking analysis of rStat1 bound to the LMP2 1/2 GAS indicates that the faster complex seen in EMSAs (Figure 6A) contains rStat1 monomer (Figure 7B, bottom panel). The binding of the Stat1 monomer is specific for the DNA sequence used since detectable levels of Stat1 could not be recovered using the 5′ mut LMP2 GAS oligonucleotides (data not shown). The apparent molecular weights of the two bands conform to those expected for a monomer or dimer of Stat1, as compared with appropriate molecular weight markers (not shown). From these experiments, it appears that a monomer of Stat1 can bind to DNA by contacting only half of the palindromic GAS site, in vitro.
How does Stat1 form a dimer without phosphotyrosine–SH2 interactions? Since two Stat1 dimers can bind to each other at adjacent GAS sites through their N-terminal domains (Xu et al., 1996), this domain may also mediate the formation of unphosphorylated Stat1 dimers. We analyzed the binding of equal amounts of Stat1, Stat1 ΔN135 (lacking the first 135 residues), Stat1 ΔN200 (lacking the first 200 residues) and Stat5/1 (the first 129 residues of Stat1 are replaced by the corresponding residues of Stat5A) to the LMP2 GAS (Figure 7C), all formed by in vitro translation. Stat1 binds to the LMP2 GAS as a dimer. Stat1 ΔN135 also binds to this element as a dimer, but the intensity is very low. Neither Stat1 ΔN200 nor Stat5/1 bind to the LMP2 GAS as dimers. In a separate experiment, we observed that Stat1 Y701F, made by translation in vitro, also forms a dimer (Figure 7C). Thus, the N-terminus of Stat1, especially residues 135–200, within the first coiled-coil domain, is required to form dimers of unphosphorylated Stat1 on the LMP2 GAS.
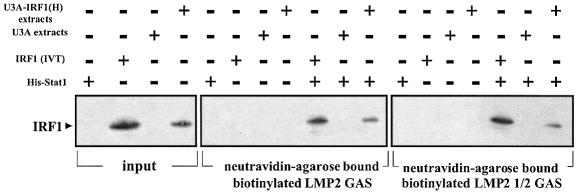
A complex of Stat1 and IRF1 binds to the LMP2 GAS
Unphosphorylated Stat1 binds to the LMP2 GAS as a dimer or monomer. Stat1 Y701F translated in vitro can also dimerize on the LMP2 GAS. However, detectable levels of Stat1 Y701F dimers on the LMP2 GAS were not observed when extracts from U3A-701 cells were analyzed in EMSAs (Figure 6A). More than twice as much rStat1 is required to detect a Stat1 monomer band in EMSA as is needed to detect rStat1 dimers. Thus the affinity of rStat1 monomers for DNA is lower than that of dimers. Since unphosphorylated Stat1 associates with IRF1 (Chatterjee-Kishore et al., 2000) and both Stat1 and IRF1 bind to the LMP2 ICS-2/GAS in vivo (Figure 4), we attempted to supershift the LMP2 GAS-bound, Stat1-containing novel complex with anti-IRF1. These attempts were unsuccessful. The Stat1–IRF1 interaction does not require any accessory factors (Chatterjee-Kishore et al., 2000). To ask whether unphosphorylated Stat1 and IRF1 can bind to DNA as a complex, we immobilized either biotinylated LMP2 GAS or LMP2 1/2 GAS oligonucleotides on neutravidin–agarose beads and saturated the agarose-bound oligonucleotides with Stat1. The beads were then incubated with cell extracts or with in vitro-translated IRF1. Since U3A cells express very low levels of endogenous IRF1, we used U3A cells reconstituted with IRF1 [U3A-IRF1(H); Kumar et al., 1997]. Detectable levels of IRF1 were not bound to either the LMP2 GAS or LMP2 1/2 GAS in the absence of Stat1 (Figure 8). LMP2 GAS- and LMP2 1/2 GAS-bound Stat1 can, however, bind to IRF1 from extracts of U3A-IRF1(H) cells. Thus a complex of Stat1 and IRF1 can bind to the LMP2 GAS. These data indicate that, although Stat1 can bind to the LMP2 GAS as a dimer in vitro, the strong Stat1–IRF1 interaction in vivo may facilitate the stabilization of a heteromeric complex of these two transcription factors that binds to the overlapping ICS-2/GAS site and thus mediates LMP2 transcription.
Fig. 8. A complex of Stat1 and IRF1 binds to the LMP2 GAS or LMP2 1/2 GAS. Streptavidin–agarose beads were saturated with biotin-labeled oligonucleotides at 4°C for 2 h. The beads were incubated with either Stat1 (2–5 µg), IRF1 (in vitro translation products), U3A extracts or U3A-IRF1(H) extracts at 4°C for 1 h. The Stat1-saturated beads were then washed with HEM buffer and incubated with either IRF1 (in vitro translation product), U3A extracts or U3A-IRF1(H) extracts. Samples were analyzed in western transfers with anti-IRF1.
Discussion
Stat1 supports constitutive LMP2 expression
Our data reveal that unphosphorylated Stat1 regulates constitutive gene expression both positively and negatively. Stat1 (Y701F) mediates transcription of the LMP 2 gene as effectively as wild-type Stat1. The ICS-2/GAS region of the LMP2 promoter, required for constitutive transcription, is occupied constitutively in all cell lines examined. We provide the first direct evidence that Stat1, unphosphorylated on Tyr701, can bind to DNA. Stat1 can bind to the LMP2 GAS element (TTCNNNTAA) as a dimer and monomer and to half the palindromic element (TTCNNNTAT) as a monomer. In U3A-701 cells, unphosphorylated Stat1 and IRF1 form a complex on the LMP2 ICS-2/GAS. In other promoters, the Stat1 partner may be different; perhaps other transcription factors that bind near GAS elements can also interact with Stat1.
Constitutive expression of many other genes is also regulated by unphosphorylated Stat1
Stat1 regulates the constitutive transcription of some caspase genes (Kumar et al., 1997) and the LMP2 gene (Chatterjee-Kishore et al., 1998). Analysis of differential gene expression in U3A and U3A-701 cells using Affymetrix gene arrays shows that the constitutive expression of many more genes depends on Stat1. The expression of more genes was decreased by Stat1 Y701F than was increased. There is already evidence that Stat1 down-regulates ligand-induced gene expression (Sharma and Iozzo, 1998; Ramana et al., 2000). However, our data now reveal that Stat1 can also down-regulate constitutive gene expression. Several of the down-regulated genes encode proteins that are involved in cell cycle progression. Treatment with IFN-α or IFN-γ can block the cell cycle by decreasing the expression of cdc2, cyclin A and cyclin B (Yamada et al., 1994; Harvat and Jetten, 1996; Tiefenbrun et al., 1996). We now show that Stat1 can also down-regulate the basal expression of these genes. Although the expression of Hsp70 is up-regulated by IFN-γ (Stephanou et al., 1999), our data show that the basal expression of Hsp70 is markedly down-regulated by Stat1 Y701F. The Bcl-xL promoter has an overlapping Stat5/Stat1-binding motif and Stat1 binds to this motif to induce leukemia-inhibiting factor (LIF)-mediated expression in cardiac myocytes (Fujio et al., 1997). rStat1 binds to this overlapping Stat5/Stat1 site as a dimer (data not shown). For some of the other genes regulated by Stat1 Y701F (e.g. cdc2 and cyclin A), the mechanism may be indirect since we can find no obvious Stat1-binding sites in the promoters of these genes.
Stat1 can up-regulate the constitutive transcription of several genes. Many of these (MHC class I, β2-microglobulin, CIITA and LMP2) are involved in the presentation of viral and tumor antigens (Gobin et al., 1997; Solheim, 1999). Increased expression of all of these genes leads to more efficient presentation of antigens in the context of class I MHC molecules.
Stat1 regulates the activities of two GAS-dependent promoters differently
Constitutive transcription of the LMP2 gene is mediated by a Stat1–IRF1 heteromer. Stat1 and Stat3 have been found to be associated in the cytoplasm of unstimulated cells (Haan et al., 2000) and HSF1 associates specifically with Stat1 in HepG2 cells (Stephanou et al., 1999). We have observed that a slowly migrating complex is formed with the GAS element of LMP2 but not of IRF1 in extracts of U3A-701 cells. The LMP2 GAS partially overlaps an ICS-2 site (White et al., 1996), in contrast to the IRF1 GAS, which does not have another known transcription factor-binding site nearby. There are other instances in which GAS elements are close to the binding sites for other transcription factors. The promoter of the MyD88 gene contains overlapping IRF1- and Stat1-binding sites (Harroch et al., 1995) and that of the CIITA gene has closely adjacent Stat1 and USF1 sites (Muhlethaler-Mottet et al., 1998).
Stat1 and IRF1 are both required for LMP2 transcription. IRF2 has also been observed to regulate LMP2 expression negatively (Chatterjee-Kishore et al., 1998). In extracts of melanoma cell lines, a complex forms on the LMP2 GAS element with a mobility similar to that of the complex with Stat1 homodimers. Cancer cells, especially melanomas, have a detectable amount of constitutively activated Stat1 due to the constitutive activation of kinases and receptors for cytokines and growth factors (Wellbrock et al., 1997). In fact, up-regulation of the epidermal growth factor (EGF) receptor has been associated with melanocytic tumor progression (Worm et al., 1995), and the binding of EGF to its receptor can signal to Stat1. It may be noted that some Stat1 homodimer binding to the LMP2 GAS is also observed in 2fTGH cells, which carry an activating N-ras mutation (Paterson et al., 1987).
We have observed that IRF1 interacts with Stat1 S727A and Stat1 p84 but neither mutant protein supports LMP2 gene transcription (data not shown). Thus the absence of transcription activation potential of Stat1 in cells expressing either Stat1 S727-A or Stat1 p84 is probably sufficient to abolish LMP2 gene transcription in these cells. Active IRF1 is a phosphoprotein, and phosphorylation is required for both DNA binding and transactivation (Pine et al., 1990; Watanabe et al., 1991; Kumar et al., 1997). Low levels of IRF1 binding to the ICS-2 region of the LMP2 promoter can be detected in extracts of U3A, 2fTGH and all the other cells used in this study (data not shown). Thus, a low level of active IRF1 may be sufficient to mediate LMP2 transcription in association with a Stat1 molecule containing an intact transactivation domain. In in vivo DNA–protein cross-linking experiments, the LMP2 ICS-2/GAS region could not be detected easily in U3A cells following immunoprecipitation with anti-IRF1. This low binding of IRF1 to the ICS/GAS region in the absence of Stat1 in vivo indicates that Stat1 also helps to recruit IRF1 to the ICS-2 sequence. Thus two routes may exist for IRF1 recruitment to the ICS-2/GAS, independent of Stat1 (still seen in U3A cells) and Stat1 dependent (observed in U3A-701 and 2fTGH cells).
Unphosphorylated Stat1 binds to DNA as a dimer in vitro
Baden et al. (1998) noted that the N-terminus of Stat4, which is highly homologous to the same region of Stat1, forms a dimer in solution. Although a similar interaction between the N-terminal domains of full-length Stats has not been observed in solution (Vinkemeier et al., 1998), a model of the structure of the N-terminus of Stat1 supports such interactions (Chen et al., 1998). The N-terminal domains of Stat1 bind to several proteins with diverse functions, such as phosphatases (Strehlow and Schindler, 1998) and transcription factors (Horvath et al., 1996), and also to each other when Stat1 dimers are bound to tandem GAS elements (Vinkemeier et al., 1996; Xu et al., 1996). We now find that the N-terminus of Stat1 is required to form unphosphorylated dimers on the LMP2 GAS element in vitro. Residues 135–200 appear to be required for binding since ΔN135 does form a dimer on DNA but ΔN200 does not. Replacement of the first 129 amino acids of Stat1 with those of Stat5 does not allow dimerization. However, it is not clear whether the complex of Stat1 Y701F and IRF1 that binds to the LMP2 ICS-2/GAS region in vivo involves a Stat1 monomer or dimer. Unphosphorylated Stat1 can also bind to the GAS as a monomer using half the palindromic binding site. This observation is also explained easily by the structure of DNA-bound Stat1 (Chen et al., 1998). Unphosphorylated Stat1 has been detected in the nucleus of rat liver cells (Ram et al., 1996) and we have also detected Stat1 Y701F in the nucleus. Thus, it is likely that unphosphorylated Stat1 has an important role in the regulation of gene expression.
Many novel functions of Stat1 remain to be elucidated
Constitutive expression of MHC class I genes in mouse T lymphocytes requires Stat1 (Lee et al., 1999). The same authors also show that transcription of the β2-microglobulin gene is down-regulated in mouse Stat1-null T lymphocytes. Our micro-array experiments show that both MHC class I and β2-microglobulin expression is up-regulated in U3A-701 cells, revealing that Stat1 regulates constitutive MHC class I and β2-microglobulin gene expression even in the absence of ligand stimulation. Thus, the data presented here and elsewhere begin to provide a greater role for Stat1 in IFN-independent regulation of the expression of several genes essential for host immune surveillance. A recent study from our laboratory (Ramana et al., 2000) has revealed the role of Stat1 in IFN-dependent down-regulation of c-myc and c-jun. Our current experiments show that Stat1 Y701F can also down-regulate the constitutive expression of several genes (e.g. Hsp70 and Bcl-xL) and up-regulate that of others (e.g. LMP2 and vinculin). Thus, Stat1 appears to be involved in both IFN-dependent and -independent positive regulation of gene expression and also in IFN-dependent and -independent negative regulation.
Materials and methods
Cells and constructs
U3A, U3A-701, U3A-IRF1(H), 2fTGH and G3A cells were grown in Dulbecco’s modified Eagle’s medium (DMEM). Namalwa, HeLa, H9 and Jurkat cells were grown in RPMI-1640. The U373-MG cells were grown in McCoy’s 5A medium. All media were supplemented with 10% fetal calf serum (FCS) and 100 µg/ml penicillin–streptomycin mix (Gibco) at 37°C under 10% CO2. Cells were treated with IFN-γ (1000 U/ml) for 15 min for EMSAs, and for 1 h for ChIP experiments. The Stat1 ΔN135, Stat1 ΔN200, Stat5/1 and pEF-luc constructs were gifts of R.Schreiber, Washington University, St Louis, R.Ransohoff, Cleveland Clinic Foundation, C.Schindler, Rockefeller University, New York and S.Ralph, Monash University, Australia, respectively.
Analysis of differential gene expression
Total RNA was extracted from cells with Trizol (Gibco-BRL). Approximately 200 µg were used to obtain cRNA according to standard Affymetrix protocols (Der et al., 1998). The cRNA was hybridized with human gene expression array chips and data were analyzed using the Affymetrix gene chip software. Expression was normalized against the Affymetrix spike RNA levels as well as against glyceraldehyde phosphate dehydrogenase (GAPDH) and actin RNA levels in the two samples. The expression of genes in U3A-701 cells was then analyzed using U3A cells as baseline.
Immunohistochemistry
Cells plated on coverslips were fixed in formaldehyde [1× phosphate-buffered saline (PBS) with 2% formaldehyde] and washed with methanol. Fixed cells were washed with PBSP [1× PBS, 0.1% bovine serum albumin (BSA), 0.02% sodium azide]. Cells were stained with anti-Stat1 (Transduction labs). Fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit antibody was used to detect Stat1. Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) to detect nuclei.
Reporter constructs and transfection procedures
The LMP2 GAS-luc (wild-type LMP2) and mt IRF-E-luc constructs were described previously (White et al., 1996). The LMP2 1/2GAS-luc construct was made by PCR-SOEing using LMP2 GAS-luc as template. The proximal primers were F1 GTAAGCTTGCCCTGCAAG GCACCGCTC and R1 GGCTCGAGATGGCACTCGGACGCCGGTC. The mutagenesis primers were F2 GATTTCGCTTTCCCCTAT ATGGCTGAGCTTC and R2 GAGAAGCTCAGCCATATAGGG GAAAGCGAA. F1 has a 5′ HindIII site and R1 has a 5′ XhoI site. The final PCR product was ligated into the XhoI and HindIII sites of the PGL3-basic vector (Promega) to produce LMP2 1/2GAS-luc. U3A, U3A-701 and 2fTGH cells were stably transfected with the pGL3basic, pEFluc, LMP2 GAS-luc, mt IRF-E-luc and LMP2 1/2 GAS-luc constructs using the Transfast reagent (Promega) according to the manufacturer’s instructions. The pBabePURO plasmid was co-transfected with all constructs to select pools of transfected cells. Luciferase activity was assayed in duplicate, in extracts of transfected cells, three times. Luciferase activity units were normalized against total protein levels assayed spectrophotometrically. Means and standard deviations were calculated using data from three luciferase assays.
In vivo genomic footprinting
Dimethyl sulfate treatments, genomic DNA preparation and LM-PCR for the ICS-2/GAS (IRF-E) region of the LMP2 promoter were described by White et al. (1996).
RNA preparation, RT–PCR, northern and S1 nuclease analyses
Total RNA was prepared using Trizol and mRNA was purified using the Oligotex mRNA kit (Qiagen). Probes for S1 nuclease and northern blots were prepared by RT–PCR using the Atlas PCR primers (Clontech) for the Bcl-xL, Hsp70, FUSE-binding protein and GAPDH genes. Primers for the LMP2 gene have been described (Chatterjee-Kishore et al., 1998). The following primers were used to amplify the CIITA and vinculin genes: CIITA (A), GTGCCTCTACCACTTCTA; CIITA (B), CAGAGA TTTGCCAGAGCC; vinculin (A), TCCTGGACTCAGGATATCGG; and vinculin (B), TCATCCTTTTCCTCTGGTGG. Approximately 5 µg of mRNA were hybridized with end-labeled PCR-derived probes corresponding to the CIITA, LMP2, vinculin or GAPDH genes using an S1 nuclease assay kit (Ambion). RNA–DNA hybrids were purified and separated in a 6% polyacrylamide gel containing 8 M urea. The sizes of the protected fragments were 677 bp for LMP2, 503 bp for CIITA, 450 bp for GAPDH and 286 bp for vinculin. For northern blots, 20 µg of total RNA were separated in 1.5% formaldehyde–formamide gels and transferred to Hybond plus membranes (Amersham). PCR-derived probes were labeled using [α-32P]dCTP and hybridized to the membrane-bound RNA overnight at 65°C in Church buffer (0.158 M NaH2PO4, 0.342 M Na2HPO4, 1 mM EDTA, 1% BSA and 7% SDS). Following stringent washing, membranes were exposed to film or phosphorimager screens. Transfers were stripped and reprobed to assay for GAPDH expression.
EMSAs
Recombinant His-Stat1 was expressed as described by Chatterjee-Kishore et al. (2000). The His6 tag was cleaved with thrombin to yield rStat1. Cells were washed with PBS and resuspended in 70 µl of HEM buffer [10 mM HEPES pH 7.9, 1 mM EDTA, 10% glycerol, 0.135 mM MgCl2, 100 mM NaF, 10 mM Na4P2O7, 1 mM phenylmethyl sulfonyl fluoride (PMSF) and pepstatin (Sigma), leupeptin (Sigma) and aprotinin (Sigma) at 0.1 mg/ml each]. Following lysis on ice for 15 min, cell suspensions were spun at 12 000 g for 15 min at 4°C. Supernatant solutions were used as total lysates in EMSAs using oligonucleotide probes end-labeled with [γ-32P]ATP. The probes used in EMSAs are listed in Figure 2. rStat1 was used at a concentration of 200 ng in EMSAs with the LMP2 GAS probe and at 500 ng with the LMP2 1/2 GAS probe. For EMSAs, 180 µl of dialysis buffer (20 mM HEPES pH 7.9, 50 mM KCl, 0.3 mM EDTA and 20% glycerol), 45 µl of reaction buffer (10 mM Na2HPO4, 1 mM EDTA), 24 µl of 80 mM MgCl2 and 8.8 µl of 3 µg/µl poly(dI–dC) were combined to make the reaction mix. For each reaction, 9 µl of reaction mix, 5 µl of cell extract containing ∼5 µg total protein or an appropriate amount of rStat1 in HEM buffer and 1 µl of end-labeled probe (∼30 000 c.p.m.) were incubated on ice for 20 min and then loaded into a 6% non-denaturing polyacrylamide gel. In EMSAs, extracts from U3A-701 cells were incubated with 1 µg of anti-Stat1 antibodies (a gift from B.R.G.Williams, Cleveland Clinic Foundation) for 20 min before adding the radioactively labeled LMP2 GAS probe.
In vitro translation and partial purification of Stat1
Stat1, Stat1 Y701F, Stat1 ΔN135, Stat1 ΔN200 and Stat5/1 were translated in vitro using the T7-based coupled transcription–translation kit (Promega) as per the manufacturer’s instructions. The translation products were mixed with heparin–agarose (Bio-Rad) and incubated for 4 h at 4°C with constant rotation of the tubes. The beads were then washed with HEM buffer three times and heparin–agarose-bound proteins were eluted with HEM buffer containing 1.5 M KCl. The eluates were dialyzed against HEM buffer overnight. Following analysis of protein concentration, the presence of Stat1 in the samples was ascertained by western analysis with antibodies directed against the C-terminus of Stat1 (Transduction Labs).
Protein–DNA and protein–protein cross-linking in vitro
Streptavidin–agarose beads (neutravidin–agarose; Pierce) were saturated with biotin-labeled LMP2 GAS or LMP2 1/2 GAS oligonucleotides at 4°C for 2 h. Beads were then washed in HEM buffer and incubated with either His-Stat1 (2–5 µg), IRF1 (in vitro translation products), U3A extracts or U3A-IRF1(H) extracts at 4°C for 1 h. The Stat1-saturated beads were then washed once with HEM buffer and incubated with either IRF1 (in vitro translation products), U3A extracts or U3A-IRF1(H) extracts for 2 h at 4°C. All samples were washed five times in HEM buffer, resuspended in SDS–PAGE loading buffer and loaded into 12% SDS–polyacrylamide gels. Proteins were transferred to PVDF membranes and analyzed in western transfers using anti-IRF1. For DMS-mediated cross-linking, neutravidin–agarose was saturated with the appropriate biotin-labeled oligonucleotides at 4°C for 2 h and washed in HEM buffer. The beads were then incubated with rStat1 (2–5 µg) at 4°C for 1 h and treated with 10 mM DMS on ice. Aliquots were removed at appropriate times, washed three times in HEM buffer and analyzed in western transfers using antibodies against the C-terminus of Stat1.
In vivo protein labeling and immunoprecipitations
Cells were labeled in vivo using [35S]methionine (Amersham) for 18 h and the proteins were extracted in HEM buffer as described for EMSAs. Aliquots containing equal amounts of radioactivity were immunoprecipitated with anti-IRF1 (Santa-Cruz Biotechnology). Immunoprecipitates were washed five times in HEM buffer and separated by 12% SDS–PAGE.
ChIP assay
DNA-bound proteins were cross-linked in vivo essentially as described by Orlando et al. (1997). Briefly, cells were treated with one-tenth volume of 11% formaldehyde per volume of medium and lysed by sonication. Chromatin was recovered by isopycnic centrifugation in a continuous CsCl gradient (1.42 g/l). Aliquots were collected from the gradient and analyzed for the presence of chromatin of ∼0.6–1.0 kb. These fractions were pooled and the chromatin was purified by dialysis. DNA-bound Stat1 and IRF1 were immunoprecipitated. Cross-links were then reversed at 65°C and the co-immunoprecipitated DNA was purified by a single step phenol–chloroform/isoamyl alcohol extraction. Purified DNA was amplified in PCR using primers for the LMP2 ICS/GAS region and for the IRF1 GAS region. The following primers were used: (i) LMP2 prom (F), GACGAGGGCTCTAAGAGTCTA and LMP2 prom (R2), GTG GATCTCGATCGGTAACCG; (ii) IRF1 prom (F), CTGCCAGAG CGAAAGGTTCTG and IRF1 prom (R), CTAAGCGTCTCCCAC GGCCTC. PCR conditions were as follows: 94°C for 5 min, followed by 35 cycles of 94°C for 1 min, 58°C for 2 min, 72°C for 3 min, followed by an additional extension time of 15 min at 72°C and a soak cycle at 4°C. PCR products were analyzed in 1% agarose gels. The expected size of the amplified product for the LMP2 ICS-2/GAS region was 280 bp and for the IRF1 GAS region was 450 bp.
Acknowledgments
Acknowledgements
We are very grateful for the assistance of Lesleyann Hawthorne in the Affymetrix analyses. This work was supported by a grant from Ares-Serono and by NIH grant CA62220.
References
- Baden H.A., Sarma,S.P., Kapust,R.B., Byrd,R.A. and Waugh,D.S. (1998) The amino-terminal domain of human STAT4. Overproduction, purification, and biophysical characterization. J. Biol. Chem., 273, 17109–17114. [DOI] [PubMed] [Google Scholar]
- Chatterjee-Kishore M., Kishore,R., Hicklin,D.J., Marincola,F.M. and Ferrone,S. (1998) Different requirements for signal transducer and activator of transcription 1α and interferon regulatory factor 1 in the regulation of low molecular mass polypeptide 2 and transporter associated with antigen processing 1 gene expression. J. Biol. Chem., 273, 16177–16183. [DOI] [PubMed] [Google Scholar]
- Chatterjee-Kishore M., van den Akker,F. and Stark,G.R. (2000) Adenovirus E1A down-regulates LMP2 transcription by interfering with the binding of Stat1 to IRF1. J. Biol. Chem., 275, 20406–20411. [DOI] [PubMed] [Google Scholar]
- Chen X., Vinkemeier,U., Zhao,Y., Jeruzalmi,D., Darnell,J.E.,Jr and Kuriyan,J. (1998) Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell, 93, 827–839. [DOI] [PubMed] [Google Scholar]
- Darnell J.E. Jr, Kerr,I.M. and Stark,G.R. (1994) Jak–STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science, 264, 1415–1421. [DOI] [PubMed] [Google Scholar]
- Decker T., Kovarik,P. and Meinke,A. (1997) GAS elements: a few nucleotides with a major impact on cytokine-induced gene expression. J. Interferon Cytokine Res., 17, 121–134. [DOI] [PubMed] [Google Scholar]
- Der S.D., Zhou,A., Williams,B.R. and Silverman,R.H. (1998) Identification of genes differentially regulated by interferon α, β, or γ using oligonucleotide arrays. Proc. Natl Acad. Sci. USA, 95, 15623–15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujio Y., Kunisada,K., Hirota,H., Yamauchi-Takihara,K. and Kishimoto,T. (1997) Signals through gp130 upregulate bcl-x gene expression via STAT1-binding cis-element in cardiac myocytes. J. Clin. Invest., 99, 2898–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobin S.J., Peijnenburg,A., Keijsers,V. and van den Elsen,P.J. (1997) Site α is crucial for two routes of IFN γ-induced MHC class I transactivation: the ISRE-mediated route and a novel pathway involving CIITA. Immunity, 6, 601–611. [DOI] [PubMed] [Google Scholar]
- Guyer N.B., Severns,C.W., Wong,P., Feghali,C.A. and Wright,T.M. (1995) IFN-γ induces a p91/Stat1α-related transcription factor with distinct activation and binding properties. J. Immunol., 155, 3472–3480. [PubMed] [Google Scholar]
- Haan S., Kortylewski,M., Behrmann,I., Muller-Esterl,W., Heinrich,P.C. and Schaper,F. (2000) Cytoplasmic STAT proteins associate prior to activation. Biochem. J., 345, 417–421. [PMC free article] [PubMed] [Google Scholar]
- Harroch S., Gothelf,Y., Revel,M. and Chebath,J. (1995) 5′ upstream sequences of MyD88, an IL-6 primary response gene in M1 cells: detection of functional IRF-1 and Stat factors binding sites. Nucleic Acids Res., 23, 3539–3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvat B.L. and Jetten,A.M. (1996) γ-Interferon induces an irreversible growth arrest in mid-G1 in mammary epithelial cells which correlates with a block in hyperphosphorylation of retinoblastoma. Cell Growth Differ., 7, 289–300. [PubMed] [Google Scholar]
- Horvath C.M., Stark,G.R., Kerr,I.M. and Darnell,J.E.,Jr (1996) Interactions between STAT and non-STAT proteins in the interferon-stimulated gene factor 3 transcription complex. Mol. Cell. Biol., 16, 6957–6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihle J.N. (1995) Cytokine receptor signalling. Nature, 377, 591–594. [DOI] [PubMed] [Google Scholar]
- Iwase S., Furukawa,Y., Kikuchi,J., Nagai,M., Terui,Y., Nakamura,M. and Yamada,H. (1997) Modulation of E2F activity is linked to interferon-induced growth suppression of hematopoietic cells. J. Biol. Chem., 272, 12406–12414. [DOI] [PubMed] [Google Scholar]
- Kumar A., Commane,M., Flickinger,T.W., Horvath,C.M. and Stark,G.R. (1997) Defective TNF-α-induced apoptosis in STAT1-null cells due to low constitutive levels of caspases. Science, 278, 1630–1632. [DOI] [PubMed] [Google Scholar]
- Lee C.K., Gimeno,R. and Levy,D.E. (1999) Differential regulation of constitutive major histocompatibility complex class I expression in T and B lymphocytes. J. Exp. Med., 190, 1451–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner P.J. and Trowsdale,J. (1998) Antigen presentation: coming out gracefully. Curr. Biol., 8, R605–R608. [DOI] [PubMed] [Google Scholar]
- Li X., Leung,S., Qureshi,S., Darnell,J.E.,Jr and Stark,G.R. (1996) Formation of STAT1–STAT2 heterodimers and their role in the activation of IRF-1 gene transcription by interferon-α. J. Biol. Chem., 271, 5790–5794. [DOI] [PubMed] [Google Scholar]
- Min W., Pober,J.S. and Johnson,D.R. (1996) Kinetically coordinated induction of TAP1 and HLA class I by IFN-γ: the rapid induction of TAP1 by IFN-γ is mediated by Stat1α. J. Immunol., 156, 3174–3183. [PubMed] [Google Scholar]
- Miyamoto M., Fujita,T., Kimura,Y., Maruyama,M., Harada,H., Sudo,Y., Miyata,T. and Taniguchi,T. (1988) Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to IFN-β gene regulatory elements. Cell, 54, 903–913. [DOI] [PubMed] [Google Scholar]
- Muhlethaler-Mottet A., Di Berardino,W., Otten,L.A. and Mach,B. (1998) Activation of the MHC class II transactivator CIITA by interferon-γ requires cooperative interaction between Stat1 and USF-1. Immunity, 8, 157–166. [DOI] [PubMed] [Google Scholar]
- Orlando V., Strutt,H. and Paro,R. (1997) Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods, 11, 205–214. [DOI] [PubMed] [Google Scholar]
- Paterson H., Reeves,B., Brown,R., Hall,A., Furth,M., Bos,J., Jones,P. and Marshall,C. (1987) Activated N-ras controls the transformed phenotype of HT1080 human fibrosarcoma cells. Cell, 51, 803–812. [DOI] [PubMed] [Google Scholar]
- Pine R., Decker,T., Kessler,D.S., Levy,D.E. and Darnell,J.E.,Jr (1990) Purification and cloning of interferon-stimulated gene factor 2 (ISGF2): ISGF2 (IRF-1) can bind to the promoters of both β interferon- and interferon-stimulated genes but is not a primary transcriptional activator of either. Mol. Cell. Biol., 10, 2448–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram P.A., Park,S.H., Choi,H.K. and Waxman,D.J. (1996) Growth hormone activation of Stat 1, Stat 3, and Stat 5 in rat liver. Differential kinetics of hormone desensitization and growth hormone stimulation of both tyrosine phosphorylation and serine/threonine phosphorylation. J. Biol. Chem., 271, 5929–5940. [DOI] [PubMed] [Google Scholar]
- Ramana C.V., Grammatikakis,N., Chernov,M., Nguyen,H., Goh,K.C. and Williams,B.R.G. (2000) Regulation of c-myc expression by IFN-γ through Stat1-dependent and Stat1-independent pathways. EMBO J., 19, 263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma B. and Iozzo,R.V. (1998) Transcriptional silencing of perlecan gene expression by interferon-γ. J. Biol. Chem., 273, 4642–4646. [DOI] [PubMed] [Google Scholar]
- Shuai K., Horvath,C.M., Huang,L.H.T., Qureshi,S.A., Cowburn,D. and Darnell,J.E.,Jr (1994) Interferon activation of the transcription factor Stat91 involves dimerization through SH2–phosphotyrosyl peptide interactions. Cell, 76, 821–828. [DOI] [PubMed] [Google Scholar]
- Sibinga N.E., Wang,H., Perrella,M.A., Endege,W.O., Patterson,C., Yoshizumi,M., Haber,E. and Lee,M.E. (1999) Interferon-γ-mediated inhibition of cyclin A gene transcription is independent of individual cis-acting elements in the cyclin A promoter. J. Biol. Chem., 274, 12139–12146. [DOI] [PubMed] [Google Scholar]
- Solheim J.C. (1999) Class I MHC molecules: assembly and antigen presentation. Immunol. Rev., 172, 11–19. [DOI] [PubMed] [Google Scholar]
- Stark G.R., Kerr,I.M., Williams,B.R.G., Silverman,R.H. and Schreiber,R.D. (1998) How cells respond to interferons. Annu. Rev. Biochem., 67, 227–264. [DOI] [PubMed] [Google Scholar]
- Stephanou A., Isenberg,D.A., Nakajima,K. and Latchman,D.S. (1999) Signal transducer and activator of transcription-1 and heat shock factor-1 interact and activate the transcription of the Hsp-70 and Hsp-90β gene promoters. J. Biol. Chem., 274, 1723–1728. [DOI] [PubMed] [Google Scholar]
- Strehlow I. and Schindler,C. (1998) Amino-terminal signal transducer and activator of transcription (STAT) domains regulate nuclear translocation and STAT deactivation. J. Biol. Chem., 273, 28049–28056. [DOI] [PubMed] [Google Scholar]
- Su W.C., Kitagawa,M., Xue,N., Xie,B., Garofalo,S., Cho,J., Deng,C., Horton,W.A. and Fu,X.Y. (1997) Activation of Stat1 by mutant fibroblast growth-factor receptor in thanatophoric dysplasia type II dwarfism. Nature, 386, 288–292. [DOI] [PubMed] [Google Scholar]
- Thor Straten P., Kirkin,A.F., Seremet,T. and Zeuthen,J. (1997) Expression of transporter associated with antigen processing 1 and 2 (TAP1/2) in malignant melanoma cell lines. Int. J. Cancer, 70, 582–586. [DOI] [PubMed] [Google Scholar]
- Tiefenbrun N., Melamed,D., Levy,N., Resnitzky,D., Hoffman,I., Reed,S.I. and Kimchi,A. (1996) α Interferon suppresses the cyclin D3 and cdc25A genes, leading to a reversible G0-like arrest. Mol. Cell. Biol., 16, 3934–3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinkemeier U., Cohen,S.L., Moarefi,I., Chait,B.T., Kuriyan,J. and Darnell,J.E.,Jr (1996) DNA binding of in vitro activated Stat1α, Stat1β and truncated Stat1: interaction between NH2-terminal domains stabilizes binding of two dimers to tandem DNA sites. EMBO J., 15, 5616–5626. [PMC free article] [PubMed] [Google Scholar]
- Vinkemeier U., Moarefi,I., Darnell,J.E.,Jr and Kuriyan,J. (1998) Structure of the amino-terminal protein interaction domain of STAT-4. Science, 279, 1048–1052. [DOI] [PubMed] [Google Scholar]
- Walter M.J., Look,D.C., Tidwell,R.M., Roswit,W.T. and Holtzman,M.J. (1997) Targeted inhibition of interferon-γ-dependent intercellular adhesion molecule-1 (ICAM-1) expression using dominant-negative Stat1. J. Biol. Chem., 272, 28582–28589. [DOI] [PubMed] [Google Scholar]
- Watanabe N., Sakakibara,J., Hovanessian,A.G., Taniguchi,T. and Fujita,T. (1991) Activation of IFN-β element by IRF-1 requires a post-translational event in addition to IRF-1 synthesis. Nucleic Acids Res., 19, 4421–4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellbrock C., Gomez,A. and Schartl,M. (1997) Signal transduction by the oncogenic receptor tyrosine kinase Xmrk in melanoma formation of Xiphophorus. Pigment Cell Res., 10, 34–40. [DOI] [PubMed] [Google Scholar]
- White L.C., Wright,K.L., Felix,N.J., Ruffner,H., Reis,L.F., Pine,R. and Ting,J.P. (1996) Regulation of LMP2 and TAP1 genes by IRF-1 explains the paucity of CD8+ T cells in IRF-1–/– mice. Immunity, 5, 365–376. [DOI] [PubMed] [Google Scholar]
- Worm M., Makki,A., Dippel,E., Czarnetzki,B.M. and Schadendorf,D. (1995) Interferon-γ downregulates epidermal growth factor receptors on human melanoma cells. Exp. Dermatol., 4, 30–35. [DOI] [PubMed] [Google Scholar]
- Wright K.L., White,L.C., Kelly,A., Beck,S., Trowsdale,J. and Ting,J.P. (1995) Coordinate regulation of the human TAP1 and LMP2 genes from a shared bidirectional promoter. J. Exp. Med., 181, 1459–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Sun,Y.-L. and Hoey,T. (1996) Cooperative DNA binding and sequence-selective recognition conferred by the STAT amino-terminal domain. Science, 273, 794–797. [DOI] [PubMed] [Google Scholar]
- Yamada H., Ochi,K., Nakada,S., Nemoto,T. and Horiguchi-Yamada,J. (1994) Changes of cell cycle-regulating genes in interferon-treated Daudi cells. Mol. Cell. Biochem., 136, 117–123. [DOI] [PubMed] [Google Scholar]