Abstract
Objective
Mutations in hematopoietic transcription factor RUNX1 cause thrombocytopenia and impaired platelet function. In a patient with a heterozygous mutation in RUNX1 we have described decreased platelet pleckstrin phosphorylation and protein kinase C-θ (PKC-θ, gene PRKCQ) associated with thrombocytopenia, impaired platelet aggregation, and dense granule secretion. Little is known regarding regulation of PKC-θ in megakaryocytes/platelets. We have addressed the hypothesis that PRKCQ is a direct transcriptional target of RUNX1.
Methods and Results
In chromatin immunoprecipitation assay using megakaryocytic cells there was RUNX1 binding in vivo to PRKCQ promoter region −1225/−1056 bp containing a RUNX1 consensus site ACCGCA at −1088/−1069 bp; electrophoretic mobility shift assay showed RUNX1 binding to the specific site. In RUNX1 overexpression studies, PKC-θ protein expression and promoter activity were enhanced; mutation of RUNX1 site showed decreased activity even with RUNX1 overexpression. Lastly, PRKCQ promoter activity and PKC-θ protein were decreased by siRNA knockdown of RUNX1.
Conclusion
Our results provide the first evidence that PRKCQ is regulated at the transcriptional level by RUNX1 in megakaryocytic cells and a mechanism for PKC-θ deficiency associated with RUNX1 haplodeficiency.
Keywords: PKC-θ, RUNX1, platelets, megakaryocytes, transcriptional regulation
Protein kinase C (PKC, gene PRKCQ) signaling is a critical aspect of megakaryocytic (MK) differentiation1, proplatelet formation,2, 3 platelet function and thrombus formation.4–8 Platelets possess several PKC isozymes including classical PKCs α, βI and βII, novel PKCs δ, η, ε and θ, and atypical PKC ζ, and λ, and the specific roles of the isozymes in platelets and thrombus formation are being defined.4–8 PKC in synergy with Ca+ elevation regulate dense and α-granule secretion upon platelet stimulation by thrombin, thromboxane A2 (TxA2) and ADP.9–11 PKC play a role in αIIbβ3 activation by promoting conformational changes required for fibrinogen binding and platelet aggregation.12, 13 The activated integrins themselves stimulate one or more of PKC isozymes leading to filopodia formation and platelet spreading.14, 15
Evidence is emerging for distinct roles of PKC isoforms in platelet activation and thrombus formation. PKC-α has been proposed as a key enzyme regulating α- and dense granule secretion, platelet aggregate formation and thrombus formation.11, 16, 17 In mouse platelets, PKC-β positively regulates outside-in αIIbβ3 signaling, platelet spreading on fibrinogen and collagen-induced thrombus formation.4, 14 Gilio et al4 showed that PKC-δ negatively regulates thrombus formation. Mouse platelets deficient in PKC-δ have enhanced filopodia formation and collagen-induced platelet aggregation;18 dense granule secretion induced by activation of GPVI was normal in one study18 but enhanced in another along with enhanced TxA2 formation.19 In the latter report, interestingly, dense granule secretion induced by PAR4 agonist was reduced indicating an agonist specific differential effect of PKC-δ. PKC-θ deficient murine platelets have revealed defective filopodia formation and spreading on fibrinogen.15, 20 PKC-θ deficient mice have been reported to have impaired hemostasis with prolonged bleeding times, and decreased agonist-stimulated platelet aggregation, dense and α-granule secretion, αIIbβ3 activation and TxA2 production.8, 21 Other studies have found that PKC-θ deficient mice have enhanced GPVI mediated α-granule secretion and inside-out activation of αIIbβ3 along with enhanced thrombus formation on collagen.20 Soriani et al15 have reported that PKC-θ plays a role in outside-in αIIβ3 signaling but not in inside-out signaling. Although these studies are conflicting in some aspects (likely due to different experimental conditions), they advance the concept that PKC-θ plays an important role in platelet function.
We have reported detailed studies on a patient22–24 with inherited thrombocytopenia, decreased platelet aggregation, secretion, and GPIIb-IIIa activation, impaired phosphorylation of pleckstrin and myosin light chain, and decreased platelet PKC-θ (protein and mRNA) associated with a mutation in RUNX1. RUNX1 is a transcription factor that plays a major role in hematopoiesis and megakaryopoiesis.25–27 It is composed of two subunits and the α-subunit (RUNX1) is the DNA binding element of the complex and recognizes the DNA sequence TGT/cGGT. CBFβ, the β subunit, stabilizes RUNX1 binding to DNA but without direct DNA contact. RUNX1 mutations are associated with familial, autosomal dominant thrombocytopenia, platelet dysfunction, and predisposition to acute leukemia.28, 29 Based on the decreased platelet PKC-θ in our patient, we hypothesized that platelet/megakaryocyte PKC-θ is regulated at the transcriptional level by RUNX1 and constitutes the mechanism for PKC-θ deficiency. Despite the important role of PKC-θ signaling in platelet function, very little is known about its transcriptional regulation in megakaryocytic cells. In the present studies, we provide the first evidence that PKC-θ is a direct transcriptional target of RUNX1. We have recently shown that RUNX1 regulates ALOX12,30 MYL931 and PF4.32 Present studies extend this to PRKCQ and underscores the complex nature of alterations in platelets/megakaryocytes in human RUNX1 haplodeficiency.
Materials and Methods
Patient Information
We have previously described22–24 the clinical presentation and studies in this 24 year old white male, documenting decreased agonist stimulated platelet aggregation, secretion, GPIIb-IIIa activation, and pleckstrin and myosin light chain (MLC) phosphorylation; platelet PKC-θ level was decreased. The patient has a single point mutation in RUNX1, in intron 3 at the splice acceptor site for exon 4 leading to a frameshift with premature termination in the conserved Runt homology domain.23
Materials
All chemicals including phorbol 12-myristate 13-acetate (PMA) were purchased from Sigma (St Louis, MO, USA) or Fisher Scientific (Pittsburgh, PA, USA). GoTaq Green PCR Master Mix, luciferase reporter vectors pGL3-Basic and pRL-TK, and Dual Luciferase Assay System were from Promega (Madison, WI, USA). PCR primers and infrared (IR) dye-labeled probes for EMSA were purchased from Integrated DNA Technologies, IDT (Coralville, IA, USA). PCR products were sequenced by capillary electrophoresis and fluorescent dye terminator detection on ABI3730xl DNA analyzer (GENEWIZ, South Plainfield, NJ, USA). Recombinant RUNX1 protein, RUNX1 expression plasmid RUNX1-pCMV6-XL4, empty expression vector pCMV6-XL4 and transfection agent Turbofectin 8.0 were from ORIGENE Technologies, Rockville, MD, USA).
Cell lines and Cell culture
Human erythroleukemia (HEL) cell line from ATCC (American Type Cell Culture, Rockville, MD, USA) was cultured in RPMI-1640 medium (Mediatech, VA, USA) in the presence of 10% fetal bovine serum and antibiotics (Mediatech, VA, USA). During induction, HEL cells were grown in 10 nM PMA.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assay was performed using HEL cells (1×108) treated with PMA for 24 h and ChIP-IT kit (Active Motif, Carlsbad, CA, USA) as described.31 Sheared fragments were immunoprecipitated with anti-RUNX1 antibody (sc-8564x, Santa Cruz Biotechnology, Santa Cruz, CA, USA) or negative control IgG antibody supplied in the kit. We amplified three regions (−665/−478 bp, −1225/−1056 bp and −1468/−1277 bp) of PRKCQ promoter with specific primers.
Electrophoretic Mobility Shift assay (EMSA)
Nuclear extracts were prepared from PMA-treated HEL cells according to Dignam et al.33 Nuclear protein-DNA interactions were performed using PKC-θ infrared (IR)-Dye labeled double-stranded oligos and Odyssey Infrared EMSA Kit (Li-Cor Biosciences, Lincoln, NE, USA). PKC-θ wild type (Wt) probes used to examine RUNX1 binding to each consensus site (in bold) and their mutants generated by deletions in RUNX1 sites (underlined) are: Wt probe I (−585/−566) with site-I- 5’CAAATGCCTGGGGTAGGTCA 3’, its mutant 5’CAAATGC CTGGGGTAGGTCA 3’; Wt probe II (−618/−599) with site-II -−’GAACGATG ACCCCAGGACAG 3’, its mutant 5’GAACGATGACCCCAGGACAG 3’; Wt probe III (−1088/−1070) with site-III-5’ATGAGCCACCGCACCTGGCC 3’, its mutant 5’ATGAGC CACCGCAC CTGGCC 3’; Wt probe IV (−1319/−1300) with site-IV-5’CAGTGCAGTGGTGA GATCTC 3’, its mutant 5’CAGTGCAGTGGTGAGATCTC 3’ and Wt probe V (−1442/−1423) with site-V-5’ACCTCTGAGGTTCTTTTTAG 3’, and its mutant 5’ACCTCTGAGGTTCTTT TTAG 3’. 3 µg of nuclear extract and 50 fmoles of IR-labelled probe were used in binding reactions performed on ice for 30 min. Oligonucleotide competitors were added 30 min before the labelled probe was added. For supershift assays, RUNX1 antibody (sc-8563x) or control IgG (Santa Cruz Biotechnology, Santa Cruz, CA, USA) was pre-incubated with nuclear protein before adding labelled probe. In addition, EMSA was performed with recombinant RUNX1 protein (200 ng) to see its binding to PKC-θ probes. Binding reaction was performed on ice for 1 h in a buffer containing 0.6 mM HEPES, pH 8.0, 1 mM DTT, 0.01% triton X-100, 2% glycerol, 5 µg/µl bovine serum albumin and 100 mM NaCl. For supershift studies, anti-RUNX1 antibody (sc-8564x) or control IgG (Santa Cruz Biotechnology, Santa Cruz, CA, USA) was pre-incubated for 30 min on ice with recombinant RUNX1 protein before the addition of labelled probe. Binding complexes were separated by electrophoresis on native 5% TBE (Tris-Borate-EDTA) gels (Bio-Rad) and detected using Odyssey Infrared Imaging System (Li-Cor Biosciences).
Construction of Luciferase Reporter Plasmids
PRKCQ promoter region (−1085/−206 bp) was amplified by a standard PCR from human genomic DNA using modified primers incorporated with restriction sites Xho I in the forward primer and Hind III in the reverse primer. The PCR product was cloned into pCR 2.1 TOPO TA cloning vector (Invitrogen, Carlsbad, CA, USA), digested with Xho I and Hind III enzymes and cloned into appropriate sites of luciferase vector, pGL3-Basic. The primer sequences of Wt construct were as follows: forward at −1085/−1068, 5’-aaactcgagGAGCCACCGCACCTGGCC-3’ with RUNX1 site (in bold) and reverse at −221/−206 5’-cacaagctt GGTTGCGCCCTGGAGC-3’ (lower cases indicate flanking sequences). Mutant (mut) construct was generated by mutating the RUNX1 site by deletions (bold and underlined) in the forward primer 5’- aaactcgag GAGCCACCGCACCTGGCC’, and using the same (−221/−206) reverse primer as described above. PCR products were confirmed by DNA sequencing prior to cloning.
RUNX1 overexpression
HEL cells (1 × 106) were co-transfected with equal amounts of Wt PRKCQ reporter construct (−1085/−206-Luc) and RUNX1expression plasmid, RUNX1-pCMV6-XL4 or empty vector pCMV6-XL4 (1 µg each) along with an internal control, pRL-TK containing Renilla luciferase gene (20 ng), using Turbofectin 8.0 transfection reagent. Wt PRKCQ construct alone was cotransfected with pRL-TK. Mutant PRKCQ reporter transfections were also performed in a similar manner. In parallel, promoterless empty vector pGL3-Basic was transfected as a control. After transfection for 3–4 h at 37°C in 5% CO2, medium containing PMA (10 nM) was added to the transfection mixture. After 48 h cells were lysed and luciferase activity was measured using Dual Luciferase Assay System (Promega). Promoter activity was expressed as firefly luciferase activity/renilla luciferase activity relative to that of the empty vector. All transfection experiments were performed three times in triplicate. RUNX1 and PKC-θ were assessed in HEL cell lysates by immunoblotting.
RUNX1 knockdown by its siRNA
HEL cells (5 × 105) were transfected with 400 nM RUNX1 siRNA pool (sc-37677) or unrelated mock siRNA (Santa Cruz Biotechnology) using siRNA transfection reagent system (Santa Cruz Biotechnology).31 After 5 hr of transfection, medium containing 20% fetal bovine serum and 50 nM of PMA was added to the cells. On the following day, the medium was replaced with that containing 10% serum and 50 nM PMA. Cells were harvested at 48 h. The effect of RUNX1 siRNA was examined by immunoblot analysis. In parallel, HEL cells were cotransfected with Wt PRKCQ construct and mock or RUNX1 siRNAs along with internal control (pRL-TK) as described above. Luciferase activity was measured in HEL cell lysates using Dual Luiferase Assay System (Promega).
Immunoblotting
Whole cell lysates (30–40 µg) from co-transfected HEL cells were subjected to sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE), transferred to Odyssey nitrocellulose membranes (Li-COR Biosciences), and probed with the antibodies against RUNX1 (sc-8563), PKC-θ (sc-1875) and actin (sc-1616R) or β-actin (sc-47778) from Santa Cruz Biotechnology. Specific protein expressions were detected with IR-labeled secondary antibodies using Odyssey Infrared Imaging System (Li-COR Biosciences).
Bioinformatics
Potential binding sites for transcription factors were analyzed by computer program TFSEARCH (http://mbs.cbrc.jp/research/db/TFSEARCH.html).
Results
Binding of RUNX1 to PRKCQ promoter by ChIP
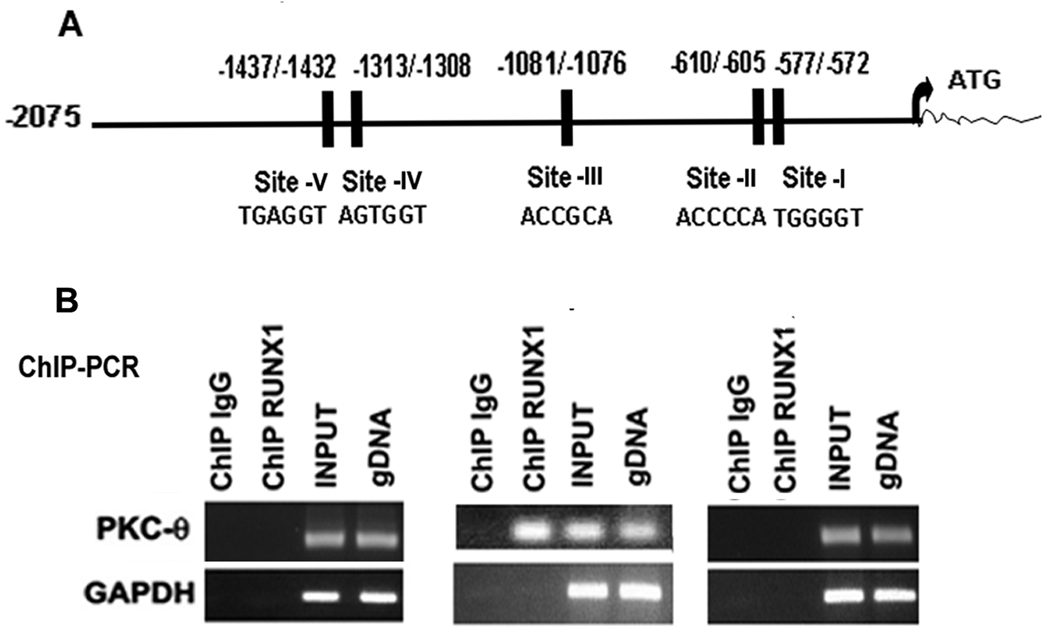
Analysis of PRKCQ promoter region (2075 bp from the ATG) using TFSEARCH revealed 5 RUNX1 consensus sites at −577/−572 (site-I), −610/−605 (site-II), −1081/−1076 (site-III) −1313/−1308 (site-IV) and −1437−−1432 (site-V) (Figure 1A). We performed immunoprecipitations on chromatin samples from PMA-treated HEL cells using an anti-RUNX1 antibody to identify an endogenous interaction between RUNX1 and PRKCQ gene. PCR primers were designed to amplify PKC-θ regions −665/−478 bp containing sites-I and -II, −1225/−1056 bp containing site -III and −1468/−1277 bp region with sites-IV and -V. These primers were used to amplify HEL cell chromatin enriched by anti-RUNX1 antibody. Only region −1225/−1056 bp with the site-III was enriched by RUNX1 antibody (Figure 1B). These studies indicate that RUNX1 binds in vivo to site-III but not to other sites under the conditions studied.
Figure 1.
Chromatin immunoprecipitation studies showing RUNX1 interaction with PRKCQ promoter. A. Promoter region (−2075/−1 bp from ATG) showing five RUNX1 consensus sites. B. Chromatin immunoprecipitation studies using HEL cells. Regions encompassing the RUNX1 sites were PCR amplified from input and immunoprecipitated samples. GAPDH was amplified as a control.
Binding of RUNX1 to consensus sites on PRKCQ promoter by EMSA
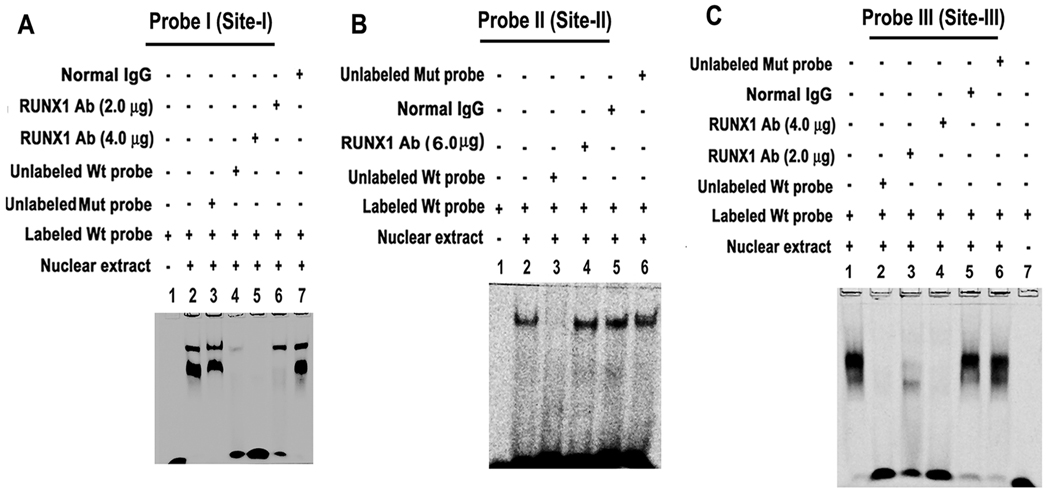
EMSA was performed using HEL cell nuclear extracts and Wt probe I that has RUNX1 consensus site-I (Figure 2A). Protein binding occurred with probe I (lane 2). Binding was not altered by competition with the mutant probe with mutation of RUNX1 site (lane 3), but was lost by competition with unlabelled Wt probe (lane 4). Binding to Wt probe was inhibited by competition with RUNX1 antibody (lanes 5 and 6 show decreasing antibody concentration), but not by IgG (lane 7).
Figure 2.
EMSA using HEL cell nuclear extracts and wild type (Wt) DNA probes containing RUNX1 sites-I (A), -II (B) and -III (C) and mutant (Mut) probes with specific mutations. RUNX1 antibody inhibited protein binding to sites-I and -III, but not to site-II.
Figure 2B shows nuclear protein binding to probe containing consensus site-II (lane 2). This binding was competed by excess unlabelled probe (lane 3), but not by RUNX1 antibody (6 µg) (lane 4). Competition studies were performed with 2, 4 and 6 µg of antibody but binding was not affected. It was not altered by normal IgG (lane 5). Lane 6 shows no effect on binding by competition with excess unlabelled mutant probe with RUNX1 site mutated. These findings suggest that the observed protein binding to the probe with site-II is not due to RUNX1, and are in line with findings with ChIP (Figure 1).
Figure 2C shows protein binding to probe III with RUNX1 consensus site-III (lane 1); this was abolished by competition with excess unlabelled probe (lane 2). Binding was inhibited by RUNX1 antibody (lanes 3–4), not by IgG (lane 5). DNA-protein binding was not altered by competition with the mutant probe (lane 6). Lane 7 shows labeled probe alone. These data indicate that RUNX1 binds to site-III. EMSA performed on probes with site-IV or site-V showed no protein binding (data not shown). Altogether, these results indicate that RUNX1 binds to site-I and site-III by EMSA.
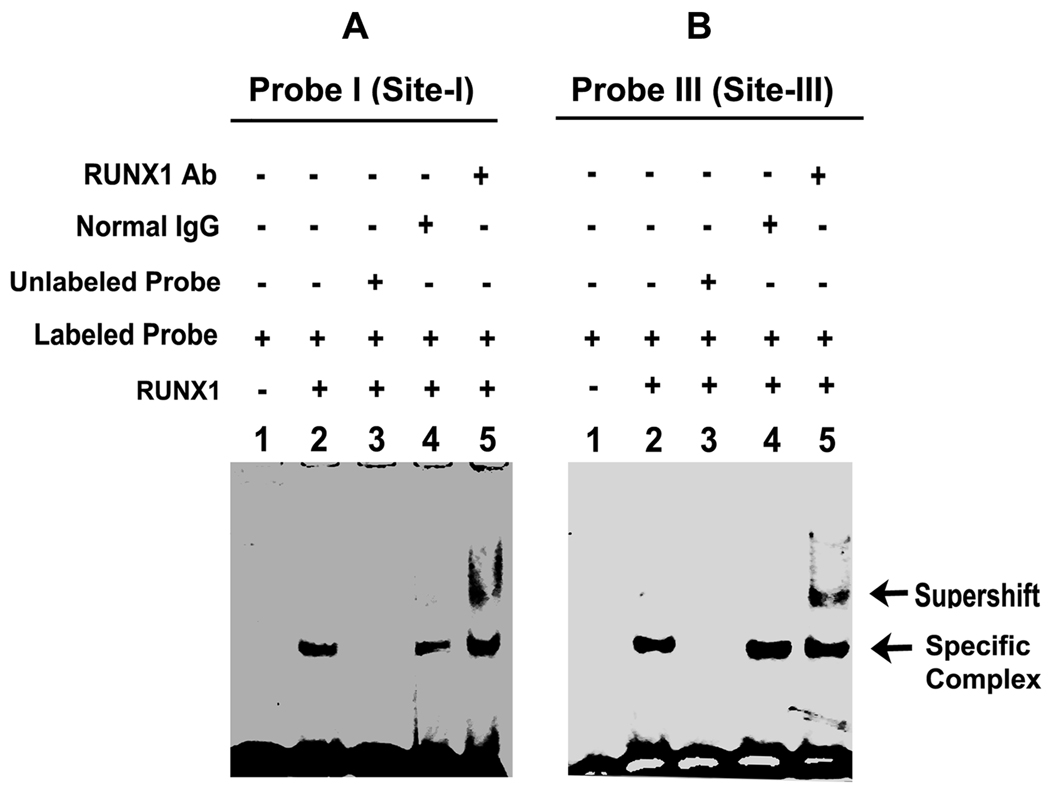
Binding studies with recombinant RUNX1
To further establish that RUNX1 binds to PKC-θ probes I and III, we performed studies using recombinant RUNX1 protein. Figure 3A shows RUNX1 binding to the probe I containing site-I (lane 2). This binding was competed by excess unlabelled probe (lane 3), unaffected by normal IgG (lane 4), but supershifted by anti- RUNX1 antibody (lane 5). Similar results were obtained with probe containing site-III (Figure 3B). These data provide further support that RUNX1 binds to site-I and site-III.
Figure 3.
EMSA using recombinant RUNX1 and PRKCQ DNA probes contain RUNX1 sites-I and -III. Protein binding to both probes was supershifted by RUNX1 antibody but not IgG.
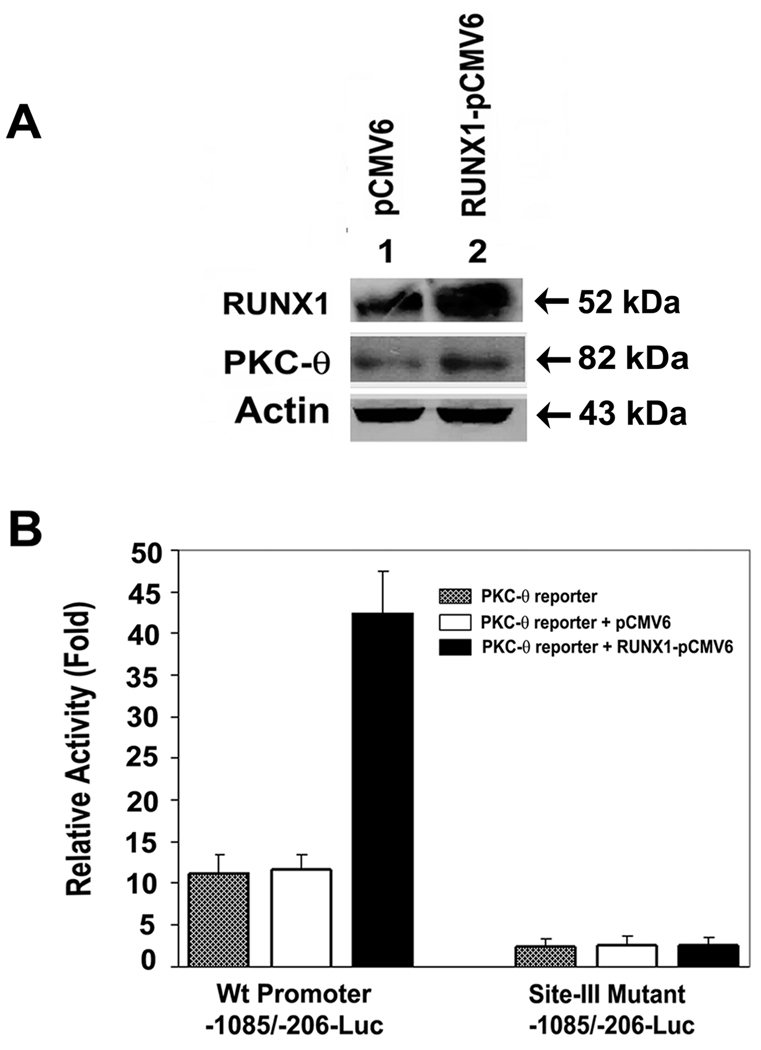
Enhancement of PRKCQ promoter activity and PKC-θ protein by overexpression of RUNX1
Immunoblot analysis of HEL cells transfected with RUNX1-pCMV6 expression plasmid revealed enhanced expression of RUNX1 and PKC-θ (Figure 4A). Promoter activity of Wt PRKCQ construct (−1085/−206-Luc) containing sites-I, -II and -III was not increased in the presence of empty vector pCMV6-XL4 (Figure 4B). It was markedly enhanced in the presence of RUNX1 expression plasmid, RUNX1-pCMV6-XL4. The ChIP studies (Figure 1) showed in vivo binding of RUNX1 to only site-III. Therefore, we studied the effect of mutating this site. Mutant promoter with site-III mutated (but intact sites I and II) showed complete loss of activity. Overexpression of RUNX1 did not increase activity indicating that site-III is involved in regulating promoter activity, but not sites -I or -II. These findings complement the results of ChIP analysis and EMSA.
Figure 4.
Effect of RUNX1 overexpression on PKC-θ protein and promoter in HEL cells. A. Immunoblotting of cell lysates. B. Shown promoter activity with cotranfections of PKC-θ promoter or its mutant (Site-III) with RUNX1 expression plasmid (RUNX1-pCMV6, black bar) or empty vector (pCMV6, open bar) or promoter region alone (stippled bar).
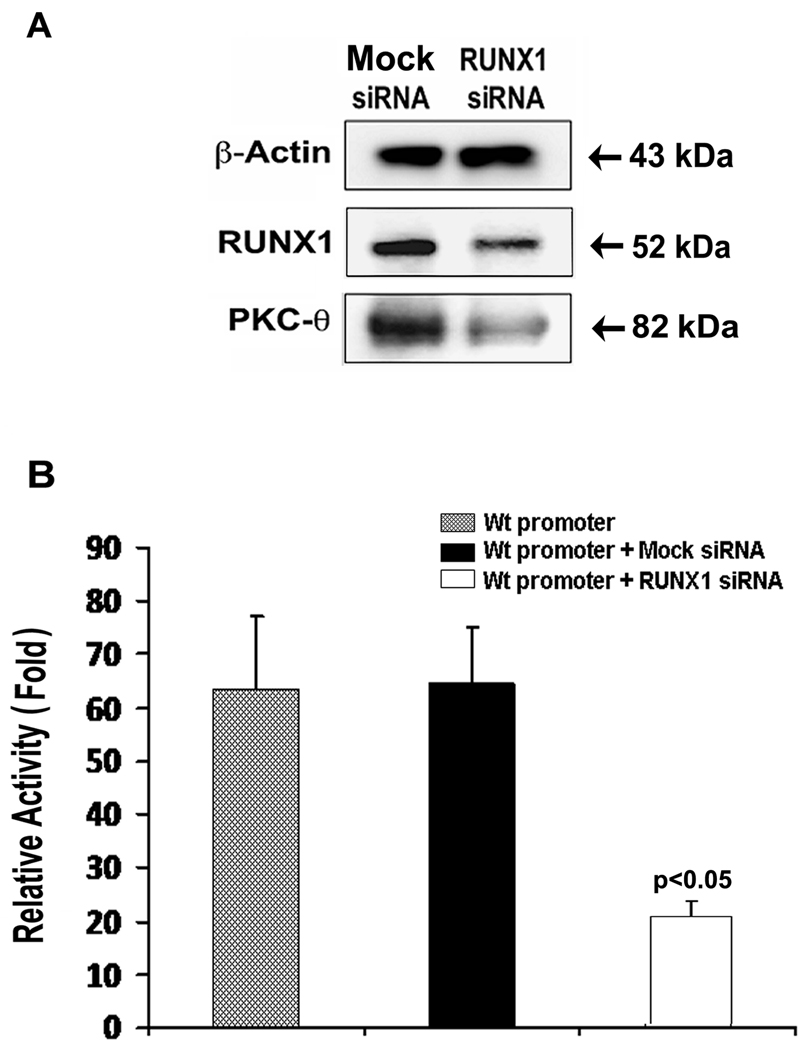
Reduction of PRKCQ promoter activity and PKC-θ protein by RUNX1 siRNA
Immunoblot analysis showed that both RUNX1 and PKC-θ were decreased by RUNX1 siRNA (Figure 5A). PRKCQ promoter (−1085/−206-Luc) activity was markedly reduced with RUNX1 siRNA (Figure 5B).
Figure 5.
Inhibition of PKC-θ protein and promoter by RUNX1 siRNA in HEL cells. A. Immunoblotting of HEL cell lysates showing β-actin, RUNX1 and PKC-θ. RUNX1 siRNA inhibited PKC-θ and RUNX1 protein and promoter activity.
Discussion
The major novel finding in our studies is that transcription factor RUNX1 regulates PRKCQ gene in megakaryocytic cells. In silico analyses of PRKCQ promoter (upto ~2075 bp) revealed five consensus sites for RUNX1 (Figure 1A). ChIP studies showed enrichment of the region (−1225/−1056) containing RUNX1 site-III but not regions with other sites (Figure 1) suggesting in vivo binding of RUNX1 to site-III. EMSA with nuclear extracts and recombinant RUNX1 (Figures 2 and 3) showed RUNX1 binding to site-III. Mutation of RUNX1 site-III abolished promoter activity indicating its functional importance; overexpression of RUNX1 increased RUNX1 and PKC-θ proteins, and promoter activity, which was lost on mutation of site-III (Figure 4). RUNX1 siRNA inhibited PKC-θ protein expression and PRKCQ promoter activity (Figure 5). Together, these findings indicate that PKC-θ is regulated at the transcriptional level by RUNX1 and that site-III is crucial for transcriptional control of PRKCQ in MK cells. The studies presented here are in HEL cells treated with PMA to induce MK differentiation, a widely used model for studies on megakaryocyte biology.34, 35 That these findings are directly relevant to platelet PKC-θ expression is supported by the findings36 in our patient with RUNX1 haplodeficiency that PKC-θ protein and mRNA are decreased in platelets, the primary cells. In addition, we have shown22 that pleckstrin phosphorylation was decreased in the platelets, which provides a strong evidence for a functional consequence of the diminished PKC-θ expression. The present studies provide a cogent explanation for decreased platelet PKC-θ associated with RUNX1 haplodeficiency. Previous studies in U937 cells have shown PKC-β to be a direct RUNX1 target;37 platelet PKC-β was normal in our patient.23
RUNX1 haplodeficiency is associated with familial thrombocytopenia, predisposition to acute leukemia, impaired megakaryopoiesis28 and impaired platelet function upon activation.22, 28, 29 Mice lacking RUNX1 have a complete absence of fetal liver derived hematopoiesis,38 and impaired megakaryocytic maturation;27 Runx1 haplodeficiency is associated with decreased platelet number.39 Multiple lines of evidence link PKC to critical aspects of megakaryocytic differentiation.1, 40 Phorbol esters activate PKC and induce progenitor cells to differentiate along megakaryocyte line and express megakaryocyte/platelet proteins.41–43 In human progenitors, PKC-θ exhibits a lineage-restricted expression being expressed in megakaryocytes and erythroblasts but not granulocytes/monocytes.44 Jacquel et al 40 have proposed that specific PKC isoforms, including PKC-θ, may not be able to induce megakaryocytic differentiation alone, but that more than one PKC isoform may be required for the differentiation process. The specific role of PKC-θ in megakaryopoiesis and in platelet formation needs to be defined. Interestingly, dominant negative inhibition of PKC-θ delays cell cycle progression in vascular endothelial cells.45
Platelet functional abnormalities constitute a hallmark of RUNX1 mutations.22, 28, 29 We postulate that the deficiency of platelet PKC-θ arising secondary to the RUNX1 haplodeficiency contributes to the functional defect shown in our patient’s platelets, including in aggregation, secretion, αIIbβ3 activation and in cytoskeletal reorganization,22, 31 all of which have been noted in studies in murine PKC-θ-deficient platelets.8, 21 Moreover, pleckstrin is a major substrate phosphorylated by PKC in platelets46, 47 and this phosphorylation was impaired in our patient.22 Pleckstrin deficient mice platelets showed marked defect in PKC-mediated exocytosis of dense and α granules, αIIbβ3 activation, actin assembly and aggregation,48 which provide further support for a potential role of PKC-θ in the observed platelet dysfunction in our patient. Although we propose that PKC-θ deficiency contributes to the functional defect in RUNX1 haplodeficiency, the magnitude of PKC-θ role remains to be delineated, particularly because RUNX1 regulates several other genes also recognized to regulate platelet responses. As shown by us, these include ALOX12 (12-lipoxygenase),30 MYL9 (myosin light chain),31 PF4 (platelet factor 4)32 and possibly others.24 Overall, the concept emerging is that RUNX1 haplodeficiency-associated platelet dysfunction and thrombocytopenia arise by the interactions involving multiple genes. Studies in RUNX1 haplodeficiency provide an opportunity to unravel the role in platelets/MK of various RUNX1-regulated genes and proteins, including those that are currently not recognized to have a role in platelets but are downregulated.
In summary, our studies reveal that PRKCQ is regulated by RUNX1 in megakaryocytes/platelets, and provide an explanation for the decreased PKC-θ expression in RUNX1 haplodeficiency. RUNX1 dysregulation of PRKCQ in megakaryocytes is an important aspect of the abnormal platelet production and function associated with human RUNX1 mutations, and an area for further investigation to unravel the mechanisms leading to defects in platelet production and function.
Acknowledgments
The authors gratefully acknowledge the assistance of Denise Tierney in manuscript preparation.
This work was supported by grants from the National Institutes of Health (R01HL85422 and R01HL56724) (AKR). GK was supported by National Institutes of Health T32 training grant HL007777.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
References
- 1.Hong Y, Martin JF, Vainchenker W, Erusalimsky JD. Inhibition of protein kinase C suppresses megakaryocytic differentiation and stimulates erythroid differentiation in HEL cells. Blood. 1996;87:123–131. [PubMed] [Google Scholar]
- 2.Jiang F, Jia Y, Cohen I. Fibronectin- and protein kinase C-mediated activation of ERK/MAPK are essential for proplatelet like formation. Blood. 2002;99:3579–3584. doi: 10.1182/blood.v99.10.3579. [DOI] [PubMed] [Google Scholar]
- 3.Rojnuckarin P, Kaushansky K. Actin reorganization and proplatelet formation in murine megakaryocytes: the role of protein kinase C-α. Blood. 2001;97:154–161. doi: 10.1182/blood.v97.1.154. [DOI] [PubMed] [Google Scholar]
- 4.Gilio K, Harper MT, Cosemans JM, Konopatskaya O, Munnix IC, Prinzen L, Leitges M, Liu Q, Molkentin JD, Heemskerk JW, Poole AW. Functional divergence of platelet protein kinase C (PKC) isoforms in thrombus formation on collagen. J Biol Chem. 2010;285:23410–23419. doi: 10.1074/jbc.M110.136176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harper MT, Poole AW. Diverse functions of protein kinase C isoforms in platelet activation and thrombus formation. J Thromb Haemost. 2009;8:454–462. doi: 10.1111/j.1538-7836.2009.03722.x. [DOI] [PubMed] [Google Scholar]
- 6.Pears CJ, Thornber K, Auger JM, Hughes CE, Grygielska B, Protty MB, Pearce AC, Watson SP. Differential roles of the PKC novel isoforms, PKC-δ and PKC-ε, in mouse and human platelets. PLoS One. 2008;3:e3793. doi: 10.1371/journal.pone.0003793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strehl A, Munnix IC, Kuijpers MJ, van der Meijden PE, Cosemans JM, Feijge MA, Nieswandt B, Heemskerk JW. Dual role of platelet protein kinase C in thrombus formation: stimulation of pro-aggregatory and suppression of procoagulant activity in platelets. J Biol Chem. 2007;282:7046–7055. doi: 10.1074/jbc.M611367200. [DOI] [PubMed] [Google Scholar]
- 8.Nagy B, Jr, Bhavaraju K, Getz T, Bynagari YS, Kim S, Kunapuli SP. Impaired activation of platelets lacking protein kinase C-θ isoform. Blood. 2009;113:2557–2567. doi: 10.1182/blood-2008-07-169268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rink TJ, Smith SW, Tsien RY. Cytoplasmic free Ca2+ in human platelets: Ca2+ thresholds and Ca-independent activation for shape-change and secretion. FEBS Letters. 1982;148:21–26. doi: 10.1016/0014-5793(82)81234-9. [DOI] [PubMed] [Google Scholar]
- 10.Walker TR, Watson SP. Synergy between Ca2+ and protein kinase C is the major factor in determining the level of secretion from human platelets. Biochem J. 1993;289:277–282. doi: 10.1042/bj2890277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshioka A, Shirakawa R, Nishioka H, Tabuchi A, Higashi T, Ozaki H, Yamamoto A, Kita T, Horiuchi H. Identification of protein kinase C-α as an essential, but not sufficient, cytosolic factor for Ca2+-induced α- and dense-core granule secretion in platelets. J Biol Chem. 2001;276:39379–39385. doi: 10.1074/jbc.M102933200. [DOI] [PubMed] [Google Scholar]
- 12.van Willigen G, Akkerman J-W. Protein kinase C and cyclic AMP regulate reversible exposure of binding sites for fibrinogen on the glycoprotein IIb–IIIa complex of human platelets. Biochem J. 1991;273:115. doi: 10.1042/bj2730115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hers I, Donath J, Litjens PE, van Willigen G, Akkerman JW. Inhibition of platelet integrin α(IIb)β(3) by peptides that interfere with protein kinases and the β(3) tail. Arterioscler Thromb Vasc Biol. 2000;20:1651–1660. doi: 10.1161/01.atv.20.6.1651. [DOI] [PubMed] [Google Scholar]
- 14.Buensuceso CS, Obergfell A, Soriani A, Eto K, Kiosses WB, Arias-Salgado EG, Kawakami T, Shattil SJ. Regulation of outside-in signaling in platelets by integrin-associated protein kinase C-β. J Biol Chem. 2005;280:644–653. doi: 10.1074/jbc.M410229200. [DOI] [PubMed] [Google Scholar]
- 15.Soriani A, Moran B, de Virgilio M, Kawakami T, Altman A, Lowell C, Eto K, Shattil SJ. A role for PKC-θ in outside-in α(IIb)β3 signaling. J Thromb Haemost. 2006;4:648–655. doi: 10.1111/j.1538-7836.2006.01806.x. [DOI] [PubMed] [Google Scholar]
- 16.Tabuchi A, Yoshioka A, Higashi T, Shirakawa R, Nishioka H, Kita T, Horiuchi H. Direct demonstration of involvement of protein kinase C-α in the Ca2+-induced platelet aggregation. J Biol Chem. 2003;278:26374–26379. doi: 10.1074/jbc.M212407200. [DOI] [PubMed] [Google Scholar]
- 17.Konopatskaya O, Gilio K, Harper MT, Zhao Y, Cosemans JM, Karim ZA, Whiteheart SW, Molkentin JD, Verkade P, Watson SP, Heemskerk JW, Poole AW. PKC-α regulates platelet granule secretion and thrombus formation in mice. J Clin Invest. 2009;119:399–407. doi: 10.1172/JCI34665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pula G, Schuh K, Nakayama K, Nakayama KI, Walter U, Poole AW. PKC-δ regulates collagen-induced platelet aggregation through inhibition of VASP-mediated filopodia formation. Blood. 2006;108:4035–4044. doi: 10.1182/blood-2006-05-023739. [DOI] [PubMed] [Google Scholar]
- 19.Chari R, Getz T, Nagy B, Jr, Bhavaraju K, Mao Y, Bynagari YS, Murugappan S, Nakayama K, Kunapuli SP. Protein kinase C-δ differentially regulates platelet functional responses. Arterioscler Thromb Vasc Biol. 2009;29:699–705. doi: 10.1161/ATVBAHA.109.184010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall KJ, Harper MT, Gilio K, Cosemans JM, Heemskerk JW, Poole AW. Genetic analysis of the role of protein kinase C-θ in platelet function and thrombus formation. PLoS One. 2008;3:e3277. doi: 10.1371/journal.pone.0003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen S, Braiman A, Shubinsky G, Ohayon A, Altman A, Isakov N. PKC-θ is required for hemostasis and positive regulation of thrombin-induced platelet aggregation and α-granule secretion. Biochem Biophys Res Commun. 2009;385:22–27. doi: 10.1016/j.bbrc.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 22.Gabbeta J, Yang X, Sun L, McLane MA, Niewiarowski S, Rao AK. Abnormal inside-out signal transduction-dependent activation of glycoprotein IIb–IIIa in a patient with impaired pleckstrin phosphorylation. Blood. 1996;87:1368–1376. [PubMed] [Google Scholar]
- 23.Sun L, Mao G, Rao AK. Association of CBFA2 mutation with decreased platelet PKC-θ and impaired receptor-mediated activation of GPIIb–IIIa and pleckstrin phosphorylation: proteins regulated by CBFA2 play a role in GPIIb–IIIa activation. Blood. 2004;103:948–954. doi: 10.1182/blood-2003-07-2299. [DOI] [PubMed] [Google Scholar]
- 24.Sun L, Gorospe JR, Hoffman EP, Rao AK. Decreased platelet expression of myosin regulatory light chain polypeptide (MYL9) and other genes with platelet dysfunction and CBFA2/RUNX1 mutation: insights from platelet expression profiling. J Thromb Haemost. 2007;5:146–154. doi: 10.1111/j.1538-7836.2006.02271.x. [DOI] [PubMed] [Google Scholar]
- 25.de Bruijn MF, Speck NA. Core-binding factors in hematopoiesis and immune function. Oncogene. 2004;23:4238–4248. doi: 10.1038/sj.onc.1207763. [DOI] [PubMed] [Google Scholar]
- 26.Mikhail FM, Sinha KK, Saunthararajah Y, Nucifora G. Normal and transforming functions of RUNX1: a perspective. J Cell Physiol. 2006;207:582–593. doi: 10.1002/jcp.20538. [DOI] [PubMed] [Google Scholar]
- 27.Ichikawa M, Asai T, Saito T, Seo S, Yamazaki I, Yamagata T, Mitani K, Chiba S, Ogawa S, Kurokawa M, Hirai H. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med. 2004;10:299–304. doi: 10.1038/nm997. [DOI] [PubMed] [Google Scholar]
- 28.Song WJ, Sullivan MG, Legare RD, Hutchings S, Tan X, Kufrin D, Ratajczak J, Resende IC, Haworth C, Hock R, Loh M, Felix C, Roy DC, Busque L, Kurnit D, Willman C, Gewirtz AM, Speck NA, Bushweller JH, Li FP, Gardiner K, Poncz M, Maris JM, Gilliland DG. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet. 1999;23:166–175. doi: 10.1038/13793. [DOI] [PubMed] [Google Scholar]
- 29.Michaud J, Wu F, Osato M, Cottles GM, Yanagida M, Asou N, Shigesada K, Ito Y, Benson KF, Raskind WH, Rossier C, Antonarakis SE, Israels S, McNicol A, Weiss H, Horwitz M, Scott HS. In vitro analyses of known and novel RUNX1/AML1 mutations in dominant familial platelet disorder with predisposition to acute myelogenous leukemia: implications for mechanisms of pathogenesis. Blood. 2002;99:1364–1372. doi: 10.1182/blood.v99.4.1364. [DOI] [PubMed] [Google Scholar]
- 30.Kaur G, Jalagadugula G, Mao G, Rao AK. RUNX1/core binding factor A2 regulates platelet 12-lipoxygenase gene (ALOX12): studies in human RUNX1 haplodeficiency. Blood. 2010;115:3128–3135. doi: 10.1182/blood-2009-04-214601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jalagadugula G, Mao G, Kaur G, Goldfinger LE, Dhanasekaran DN, Rao AK. Regulation of platelet myosin light chain (MYL9) by RUNX1: implications for thrombocytopenia and platelet dysfunction in RUNX1 haplodeficiency. Blood. 2010;116:6037–6045. doi: 10.1182/blood-2010-06-289850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aneja K, Jalagadugula G, Mao G, Singh A, Rao AK. Mechanism of platelet factor 4 (PF4) deficiency with RUNX1 haplodeficiency: RUNX1 is a transcriptional regulator of PF4. J Thromb Haemost. 2010 doi: 10.1111/j.1538-7836.2010.04154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cupit LD, Schmidt VA, Gnatenko DV, Bahou WF. Expression of protease activated receptor 3 (PAR3) is upregulated by induction of megakaryocyte phenotype in human erythroleukemia (HEL) cells. Exp Hematol. 2004;32:991–999. doi: 10.1016/j.exphem.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 35.Long MW, Heffner CH, Williams JL, Peters C, Prochownik EV. Regulation of megakaryocyte phenotype in human erythroleukemia cells. J Clin Invest. 1990;85:1072–1084. doi: 10.1172/JCI114538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gabbeta J, Vaidyula VR, Dhanasekaran DN, Rao AK. Human platelet Gαq deficiency is associated with decreased Gαq gene expression in platelets but not neutrophils. Thromb Haemost. 2002;87:129–133. [PubMed] [Google Scholar]
- 37.Hug BA, Ahmed N, Robbins JA, Lazar MA. A chromatin immunoprecipitation screen reveals protein kinase Cbeta as a direct RUNX1 target gene. J Biol Chem. 2004;279:825–830. doi: 10.1074/jbc.M309524200. [DOI] [PubMed] [Google Scholar]
- 38.Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe AH, Speck NA. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci U S A. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun W, Downing JR. Haploinsufficiency of AML1 results in a decrease in the number of LTR-HSCs while simultaneously inducing an increase in more mature progenitors. Blood. 2004;104:3565–3572. doi: 10.1182/blood-2003-12-4349. [DOI] [PubMed] [Google Scholar]
- 40.Jacquel A, Herrant M, Defamie V, Belhacene N, Colosetti P, Marchetti S, Legros L, Deckert M, Mari B, Cassuto JP, Hofman P, Auberger P. A survey of the signaling pathways involved in megakaryocytic differentiation of the human K562 leukemia cell line by molecular and c-DNA array analysis. Oncogene. 2006;25:781–794. doi: 10.1038/sj.onc.1209119. [DOI] [PubMed] [Google Scholar]
- 41.Ballen KK, Ritchie AJ, Murphy C, Handin RI, Ewenstein BM. Expression and activation of protein kinase C isoforms in a human megakaryocytic cell line. Exp Hematol. 1996;24:1501–1508. [PubMed] [Google Scholar]
- 42.Nagata KI, Okano Y, Nozawa Y. Protein kinase C isozymes in human megakaryoblastic leukemia cell line, MEG-01: possible involvement of the isozymes in the differentiation process of MEG-01 cells. Br J Haematol. 1996;93:762–771. doi: 10.1046/j.1365-2141.1996.d01-1714.x. [DOI] [PubMed] [Google Scholar]
- 43.Zauli G, Bassini A, Catani L, Gibellini D, Celeghini C, Borgatti P, Caramelli E, Guidotti L, Capitani S. PMA-induced megakaryocytic differentiation of HEL cells is accompanied by striking modifications of protein kinase C catalytic activity and isoform composition at the nuclear level. Br J Haematol. 1996;92:530–536. doi: 10.1046/j.1365-2141.1996.00384.x. [DOI] [PubMed] [Google Scholar]
- 44.Oshevski S, Le Bousse-Kerdiles MC, Clay D, Levashova Z, Debili N, Vitral N, Jasmin C, Castagna M. Differential expression of protein kinase C isoform transcripts in human hematopoietic progenitors undergoing differentiation. Biochem Biophys Res Commun. 1999;263:603–609. doi: 10.1006/bbrc.1999.1425. [DOI] [PubMed] [Google Scholar]
- 45.Tang S, Morgan KG, Parker C, Ware JA. Requirement for protein kinase C-θ for cell cycle progression and formation of actin stress fibers and filopodia in vascular endothelial cells. J Biol Chem. 1997;272:28704–28711. doi: 10.1074/jbc.272.45.28704. [DOI] [PubMed] [Google Scholar]
- 46.Tyers M, Rachubinski RA, Stewart MI, Varrichio AM, Shorr RG, Haslam RJ, Harley CB. Molecular cloning and expression of the major protein kinase C substrate of platelets. Nature. 1988;333:470–473. doi: 10.1038/333470a0. [DOI] [PubMed] [Google Scholar]
- 47.Haslam RJ, Koide HB, Hemmings BA. Pleckstrin domain homology. Nature. 1993;363:309–310. doi: 10.1038/363309b0. [DOI] [PubMed] [Google Scholar]
- 48.Lian L, Wang Y, Flick M, Choi J, Scott EW, Degen J, Lemmon MA, Abrams CS. Loss of pleckstrin defines a novel pathway for PKC-mediated exocytosis. Blood. 2009;113:3577–3584. doi: 10.1182/blood-2008-09-178913. [DOI] [PMC free article] [PubMed] [Google Scholar]