Abstract
In cell-free extracts of Xenopus eggs that support the assembly of replication-competent nuclei, we found that lamin B3 specifically associates with four polypeptides (termed SLAPs, soluble lamin associated proteins). Here, one SLAP is identified as the nuclear pore complex protein Nup153, one member of the F/GXFG motif-containing nucleoporins. In vitro translated Nup153 and lamin B3 co-immunoprecipitate, and lamin B3 interacts specifically with the C-terminal domain of Nup153. During nuclear envelope assembly, other F/GXFG-containing nucleoporins are incorporated into the nuclear envelope preceding lamina assembly. Incorporation of Nup153 occurs at the same time as lamina assembly. When lamina assembly is prevented using the dominant-negative mutant XlaminBΔ2+, Nup153 does not appear at the nuclear envelope, while other F/GXFG-containing nucleoporins and Nup93 are recruited normally. When the lamina of pre-assembled nuclei is disrupted using the same dominant-negative mutant, the distribution of other nucleoporins is unaffected. However, Nup153 recruitment at the nuclear envelope is lost. Our results indicate that both the recruitment and maintenance of Nup153 at the pore are dependent upon the integrity of the lamina.
Keywords: nuclear envelope/nuclear lamina/Nup153/Xenopus
Introduction
The bi-directional movement of macromolecules across the nuclear envelope (NE) is a multi-step process mediated by nuclear pore complexes (NPCs) (Melchior and Gerace, 1995). Each NPC is cylindrical in the plane of the NE and spans the inner and outer membranes of the NE. Analysis of the basic structure of the NPC, which is embedded in the NE, indicates that it is composed of two ring structures, a spoke ring complex and a central transporter. The first ring structure or cytoplasmic ring lies above the outer nuclear membrane, has a mass of 32 MDa and is decorated with eight 30- to 50-nm-long filaments. The second ring structure or nuclear ring has a mass of 21 MDa and is capped by a basket-like structure, termed the nuclear pore basket. Between the cytoplasmic and nuclear ring structures lies the spoke ring complex consisting of eight spokes, which appear to anchor the NPC to the nuclear membrane. The central transporter is housed within the spoke ring complex and has been modelled as an hourglass-shaped cylinder (Akey and Radermacher, 1993; Goldberg and Allen, 1996; Kiseleva et al., 1998).
It has been suggested that the filaments on the cytoplasmic ring mediate the binding of karyophilic proteins to the nuclear pore. Protein components of the cytoplasmic ring filaments have been identified and include Nup214/CAN, Nup84 (Bastos et al., 1996) and the 358 kDa Ran-binding protein RanBP2 (Wu et al., 1995; Delphin et al., 1997). The nuclear pore basket comprises eight 100-nm-long filaments, joined distally by a 30- to 50-nm-diameter terminal ring. The nuclear pore basket appears to function as an initial docking site for export cargo (Bastos et al., 1996; Kiseleva et al., 1996) and as a terminal docking site for import cargo (Rutherford et al., 1997; Shah et al., 1998). Three protein components of the nuclear pore basket have been identified, namely Nup153, Nup98 and Nup93 (Sukegawa and Blobel, 1993; Radu et al., 1995; Grandi et al., 1997). Nup153 has been reported as being located exclusively at the terminal ring of the nuclear pore basket (Panté and Aebi, 1994), although other groups have reported its presence throughout the basket (Cordes et al., 1993) or suggested that it is a mobile element (Nakielny et al., 1999). Nup98 and Nup93 are located throughout the nuclear pore basket (Radu et al., 1995; Grandi et al., 1997).
Nup153 is one of a family of O-linked N-acetylglucosamine NPC glycoproteins and is also a member of the F/GXFG family of nuclear pore proteins (Radu et al., 1995). Nup153 was originally cloned from a rat liver cDNA library. It has an estimated mass of 152.8 kDa and is distinguished by a novel 4-fold repeat of a Cys2–Cys2-type zinc finger motif. Recombinant Nup153 binds DNA in a zinc-dependent fashion and based upon its location within the nucleus and its DNA binding properties, Nup153 has been implicated in RNA export (Sukegawa and Blobel, 1993). Consistent with this hypothesis, overexpression of Nup153 in BHK cells results in inhibition of RNA transport (Bastos et al., 1996), while injection of anti-Nup153 antibodies into Xenopus oocytes blocks the export of snRNA, mRNA and 5S RNA (Ullman et al., 1999). In addition, Nup153 binds to import cargo, indicating that it is involved directly in nuclear import (Shah et al., 1998). Nup153 has also been reported to shuttle between the nucleoplasmic and cytoplasmic faces of the NPC and this behaviour may be relevant to its function (Nakielny et al., 1999).
The nucleoplasmic ring of the NPCs interacts with nuclear lamina filaments (Goldberg and Allen, 1996). Using cell-free extracts of Xenopus eggs, which support the assembly of a nucleus capable of supporting DNA replication in vitro (Blow and Laskey, 1986; Hutchison et al., 1987), we have investigated the function of the nuclear lamina. Using physical and functional depletion of lamins from Xenopus egg extracts, we and others have demonstrated that nuclear lamina assembly is required for DNA replication (Newport et al., 1990; Meier et al., 1991). Nuclei that lack a lamina are capable of active nuclear transport of replication proteins such as proliferating cell nuclear antigen (PCNA), but fail to accumulate those proteins at replication centres and do not support semi-conservative DNA replication (Jenkins et al., 1993). Ultrastructural examinations of these nuclei have revealed that for the most part the organization of NPCs is normal (Goldberg et al., 1995), which is consistent with our earlier report that these nuclei import karyophilic proteins. However, in the absence of a lamina, a proportion of all nuclear pores are aberrantly assembled in that the nuclear pore basket appears on top of the cytoplasmic ring (Goldberg et al., 1995). In these circumstances, the condition of the nuclear pore basket on the nucleoplasmic ring is unknown. However, if the NPC basket is absent, incorrectly assembled or incomplete, directional transport within nuclei may be impaired. Therefore, a simple hypothesis for the failure of PCNA to be targeted to replication centres in nuclei that lack a lamina is that in all nuclear pores, elements of the nuclear pore basket are assembled incorrectly. As a first test of this hypothesis, we have characterized the association between the major lamin isoform in egg extracts (lamin B3) and NPC proteins.
In this paper, we demonstrate the existence of a novel complex containing lamin B3 and Nup153. We show that Nup153 is incorporated into the NE at the same time as lamina assembly and after assembly of other F/GXFG nucleoporins. Using dominant-negative mutants of lamin B1 (Ellis et al., 1997) we have constructed nuclei that lack a lamina and thus demonstrate that the accumulation of Nup153 at the NE is linked to the establishment of a stable lamina.
Results
Lamin B3 forms a complex with Nup153 in vitro
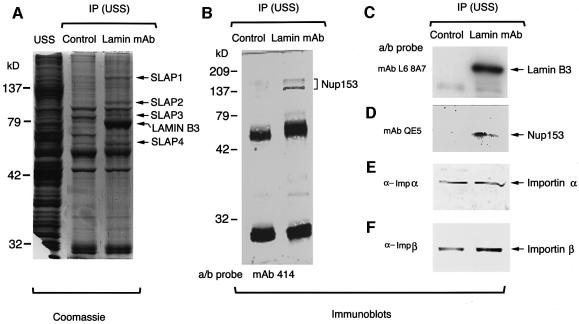
In previous reports we have shown that Xenopus egg extracts that have been depleted of lamin B3 assemble sperm pronuclei that lack a lamina (Jenkins et al., 1993; Goldberg et al., 1995; Zhang et al., 1996). These nuclei accumulate some karyophilic proteins at apparently normal rates (Jenkins et al., 1993) and possess regularly spaced NPCs (Goldberg et al., 1995), suggesting that NE assembly is independent of lamina assembly. However, ultrastructural investigations of the surfaces of these nuclei with field emission in-lens scanning electron microscopy (FEISEM) have revealed that at least a proportion of NPCs (between 5 and 10%) are obviously abnormal. The abnormal NPCs possess a nuclear pore basket at their cytoplasmic face (Goldberg et al., 1995). Since the lamina interacts directly with nuclear pores (Dwyer and Blobel, 1976) and lamina assembly influences nuclear pore assembly (Goldberg et al., 1995), we wished to determine whether or not lamin B3 forms physical associations with nucleoporins. Lamin B3 was immunoprecipitated from a cytosolic fraction of Xenopus eggs (USS) with the monoclonal antibody (mAb) L6 5D5 (Stick, 1988). The immunoprecipitate was resolved by 8% SDS–PAGE, which was stained with Coomassie Blue. As controls, total USS or immunoprecipitates obtained with the anti-PCNA mAb PC10 were resolved in adjacent lanes (Figure 1A). Lamin B3 was readily detected in L6 5D5 immunoprecipitates as a single large band with an Mr of 78 kDa (Figure 1A, right-hand lane). In addition to lamin B3, three other bands were consistently detected by Coomassie staining in L6 5D5 immunoprecipitates, which were absent from control PC10 immunoprecipitates. These bands migrated with Mr values of ∼160, 102 and 90 kDa, respectively, and were termed soluble lamin associated protein (SLAP) 1–3, respectively (Figure 1A, arrows). A fourth band (SLAP4) migrating with an Mr of 56 kDa was also detected in L6 5D5 immunoprecipitates, which was present at greatly reduced levels in PC10 immunoprecipitates.
Fig. 1. Co-immunoprecipitation of Nup153 with lamin B3. Lamin B3 or PCNA was immunoprecipitated from Xenopus cytosol (USS, prepared by centrifugation of LSS at 200 000 g for 4 h) using mAb L6 5D5 or mAb PC10, respectively. Immunoprecipitates were resolved on 8% SDS–PAGE along with USS and either stained with Coomassie Blue (A) or transferred to nitrocellulose and blotted with mAb 414 (B), mAb L6 8A7 (C), mAb QE5 (D), rabbit anti-importin α (E) or rabbit anti-importin β (F). The positions of SLAPs, together with lamin B3, are indicated by arrows in (A).
In order to establish whether any SLAPs were nucleoporins or functionally related molecules, immunoblotting experiments were performed. L6 5D5 (lamin mAb) or PC10 (control) immunoprecipitates were resolved by 8% SDS–PAGE, transferred to nitrocellulose filters and probed with one of the following antibody reagents: mAb 414 (which detects F/GXFG nucleoporins); mAb QE5 (which detects Nup153, p214/CAN and p62); mAb L6 8A7 (which detects Xlamins B1, B2 and B3); mAb PC7 (which detects PCNA); rabbit anti-importin α and rabbit anti-importin β. As expected, the lamin mAb L6 8A7 detected a single band migrating at 78 kDa in the lamin mAb L6 5D5 immunoprecipitates but not control immunoprecipitates (Figure 1C). Similarly, mAb PC7 detected a band migrating at 36 kDa in control immunoprecipitates but not L6 5D5 immunoprecipitates (not shown). mAbs 414 and QE5 both detected a major band migrating with an approximately identical Mr to SLAP1 in L6 5D5 immunoprecipitates but not control immunoprecipitates (Figure 1B and D). Based upon its relative mobility and the known reactivities of mAbs 414 and QE5, we inferred that this band represented Nup153. In immunoblotting assays with mAb 414 as well as with specific peptide antibodies against Nup153 (see below), the major 150 kDa band, together with several slower migrating forms, was typically detected. These probably represent post-translationally modified forms of the protein, as Nup153 is a substrate for a number of protein kinases in vitro (C.Smythe, unpublished observations) and in vivo (Favreau et al., 1996). Neither mAb 414 (Figure 1B) nor mAb QE5 (not shown) detected additional nucleoporins in L6 5D5 immunoprecipitates, although p62, p214/CAN and NUP358 were readily detected by mAb 414 in USS (not shown). Importin α (Figure 1E) and importin β (Figure 1F) were detected in approximately equal quantities in both L6 5D5 and PC10 immunoprecipitates. Several conclusions can be drawn from these experiments. First, the nuclear localization signal (NLS) import mediators importin α and importin β are associated with both lamin B3 and PCNA in USS. Since both importins were present in each immunoprecipitate, we concluded that neither importin was one of the SLAPs (which are not present in PC10 immunoprecipitates). Secondly, Nup153 is specifically complexed with a fraction of lamin B3 in USS. This complex does not contain additional F/GXFG nucleoporins. Thirdly, SLAP1 and Nup153 migrate with almost identical Mr values, suggesting that SLAP1 is Nup153.
To confirm that lamin B3 interacts with Nup153, in vitro translation assays were performed. cDNAs encoding lamin B3, lamin A and Nup153, together with the LAP family member emerin as a control, were individually transcribed and translated in the presence of [35S]methionine. Approximately equimolar amounts of each translation product were mixed in various combinations and subjected to immunoprecipitation with antibodies linked to Dynabeads. Immunoprecipitates were analysed by SDS–PAGE and autoradiography. In each case, labelled species corresponded to the expected Mr, although a significant number of incomplete translation products were observed with Nup153 (Figure 2). When lysates containing Nup153 were immunoprecipitated with the F/GXFG mAb 414, the immunoprecipitates contained an ∼150 kDa band as expected, together with a series of smaller fragments (Figure 2, lanes 1, 2, 3 and 7). mAb 414 immunoprecipitates from lysates containing both Nup153 and lamin B3 showed an additional doublet (Figure 2, lane 1) migrating with a mobility expected of unmodified and prenylated lamin B3 (Vorburger et al., 1989; Firmbach-Kraft and Stick, 1995). mAb 414 did not immunoprecipitate any lamin B3 when it was the only translation product present (not shown). Similarly, lamin A could also be co-immunoprecipitated with Nup153 (Figure 2, lane 2). In contrast, in mAb 414 immunoprecipitates from lysates containing Nup153 and emerin, full-length Nup153 and related fragments were the only labelled species detected (Figure 2, lane 3). To confirm that translated emerin was capable of interaction, lysates containing emerin mixed together with either lamin A, lamin B3 or Nup153 were subjected to immunoprecipitation with the emerin antibody MANEM3. We found that lamins A and B3, but not Nup153, were recovered in emerin immunoprecipitates (Figure 2, lanes 4–6). Taken together, these data indicate that Nup153 forms a complex with lamin B3 and lamin A, but not with emerin.
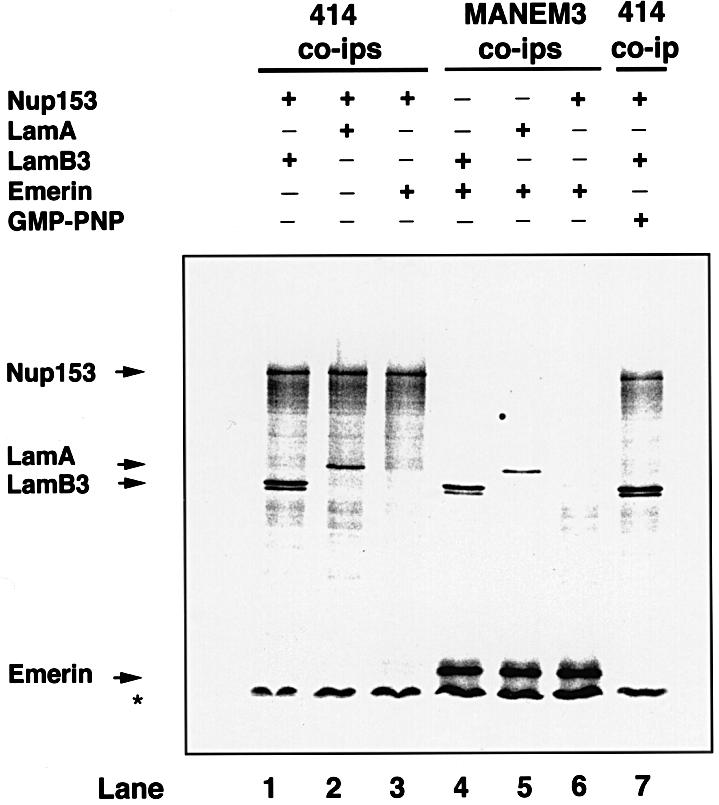
Fig. 2. Co-immunoprecipitation of lamin B3 and Nup153 from rabbit reticulocyte lysates. HA-tagged Nup153, lamin B3, lamin A or emerin was transcribed and translated in rabbit reticulocyte lysates in the presence of [35S]methionine. Translation products were mixed in the absence or presence of GMP-PNP (10 mM) as indicated, immunoprecipitated with F/GXFG mAb 414 or the anti-emerin mAb MANEM3, then resolved on 8% SDS–PAGE and detected by fluorography. The relative mobilities of full-length Nup153, lamin A (LamA), lamin B3 (LamB3) and emerin are indicated with arrows. The asterisk indicates the position of the dye front.
Nup153 associates specifically with import cargo and this association is mediated by importin β (Shah et al., 1998). Since lamin B3 co-immunoprecipitates from egg extracts with importin α and β (see above) and rabbit reticulocyte lysates contain both importins, we wished to exclude the possibility that the association between Nup153 and lamin B3 was as part of an import cargo. Therefore, we performed immunoprecipitation reactions in the presence or absence of guanylyl-imido diphosphate (GMP-PNP), which dissociates import complexes (Shah et al., 1998). Lamin B3 (but not emerin) co-immunoprecipitated with Nup153 in mixed lysates, both in the presence and absence of GMP-PNP, indicating that the association between the two proteins was not mediated by importin β (Figure 2, lanes 1 and 7). As expected (Shah et al., 1998), GMP-PNP dissociated Nup153 from importin β complexes (data not shown).
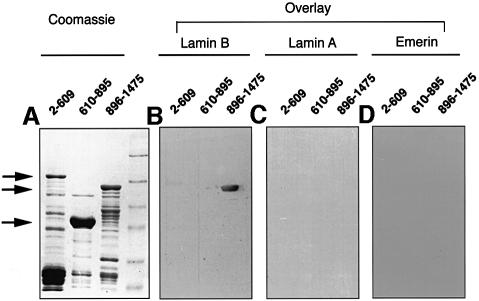
We wished to obtain additional evidence for the interaction between Nup153 and lamin B3. One approach would be to examine the association between recombinant purified Nup153 and lamin B3; however, the expression of both proteins in soluble forms currently presents insurmountable difficulties. In an alternative approach, we performed blot overlay assays. Recombinant Nup153 was expressed in Escherichia coli as a series of glutathione S-transferase (GST) fusion constructs (corresponding to residues 2–609, 610–895 and 896–1475). These peptides were resolved on SDS–PAGE and stained with Coomassie Blue (Figure 3A). Alternatively, they were transferred to nitrocellulose and probed with either [35S]methionine-labelled lamin B3, lamin A or emerin. Lamin B3 associated strongly with the fragment corresponding to the C-terminal F/GXFG-rich domain of Nup153 (residues 896–1475), but not to any other part of the protein (Figure 3B). In contrast, lamin A and emerin did not react with any of the Nup153 fragments (Figure 3C and D). Since lamin A, emerin and lamin B3 all have NLS sequences, these data strongly suggest that the interaction between Nup153 and lamin B3 is not linked to nuclear protein import.
Fig. 3. Interaction of lamin B3 with the C-terminus of Nup153 in blot overlay assays. Nup153 was expressed as three fragments (residues 2–609, 610–895 and 896–1475, respectively) in E.coli and partially purified. The fragments were resolved on SDS–PAGE and either stained with Coomassie Blue (A) or transferred to nitrocellulose and overlayed with [35S]methionine-labelled lamin B3 (B), [35S]methionine-labelled lamin A (C) or [35S]methionine-labelled emerin (D). After extensive washing, the nitrocellulose filters were dried and autoradiographed. Arrows show the positions of the full-length recombinant peptides. Molecular weight markers are 97.5, 66.3, 45, 31, 21.5 and 14.5 kDa, respectively.
Peptide antibodies against a conserved domain of Nup153 decorate the NE in human and Xenopus
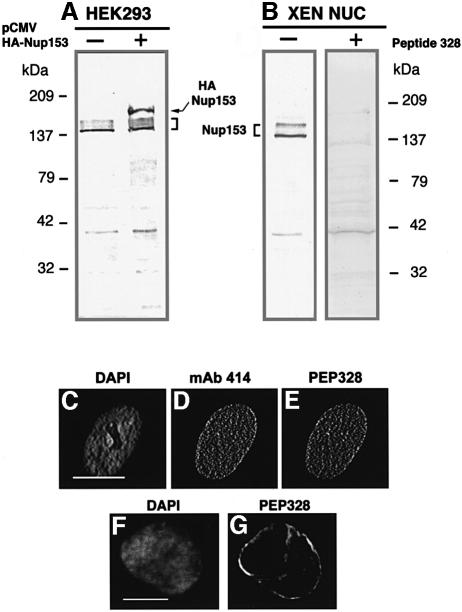
Since lamin B3 and Nup153 associate physically, we wished to investigate whether or not lamina assembly influences the incorporation of Nup153 into the NE. A prerequisite for these investigations was the generation of reliable antibody reagents that specifically detect Xenopus Nup153. The amino acid sequence of Xenopus Nup153 was not available at the time that these investigations were initiated, therefore we compared the amino acid sequences of human and rat Nup153 for regions of extensive homology. A region of homology between residues 1456 and 1475 was identified and a peptide corresponding to this region of human Nup153 (PEP328) was synthesized. The recent publication of the amino acid sequence of Xenopus Nup153 also reveals extensive homology with human Nup153 within the same domain (Shah et al., 1998). A polyclonal sheep antiserum against PEP328 was obtained and affinity purified. The reactivity of anti-PEP328 antibodies was determined in immunoblot assays as follows. Recombinant haemagglutinin (HA)-tagged human Nup153 was expressed in the human embryonic kidney cell line HEK293. Whole-cell extracts of HEK293 expressing Nup153 were prepared and resolved by 8% SDS–PAGE alongside cell extracts from untransfected HEK293 cells. Resolved proteins were transferred to nitrocellulose and blotted with pre-immune serum or anti-PEP328. No bands were detected on immunoblots probed with pre-immune serum (not shown). In contrast, a series of bands was detected at ∼160 kDa (corresponding to the expected position of endogenous Nup153) in transfected and untransfected 293 cells with anti-PEP328 antibodies (Figure 4A). In addition, in transfected 293 cells a second band was detected that migrated with the expected Mr of HA-tagged Nup153 (Figure 4A), which was recognized by an anti-HA mAb (data not shown). A number of faster migrating bands were detected with this antibody, but at greatly reduced intensity. These results suggested that anti-PEP328 antibodies specifically detect recombinant and endogenous Nup153 in immunoblot assays. To determine whether anti-PEP328 antibodies also detect Xenopus Nup153, sperm pronuclei were assembled in vitro, isolated and resolved on SDS–PAGE, and blotted with either pre-immune serum or anti-PEP328 antibodies. Pre-immune serum weakly detected a single band migrating at 42 kDa in sperm pronuclei (not shown). In contrast, anti-PEP328 antibodies reacted strongly with a series of bands migrating at ∼160 kDa (Figure 4B) and weakly with the 42 kDa band. To confirm that the reactivity with the ∼160 kDa bands was specific, the anti-PEP328 antibodies were pre-treated with an excess of peptide followed by immunoblotting on sperm pronuclei. Weak reactivity against a low molecular weight (∼42 kDa) band was detected, but the reaction with the ∼160 kDa bands was eliminated (Figure 4B).
Fig. 4. Anti-PEP328 antibodies react specifically with Nup 153 and detect it at nuclear pores. Extracts of untransfected HEK293 cells (A, –), HEK293 cells transfected with HA-tagged rat Nup 153 (A, +) and isolated Xenopus sperm pro-nuclei (B) were resolved by 8% SDS–PAGE and transferred to nitrocellulose filters. The filters were blotted with either sheep anti-PEP328 (A, all lanes and B, left-hand lane) or sheep anti-PEP328 that had been pre-absorbed with PEP328 (1 mg/ml, B, right-hand lane). (C–E) HDFs were fixed after extraction with CSK-Triton and RSB-MagiK and co-stained with DAPI (C), mAb 414 (D) and anti-PEP328 (E). The distribution of sheep immunoglobulins was revealed with TRITC-conjugated rabbit anti-sheep Ig while the distribution of mouse immunoglobulins was revealed with FITC-conjugated goat anti-mouse Ig. (F and G) Xenopus sperm pronuclei were extracted with CSK-Triton and RSB-MagiK, fixed with EGS and stained with DAPI (F) and sheep anti-PEP328 followed by TRITC-conjugated rabbit anti-sheep Ig (G). In (C) and (F), the distribution of DNA is revealed with DAPI. Scale bars = 10 µm.
Results from immunoblotting indicated that anti-PEP328 specifically detected Nup153. We therefore used the antibody to investigate the cellular distribution of Nup153 by indirect immunofluorescence. Human dermal fibroblasts (HDFs) were fixed and double indirect immunofluorescence microscopy was performed using anti- PEP328 antibodies and an mAb against F/GXFG-containing-nucleoporins (mAb 414). Staining with mAb 414 revealed a punctate distribution over the NE of HDFs (Figure 4C and D). Note that the apparent distribution of NPCs over the surface of nuclei in fibroblast cultures is common, since these cells adopt a highly flattened morphology in which nuclei are <2 µm in depth. An almost identical pattern was detected with anti-PEP328 in these co-stained cells (Figure 4E), indicating that the same structures were stained. In cells that were co-stained with control mAb PC10 and anti-PEP328 antibodies, no specific co-localization was detected (data not shown). To assess the usefulness of the antibodies in detecting Nup153 in Xenopus embryonic nuclei, we assembled nuclei in extracts of Xenopus eggs, then extracted, fixed and stained them with anti-PEP328 antibodies. The antibodies gave rise to a pronounced envelope staining pattern (Figure 4F and G), which was almost identical to that obtained with mAb 414 (see below). This staining pattern was eliminated if the antibody was pre-absorbed with PEP328 peptide, indicating that anti-PEP328 antibodies specifically detect Nup153 in these nuclei (data not shown).
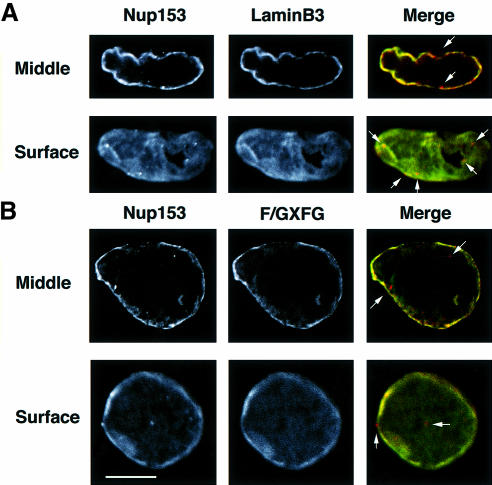
To characterize further the distribution of Nup153 as detected with our antibody reagent, confocal microscopy was performed on fully formed nuclei prepared for double immunofluorescence. The reason for double labelling was that in vitro assembled nuclei are larger and more fragile than somatic cell nuclei, and for immunofluorescence studies are collected by cytospin. This can result in some distortion of the nuclear membrane, often giving rise to the false impression that NE proteins are located in the nucleoplasm. Therefore, optical sectioning with a confocal microscope is essential to describe the localization of NE antigens accurately. Typical confocal micrographs are displayed in Figure 5. Optical sections in each fluorescent channel were collected sequentially, either through the middle or at the surface of a nucleus. In this way it was possible to demonstrate that both lamin B3 (Figure 5A, LaminB3) and the majority of F/GXFG antigens (Figure 5B, F/GXFG) are located exclusively at the NE as expected. Similarly, we found that the majority of Nup153 staining was located at the NE (Figure 5A and B, Nup153). However, while the majority of Nup153 is located at the NE, dot-like staining was also detected in every nucleus examined (a total of 23). The dot-like structures were always located very close to the NE (as revealed clearly in the surface images) but did appear on both sides of the NE (Figure 5A and B, arrows). Note that in some mid sections, some Nup153-positive dots appear to be some distance from the general rim staining of the NE. These are likely to be due to local folds in the NE, which could be detected as diffuse staining of either lamin B3 or F/GXFG epitopes in merged images (Figure 5). Taken together, these data suggest that the majority of Nup153 detected by our antibody is located at the NE.
Fig. 5. Confocal microscope analysis of Nup153, lamin B3 and F/GXFG nucleoporin distribution in sperm pronuclei. Sperm pronuclei were assembled in vitro and collected on coverslips, extracted with CSK-Triton followed by RSB-MagiK and fixed. Fixed nuclei were co-stained with anti-Nup153 antibody (Nup153) and mAb L6 5D5 (LaminB3) in (A) or anti-Nup153 antibody (Nup153) and mAb 414 (F/GXFG) in (B). The distribution of mAbs was revealed with FITC-conjugated goat anti-mouse antibodies. The distribution of anti-Nup153 was revealed with TRITC-conjugated donkey anti-sheep antibodies. Nuclei were imaged on a Zeiss LCM410 equipped with a PlanApochromat ×63/1.40 N/A oil immersion lens. Optical sections were collected sequentially in each fluorescent channel at 0.4 µm intervals. Images were projected either at the middle of a nucleus or at the surface of a nucleus. Micrographs are presented either as single fluorescent (black and white) or merged images. Arrows show the positions of dot-like structures stained with anti-Nup153 antibodies. Scale bar = 20 µm.
The accumulation of Nup153 in the NPCs is concomitant with the appearance of the lamina during Xenopus NE assembly
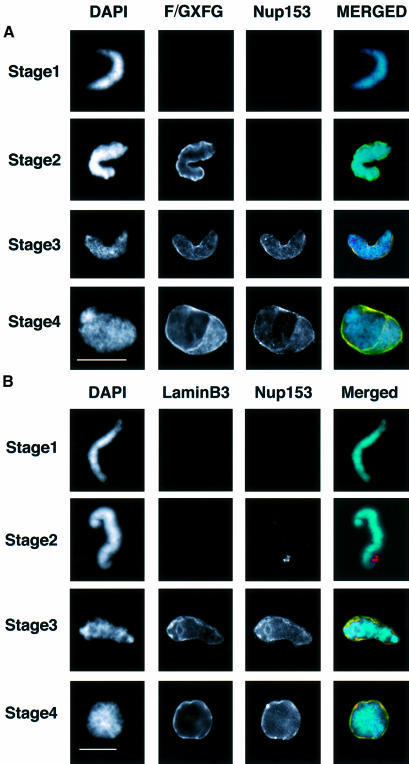
Having obtained an antibody that reliably detects Nup153 in Xenopus, we used this reagent to investigate the timing of Nup153 incorporation into the NE. Sperm pronuclei were assembled in Xenopus egg extracts. At 20 min time intervals, nuclei were collected on glass coverslips, extracted, fixed and co-stained with mAb 414 (F/GXFG) and anti-PEP328 antibodies (anti-Nup153; Figure 6A) or mAb L6 5D5 (anti-lamin B3) and anti-PEP328 antibodies (Figure 6B). In previous publications, it has been reported that sperm pronuclei assembled in vitro undergo discrete morphological and volume changes during the acquisition of a functional NE. This includes initial decondensation of chromatin (stage 1), followed by more extensive decondensation upon the acquisition of first a transport system (stage 2) and finally a lamina (stages 3 and 4) (e.g. Hutchison et al., 1988; Wiese et al., 1997). Typically, this series of changes occurs over a period of 60 min in vitro. As expected, stage 1 nuclei were not stained by any of the antibodies used in the study. Stage 2 nuclei were stained brightly with mAb 414, but were not stained with either anti-lamin B3 or anti-Nup153 antibodies. Stage 3 and 4 nuclei were stained readily by all three antibodies. The staining patterns of mAb 414 and anti-lamin B3 compared with anti-Nup153 were almost identical. The apparent absence of Nup153 staining compared with other F/GXFG-containing proteins at early stages might conceivably be due to masking of Nup153 epitopes during NE assembly. We therefore isolated nuclei at early and late stages during assembly and performed western blotting with mAb 414 and anti-nup153 antibodies. This showed that at early time points, the predominant F/GXFG nucleoporin detected in isolated reforming nuclei was the most abundant F/GXFG p62 (Figure 6C). In keeping with the results obtained by immunofluorescence microscopy, Nup153 was undetectable at early stages of reassembly, and was only detected at later stages. Taken together, these data indicated that Nup153 recruitment occurs at about the same time as the incorporation of lamin B3 into the lamina.
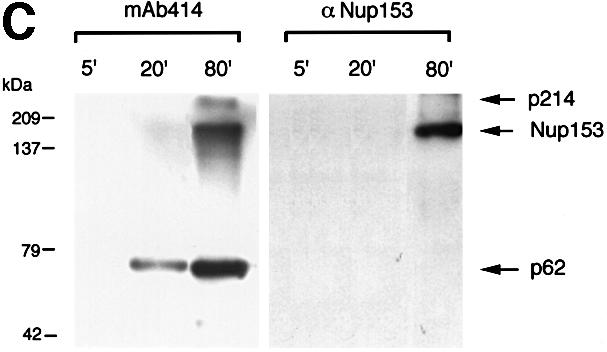
Fig. 6. Investigation of the timing of accumulation of nucleoporins, lamins and Nup 153 at the NE in assembling sperm pronuclei. Sperm pronuclei were assembled in egg extracts. (A and B) At 20 min intervals throughout the incubation, nuclei were collected on coverslips, extracted with CSK-Triton followed by RSB-MagiK and fixed. Fixed nuclei were co-stained with mAb 414 (F/GXFG) and anti-Nup153 antibody (Nup153, A) or mAb L6 5D5 (lamin B3) and anti-Nup153 antibody (Nup153, B). The distribution of mAbs was revealed with FITC-conjugated goat anti-mouse antibodies. The distribution of anti-Nup153 was revealed with TRITC-conjugated donkey anti-sheep antibodies. The distribution of chromatin was detected with DAPI. Images were collected with a CCD camera mounted on a Zeiss Axiovert 10 epifluorescence microscope. To determine the relative distributions of chromatin, F/GXFG nucleoporins, lamin B3 and Nup153, images from all three fluorescent channels were merged. Stages 1–4 illustrate morphologies typically associated with NE assembly. Scale bars = 20 µm. (C) At the times indicated, in vitro assembled nuclei were isolated as described in Materials and methods and proteins from 7.5 × 103 nuclei were subjected to electrophoresis on a 10% SDS–PAGE minigel, transferred to nitrocellulose using conditions to maximize efficiency of transfer of both high and low molecular weight proteins. Filters were blotted either with mAb 414 (left-hand panel) or anti-Nup153 antibody (right-hand panel). Under these conditions, Nup153 appears as a single band.
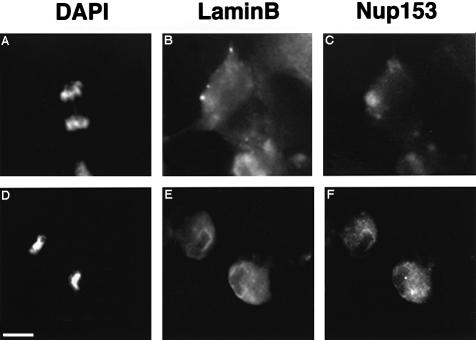
We extended our observations concerning the recruitment of Nup153 and lamin B3 to the NE in the Xenopus cultured cell line XLK-2. Cells were fixed using 4% paraformaldehyde and 0.1% Triton X-100 and analysed by double indirect immunofluorescence microscopy. We found that in anaphase cells, both B-type lamins and Nup153 showed diffuse cytoplasmic staining, indicating the relative absence of these proteins at the nuclear periphery at this stage (Figure 7A–C). Perichromatin labelling of both Nup153 and lamin B3 was first observed in early G1 phase cells (Figure 7D–F). However, considerable cytoplasmic staining remained at this stage, consistent with the view that recruitment of Nup153 and B-type lamins to the NE is a late event in these cells.
Fig. 7. The recruitment of Nup153 and lamin B3 during NE reassembly in Xenopus XLK-2 cells. Cells were fixed and co-stained with DAPI (A and D), mAb L6 5D5 (lamin B3, B and E) and anti-Nup153 antibody (Nup153, C and F) followed by secondary antibodies as described in Figure 5. Fluorescent images of cells in anaphase (A–C) and early G1 phase (D–F) were collected as described in Figure 5. Scale bar = 5 µm.
The association of Nup153 with the NE is dependent upon lamina assembly
Since the incorporation of Nup153 into the NE occurs at the same time as lamina assembly and Nup153 associates with lamin B3 in vitro, we wished to investigate whether or not Nup153 localization is dependent upon lamina function. For this investigation we used a dominant-negative mutant of lamin B1 (termed Xlamin B1 Δ2+) to modulate lamina assembly. We chose to employ this mutant protein for two reasons. First, physical depletion of lamin B3 with antibodies may lead to co-depletion of relevant populations of Nup153 and in any case never results in complete depletion of lamin B3 (Jenkins et al., 1993). Secondly, the mutant protein is effective in both preventing lamina assembly completely and in disassembling the lamina in pre-formed nuclei (Ellis et al., 1997).
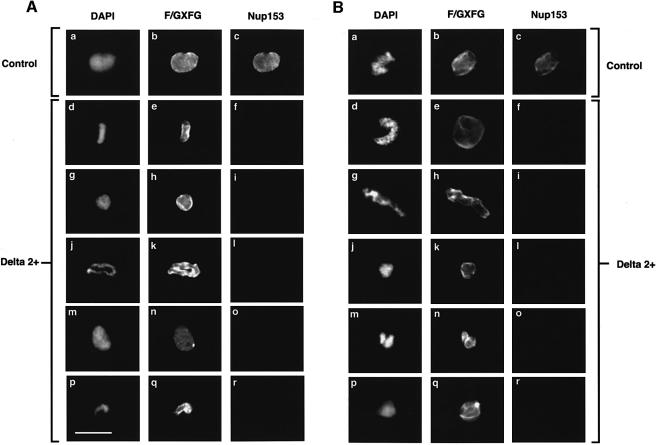
Sperm pronuclei were assembled in extracts supplemented with Xlamin B1 Δ2+ or control buffer, then fixed, extracted and either stained with mAb L6 5D5 or co-stained with mAb 414 and anti-PEP328. Nuclei assembled in control extract displayed typical envelope staining with lamin mAb L6 5D5 (not shown) and were also stained extensively with mAb 414 and anti-Nup153 (Figure 8A, a–c and Figures 4, 5 and 6). As expected (Ellis et al., 1997), nuclei assembled in the presence of Xlamin B1 Δ2+ did not stain with the mAb L6 5D5 (not shown), but in most cases did display strong envelope staining with mAb 414, confirming that NPC formation is independent of lamina assembly (Figure 8A, b, e, h, k, n and q). However, the same nuclei were not stained with anti-Nup153 antibodies (Figure 8A, c, f, i, l, o and r). These results indicate that incorporation of Nup153 but not other F/GXFG nucleoporins into the NE is prevented if lamina assembly is inhibited.
Fig. 8. The incorporation of Nup153 into the NE in nuclei treated with the dominant-negative lamin mutant Xlamin B1 Δ2+. (A) Sperm pronuclei were assembled in egg extract for 90 min in either the presence (Delta 2+) or absence (Control) of Xlamin B1 Δ2+. The nuclei were then collected on glass coverslips, extracted and fixed as described for Figure 5. The nuclei were stained with mAb 414 (F/GXFG, panels b, e, h, k, n, q) and anti-Nup153 (Nup153, panels c, f, i, l, o, r). The distribution of mAb 414 was revealed with FITC-conjugated goat anti-mouse Ig while the distribution of anti-Nup153 was revealed with TRITC-conjugated donkey anti-sheep Ig. The distribution of chromatin in each image is revealed with DAPI (panels a, d, g, j, m, p). (B) Alternatively, sperm pronuclei were assembled in egg extracts over 90 min before Xlamin B1 Δ2+ (Delta 2+) or control buffer (Control) was added to the incubation. Ninety minutes after the addition of the mutant protein, nuclei were processed for immunofluorescence as described in (A). The nuclei were stained with mAb 414 (F/GXFG, panels b, e, h, k, n, q), anti-Nup153 (Nup153, panels c, f, i, l, o, r) and DAPI (panels a, d, g, j, m, p). Scale bar = 20 µm.
Next we investigated whether or not a stable lamina is required to maintain the association of Nup153 with the NE. To do this, nuclei were allowed to assemble in egg extract for 90 min prior to any additions. Xlamin B1 Δ2+ or an equivalent volume of buffer was then added to the extracts and incubated for a further 90 min. The nuclei were then extracted, fixed and either stained with mAb L6 5D5 or co-stained with mAb 414 plus anti-PEP328. Again, nuclei incubated in extracts supplemented with buffer alone (Figure 8B, a–c) displayed identical staining patterns to the controls shown in Figure 8A, a–c. As expected (Ellis et al., 1997), nuclei incubated in extracts supplemented with Xlamin B1 Δ2+ were not stained with L6 5D5, indicating complete disassembly of the lamina (not shown). Staining of the envelope with mAb 414 was unchanged. In contrast, staining with anti-PEP328 was either significantly reduced or absent (Figure 8B, f, i, l, o and r). These data suggest that the stable association of Nup153 with the NE is dependent upon lamina integrity.
Taken together, the results in Figure 8A and B indicate that the establishment and maintenance of Nup153 at the NE is dependent on the presence of an intact lamina. That disruption of lamina integrity resulted in the loss of a protein, believed to reside on the pore basket, raised the possibility that the stability of the entire pore basket was dependent on an intact lamina. In this scenario, loss of Nup153 from the pore following lamina disruption might simply be the indirect consequence of a general loss in pore basket stability in lamin-deficient nuclei. In order to test this, we investigated whether the recruitment of another nuclear pore basket protein, Nup93, was affected by the loss of lamina function induced by Xlamin B1 Δ2+ by co-staining nuclei prepared for the experiment described above in Figure 8A and B with mAb 414 and anti-Nup93 antibodies. We found that unlike Nup153, Nup93 recruitment was unaffected either by the prevention of lamina formation (Figure 9A) or by disassembling the lamina in pre-formed nuclei (Figure 9B). These results indicate that the loss of Nup153 from the nuclear pore in nuclei lacking a lamina is not simply due to loss of nuclear pore basket stability.
Fig. 9. The incorporation of Nup93 into the NE occurs independently of the lamina. (A) Sperm pronuclei were assembled in egg extract for 90 min in either the presence (Delta 2+) or absence (Control) of Xlamin B1 Δ2+. The nuclei were then collected on glass coverslips, extracted and fixed as described for Figure 5. The nuclei were stained with mAb 414 (F/GXFG, panels b, e, h) and anti-Nup93 (Nup93, panels c, f, i,). The distribution of mAb 414 was revealed with FITC-conjugated goat anti-mouse Ig while the distribution of anti-Nup93 was revealed with TRITC-conjugated goat anti-rabbit Ig. The distribution of chromatin in each image is revealed with DAPI (panels a, d, g). (B) Alternatively, sperm pronuclei were assembled in egg extracts over 90 min before Xlamin B1 Δ2+ (Delta 2+) or control buffer (Control) was added to the incubation. Ninety minutes after the addition of the mutant protein, nuclei were processed for immunofluorescence as described in (A). The nuclei were stained with mAb 414 (F/GXFG, panels b, e, h), anti-Nup93 (Nup93, panels c, f, i) and DAPI (panels a, d, g). Scale bar = 20 µm.
Discussion
In this paper we report that lamin B3 and Nup153 co-immunoprecipitate from Xenopus egg extracts. In addition to Nup153, lamin B3 is also complexed to at least three other proteins termed SLAP2, SLAP3 and SLAP4. A complex between lamin B3 and HA-tagged Nup153 is also formed following linked transcription and translation of these proteins in rabbit reticulocyte lysates. Finally, in blot overlay assays, lamin B3 but not lamin A or emerin associates with the C-terminal (F/GXFG-rich) domain of Nup153. The complex between lamin B3 and Nup153 appears to indicate a functional dependency specifically between these two proteins since the stable association of Nup153, but not Nup93, and the NE is dependent upon the presence of a lamina. Our data indicate either that stable lamina assembly is required for the recruitment of Nup153 to the pore or, alternatively, that the soluble lamin B3 pool acts to regulate the appearance of Nup153 at this site.
B-type lamins form a novel type of interaction with Nup153
Nup153 is known to form a stable complex with at least three types of transport molecule, including NLS/importin αβ cargo (Shah et al., 1998), a RanGTP modulated shuttling complex with transportin 1 (Nakielny et al., 1999) and mRNA in nuclear export complexes (Bastos et al., 1996). Since lamin B3 contains an NLS sequence, it was possible that the Nup153–lamin B3 association was the consequence of both molecules participating in import complexes mediated by importins. Several lines of evidence presented here suggest that Nup153–lamin B3 is in fact a novel type of complex. First, in rabbit reticulocyte lysates, Nup153 and lamin B3 remain associated in the presence of GMP-PNP, conditions that dissociate importin complexes (Shah et al., 1988 and data not shown). Secondly, emerin, which also contains an NLS, does not associate with Nup153. Finally, PCNA co-immunoprecipitates with importins α and β from egg extracts but does not associate with Nup153. In contrast to lamin B3, which interacted specifically with the C-terminal fragment of Nup153 in blot overlay assays, lamin A and emerin did not interact in this assay with any fragment. This suggests that the observed association between lamin A and Nup153 in reticulocyte lysates is functionally distinct from that of lamin B3, and may reflect the presence of lamin A in import complexes.
Immunoprecipitation of lamin B3 from egg extracts leads to co-precipitation of at least four proteins (termed SLAPs). On the basis of its size and apparent cross-reactivity with known antibodies in an SDS–PAGE analysis, we conclude that SLAP1 is Nup153. It is clear that lamin B3 immunoprecipitates contain Nup153; however, the possibility exists that SLAP1 is another protein that co-migrates with Nup153. The identity of SLAPs 2–4 is not known at present, therefore we cannot formally rule out the possibility that one or more of these proteins mediates associations between lamin B3 and Nup153. Although lamin B3 and Nup153 also interact when translated in rabbit reticulocyte lysates, the possibility remains that the lysates contain homologues of SLAPs 2–4 that mediate the interaction. The results of solid phase overlay assays do suggest a direct interaction between lamin B3 and Nup153. In contrast to transportin, which associates with Nup153 via an N-terminal region (Nakielny et al., 1999), lamin B3, like importin β (Shah et al., 1998), is complexed via the C-terminal F/GXFG-rich domain of Nup153. Taken together, our data indicate that in nuclear assembly-competent egg extracts, Nup153 and lamin B3 form a molecular complex that is distinct from the import/export cargo complexes described previously (Shah et al., 1998; Nakielny et al., 1999). This raises the question of whether Nup153 interacts with lamin B3 within the context of the lamina. In one ultrastructural study, Nup153 was only found in the terminal ring of the NPC basket. In this position, Nup153 could not interact directly with lamin filaments (Panté and Aebi, 1993). However, Nup153 has also been localized throughout the legs of the pore basket (Cordes et al., 1993) and recently it has been proposed that Nup153 acts as a mobile element, shuttling through the pore (Nakielny et al., 1999). In those circumstances, Nup153 might interact either stably or transiently with lamin filaments. However, as the lamina is believed to exist as a dense meshwork of filaments extending across the nucleoplasmic surface of the envelope, while Nup153 resides predominantly at nuclear pores, only a subset of lamin polypeptides might be expected to interact with Nup153. Future studies will be directed toward understanding the nature and basis of the interaction between Nup153 and lamin B3.
Disruption of lamina assembly abolishes Nup153 recruitment in Xenopus
We used a dominant-negative mutant of Xenopus lamin B1 to interfere with the normal assembly of the nuclear lamina. We found that in this circumstance, Nup153, unlike other nucleoporins, failed to locate at the NE. One interpretation of these data would be that lamin B3–Nup153 interactions are necessary for the stability of Nup153 at the pore. Introduction of the mutant blocks lamina assembly and destabilizes pre-existing lamina, which would in turn result in the loss of NE-associated Nup153. An alternative view is that the lamin–Nup153 interaction reflects a stable, soluble, pre-assembly complex and that incorporation of lamin B3 subunits into the lamina dictates or controls the rate of release of Nup153 to a lower affinity site at the nuclear pore. In this scenario, the introduction of mutant Xlamin B1 results in the stabilization of unassembled, soluble lamin forms (Ellis et al., 1997), which would retain a high affinity for Nup153, blocking its recruitment to the pore. The results obtained here demonstrate that lamin B3 interacts with the C-terminal fragment of Nup153. Previous work (Enarson et al., 1998) has shown that targeting of Nup153 to the pore involves sequences in the N-terminal region of Nup153. Thus, lamin B3 cannot simply prevent Nup153 targeting to the nuclear pore by direct competition for binding. We have attempted to examine the effects of the Xlamin B Δ2+ protein in somatic cell nuclei; however, it is clear that B-type lamins are not eliminated from the lamina following expression of this mutant in somatic cells (A.Vaughan, M.Alvarez-Reyes, J.M.Bridger, J.L.V.Broers, F.C.S.Raemakers, M.Wenhert, G.E.Morris, W.G.F.Whitfield and C.J.Hutchison, submitted). Clearly, distinguishing between these and other models will be the subject of future work and will necessarily involve the identification of SLAPs 2–4.
It is clear that in Xenopus egg extracts the reassembly of the NE occurs as a series of steps, which involves the co-ordinated recruitment of distinct membrane precursors (Drummond et al., 1999) as well as soluble components to generate a double membrane perforated by pores and supported by the lamina. FEISEM analysis of Xenopus NE assembly has revealed a range of morphologically distinct NPC intermediates. These have been suggested to reflect a sequential assembly process, which is initiated by the appearance of depressions or holes in the envelope membrane and onto which different elements of NPC structure are added in a defined order (Goldberg et al., 1997). It is clear that lamina assembly is a relatively late event in this sequence. Lamina assembly is dependent upon active nuclear protein import (Newport et al., 1990) and NPC formation still occurs in nuclei that lack a lamina (Jenkins et al., 1993; Goldberg et al., 1995). The data presented here indicate that in Xenopus the association of the bulk of Nup153 with the NE is also a relatively late event in extracts and in XLK-2 cells, occurring concomitantly with, and dependent upon, lamina assembly.
Bodoor et al. (1999) have reported that during NE reassembly in NRK cells, recruitment of Nup153 to the chromatin periphery occurs much earlier in anaphase and may be independent of membrane recruitment. The basis for the difference is currently unknown and may simply reflect cell type differences. For example, Haraguchi et al. (2000) have reported on the timing of lamin B receptor (LBR), emerin and Nup153 recruitment in living HeLa cells. They found that emerin and LBR recruitment occurs concomitantly with the recruitment of the bulk of Nup153, although they report that some Nup153 may be detected at earlier times. It is conceivable that, in the studies reported here, the early recruitment of low levels of Nup153 is below the level of detection of our reagents. It is also possible that the relative affinities of Nup153 and membrane components for chromatin may be different in different cell types. Thus, in Xenopus, the rapid recruitment of membrane to the chromatin periphery may preclude an initial, early appearance of Nup153, and possibly other nucleoporins, at that site. This early establishment of a double membrane might therefore force one of several possible pathways of NPC assembly. One prediction of this model would be that, in circumstances where the dynamics of membrane recruitment were reduced, an alternative pathway of pore assembly might be initiated. The appearance of pore-like structures on chromatin in the absence of any NE membrane has indeed been observed under precisely such conditions, using a reconstituted Xenopus cell-free system in which the NE membrane precursor pool was deliberately reduced (Sheehan et al., 1988).
The nuclear pore basket is part of a complex of filamentous structures that emanates from the nucleoplasmic face of the NPCs. Extending further into the nucleoplasm are additional filamentous structures containing the protein Tpr (Cordes et al., 1997; Shah et al., 1998). In a recent report, a novel protein component of filaments emanating from the nucleoplasmic face of NPCs was described. This protein, termed PBC68, has been described as a component of the nuclear pore basket. Significantly, the association of PBC68 with nuclear pores after mitosis occurs only after lamin-associated polypeptide-2β (LAP2β) and lamin B are incorporated into the NE (Theodoropoulos et al., 1999). These data, together with the evidence presented here, suggest that the assembly of structures at the nucleoplasmic face of the NPC may well be dependent upon prior assembly of the lamina. It will be of interest to establish whether any of the Xenopus SLAPs (Mr 102, 90 and 56 kDa) are homologues of PBC68.
Why might a lamina be required for the stable association of Nup153 with the NE?
It has recently been reported that the N-terminus of Nup153 contains an M9-type NLS (Nakielny et al., 1999), which interacts with transportin in a RanGDP-dependent fashion (Nakielny et al., 1999). Furthermore, it has been suggested that Nup153 may be a mobile component of the NPC, shuttling between the nucleoplasmic and cytoplasmic faces of the NPC (Nakielny et al., 1999). Previous studies on in vitro assembled nuclei clearly demonstrated that nucleoporins in different parts of the NPC could be resolved using confocal microscopy (Shah et al., 1998). In the present investigation, we consistently detected dot-like structures containing Nup153 lying very close to both faces of the NE, which did not co-localize with the majority of F/GXFG nucleoporins. These structures may reflect a subset of pores in which Nup153 is actively shuttling and/or a subset that may be detected by this antibody. As the absence of a lamina abolishes the appearance of Nup153 but not other, presumed, stable nucleoporins at the NE, it is conceivable that the lamina may have a function in controlling Nup153 mobility.
Conclusions
We have described a novel complex between the F/GXFG nucleoporin Nup153 and the type V intermediate filament protein lamin B3. This complex appears to have functional significance in terms of NPC assembly and may influence the way in which NPCs function in import and export.
Materials and methods
Antibodies
Anti-Nup153 antisera were generated in sheep as follows: the primary sequences of rat and human Nup153 were compared and a region of precise homology corresponding to residues 1456–1475 in the human sequence was selected. Ten milligrams of peptide corresponding to the sequence were synthesized, purified and coded PEP328. Two milligrams of the peptide were conjugated separately to bovine serum albumin (BSA) and limpet haemocyanin with glutaraldehyde. These conjugates were used as immunogens in sheep. Sheep serum was collected prior to immunization and then 10 days after each inoculation. After the third inoculation, the animal was killed. Antibody against the peptide was isolated from serum by affinity chromatography using purified peptide conjugated to CNBr-activated Sepharose 4B as described (Harlow and Lane, 1988).
mAb 414 is a monoclonal antibody that reacts with F/GXFG nucleoporins (Aris and Blobel, 1989). mAb QE5 is a monoclonal antibody that reacts with p62, Nup 153 and p214/CAN (Panté et al., 1994) and was generously provided by Dr Brian Burke, University of Calgary, Canada. mAb L6 8A7 is a monoclonal antibody that reacts with Xenopus lamins B1, B2 and B3 (Stick and Hausen, 1985), while mAb L6 5D5 reacts specifically with Xenopus lamin B3 (Stick, 1988). Both reagents were generously provided by Dr Reimer Stick, University of Göttingen, Germany. mAb PC10 is a monoclonal antibody against PCNA (Waseem and Lane, 1990) and was a generous gift from Professor D.P.Lane, University of Dundee, UK. PEP45 and PEP65 are rabbit polyclonal antibodies against importins α and β, respectively, and were generously provided by Dr Dirk Görlich, University of Heidelberg, Germany. MANEM3 antibodies against human emerin were a generous gift from Professor Glenn Morris, NE Wales Institute, Wrexham, UK. Rabbit anti-Nup93 antibodies were generously provided by Dr Douglass Forbes (University of California, San Diego, CA).
Preparation of egg extracts
Xenopus egg extracts (LSS) were prepared according to the method described by Hutchison (1994). The extracts were snap frozen and stored directly in liquid nitrogen. Egg cytosol (termed USS) was prepared from LSS by high speed ultracentrifugation at 200 000 g for 4 h in a Beckman TLS 55 rotor (Drummond et al., 1999).
Immunoprecipitation assays
mAb L6 5D5 or mAb PC10 was conjugated to paramagnetic Dynabeads as described previously (Goldberg et al., 1995). Samples (2 ml) of egg cytosol were incubated with 900 µl of either L6 5D5– or PC10–Dynabeads for 1 h at room temperature. The Dynabeads were recovered from the cytosol and washed twice with a 10× volume of 50 mM Tris–HCl pH 8.0, 5 mM dithiothreitol (DTT), then twice with a 10× volume of 0.5 M KCl, 50 mM Tris–HCl pH 8.0, 5 mM DTT and once with a 10× volume of 1 M urea, 50 mM Tris–HCl pH 8.0, 5 mM DTT. After each washing step the Dynabeads were recovered using a magnetic particle separator. The final pellet was suspended in SDS sample buffer, boiled for 3 min and stored at –20°C. For Coomassie gels, the entire immunoprecipitate was resolved on a single lane of each gel. For immunoblotting experiments, the equivalent of 100 µl of extract was resolved in each lane of a gel.
Preparation of Xenopus sperm pronuclei for indirect immunofluorescence microscopy
Extracts recovered from liquid N2 were thawed, supplemented with an energy regeneration system (Hutchison, 1994) and incubated with demembranated sperm at a concentration of 2 × 105/100 µl of extract. Incubations were performed at 21°C. For immunofluorescence studies, samples were first isolated on glass coverslips. The samples were then extracted sequentially with CSK buffer [10 mM PIPES–NaOH pH 6.8, 10 mM KCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride (PMSF) and 10 µg/ml aprotinin] containing 0.5% Triton X-100, followed by RSB-Magik (42.5 mM Tris–HCl pH 7.3, 8.5 mM NaCl, 2.6 mM MgCl2, 0.05 mM PMSF and 10 µg/ml aprotinin) containing 1% Tween-20 and 0.5% sodium deoxycholate (both v/v) (Zhang et al., 1996). The nuclei were then fixed with ethylene glycol bis succinimidyl succinate (EGS) prior to immunostaining (Hutchison, 1994).
Preparation of Xenopus sperm pronuclei for immunoblotting
In vitro formed nuclei were isolated as described (Hutchison, 1994). Briefly, extracts recovered from liquid N2 were thawed, supplemented with an energy regeneration system (Hutchison, 1994) and incubated with demembranated sperm at a concentration of 2 × 105/100 µl of extract. Incubations were performed at 21°C. At the indicated times, 60 µl of extract were diluted in 1 ml of nuclear isolation buffer, NIB (Hutchison, 1994), underlayered with NIB containing 60% Percoll and centrifuged for 10 min at 3000 g. Material at the interface was removed into 1 ml of NIB, underlayered with 200 µl of NIB containing 30% sucrose and centrifuged at 4000 g for 10 min. The pellet was resuspended in 10 µl of NIB and frozen at –80°C until required.
In vitro translation assays
For in vitro transcription and translation, the following cDNA were used: Xenopus lamin B3 cloned into pET3 (Stick, 1988), human lamin A cloned into pET7 (Moir et al., 1991), human emerin cloned into pET29-b (Ellis et al., 1998) and human Nup153 cloned into pET7 (Bastos et al., 1996). In vitro translations were performed with a T7 quick coupled transcription–translation system as described (Jones and Smythe, 1996). Quick master mix (40 µl) was supplemented with 2 µCi of [35S]methionine (specific activity 1 mCi/µl; Amersham) and 1 µg of DNA and nuclease-free H2O to a final volume of 50 µl. The reactions were performed at 30°C for 90 min. Freezing in liquid nitrogen terminated the reactions. The lysates were then used either directly for SDS–PAGE or for immunoprecipitation. For immunoprecipitation assays, 30 µl of lysate were incubated with the equivalent of 20 µl of mAb 414 coupled to 30 µl of goat anti-mouse–Dynabeads or 30 µl of mAb MANEM3 supernatant coupled to 30 µl of Dynabeads and incubated for 1 h at room temperature with end-over-end rotation. The Dynabeads were collected and washed three times with phosphate-buffered saline (PBS), then once with PBS containing 0.3 M KCl and 0.1% NP-40, and prepared for SDS–PAGE and autoradiography. For immunoprecipitation reactions in the presence of GMP-PNP, lysates were supplemented with the nucleotide analogue (Calbiochem) to a final concentration of 10 mM and incubated for 10 min at 37°C prior to addition of the antibody. In addition, the Dynabead–antibody complex was also supplemented with 10 mM GMP-PNP. During washing steps, GMP-PNP was present in PBS and PBS containing 0.3 M KCl to a final concentration of 2 mM.
SDS–PAGE and immunoblotting
Analytical 8% SDS–polyacrylamide gels were cast in a Bio-Rad gel apparatus. Alternatively, 10% gels were purchased from Novex. Each lane was loaded with the equivalent of 10 µl of Dynabeads or 2 µl of egg cytosol. The gels were resolved at 30 mA (200 mV) for 2 h with cooling. Resolved gels were stained for 1 h at room temperature with Coomassie Blue and destained overnight. Alternatively, the equivalent of 10 µl of Dynabeads or the equivalent of 2 × 105 whole-cell extracts or isolated nuclei was resolved on 8% gels, transferred to nitrocellulose and immunoblotted according to published protocols (Jenkins et al., 1993). In Figure 6C, transfer of protein to nitrocellulose was carried out for 8 h and 25 V using a Bio-Rad miniblotter, in order to achieve efficient transfer of both low and high molecular weight proteins. The following dilutions were used for primary antibodies: mAb 414 was diluted 1:1000 in PBS containing 1% newborn calf serum (PBS–NCS); mAb QE5 was diluted 1:500 in PBS–NCS; mAb L6 8A7 was diluted 1:500 in PBS–NCS; PEP45 and PEP65 were both diluted 1:2000 in PBS–NCS; PEP328 and PEP337 were diluted 1:10 in PBS–NCS. Secondary antibodies were alkaline phosphatase-conjugated goat anti-mouse Ig, goat anti-rabbit Ig or rabbit anti-sheep Ig, all purchased from Bio-Rad and diluted according to the manufacturer’s recommendation.
Blot overlay assays
GST–Nup153 fusion constructs encoding residues 2–609 (GST– Nup153-N), 610–895 (GST–Nup153-M) and 896–1475 (GST–Nup153-C) were generous gifts from Sarah Nakielny and Gideon Dreyfuss, Howard Hughes Medical Institute, University of Pennsylvania School of Medicine, PA. Recombinant GST–Nup153 fragments were expressed in BL21 (DE3) cells and purified as described (Nakielny et al., 1999). As reported in this paper, extensive degradation of GST–Nup153-N occurred despite the inclusion of protease inhibitors in all buffers. Five micrograms of each protein were loaded in sequential lanes of 8% SDS–PAGE gels and resolved. The resolved proteins were transferred to nitrocellulose membrane. The membrane was then washed briefly in Blot Rinse Buffer (10 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1% Tween-20; BRB) and then blocked by incubation for 1 h at room temperature in BLOTTO (BRB containing 5% dried skimmed milk). The filters were rinsed in BRB and then incubated in 5 ml of BRB containing 1% NCS, supplemented with either 1.5 µCi of either [35S]methionine-labelled lamin A, [35S]methionine-labelled lamin B3 or [35S]methionine-labelled emerin (final dilutions >1:100) and incubated overnight at 4°C. This protocol promotes the dissociation of low affinity complexes in solution and allows the detection of pseudo-first order interactions between soluble proteins and immobilized ligands. [35S]Met proteins were prepared by coupled transcription–translation according to Jones and Smythe (1996). The filters were washed five times in BRB, dried, and exposed to Fuji XRX autoradiograph film for 8, 18 and 24 h at –70°C.
Addition of dominant-negative mutants of Xlamin B1 to extracts
XlaminB1Δ2+ was used essentially as described (Ellis et al., 1997). Briefly, the fused protein was expressed, solubilized and purified in 4 M urea. Prior to use, the protein was dialysed into renaturation buffer (500 mM NaCl, 25 mM Tris–HCl pH 8.0, 1 mM DTT) for 2 h at room temperature. The dialysed protein was added to extracts at a final concentration of 80 µg/ml. To prevent lamina assembly, the fusion protein was pre-incubated with the extract for 30 min at room temperature prior to the addition of demembranated sperm. To disrupt a pre-formed lamina, demembranated sperm heads were incubated in egg extracts for 90 min prior to the addition of the fusion protein. The incubation was then continued for a further 90 min at 21°C to achieve complete lamina disassembly. The extracts were processed for indirect immunofluorescence as described above.
Preparation of human dermal fibroblasts for indirect immunofluorescence microscopy
HDFs were grown on coverslips as described previously (Kill et al., 1991). Prior to fixation, the cells were extracted by a modification of the procedure described by Dyer et al. (1997). Briefly, cells were washed in CSK buffer and then extracted with CSK containing 0.5% Triton X-100 for 10 min at 4°C. The cells were then rinsed three times in RSB before further extraction with RSB containing 1% Tween-20 and 0.5% sodium deoxycholate (both v/v) for 10 min at 4°C. The cells were then fixed in methanol:acetone (pre-chilled to –20°C) for 10 min at 4°C and processed for indirect immunofluorescence microscopy.
Transfection of HEK293 cells with HA-tagged Nup153
cDNA encoding HA-tagged Nup153 was generously provided by Dr Brian Burke. HA-tagged Nup153 was expressed in HEK293 cells according to the method described by Alessi et al. (1996). Briefly, 293 cells were cultured at 37°C in 10-cm-diameter dishes in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum. Twenty-four hours after subculture, the cells were transfected with 1 µg/ml DNA per plate using calcium phosphate precipitation. Aliquots of 10 µg of plasmid DNA in 0.45 ml of sterile water were added to 50 µl of sterile 2.5 M CaCl2 and then to 0.5 ml of sterile buffer composed of 50 mM N,N-bis[2-hydroxyethyl]-2-aminoethanesulfonic acid–HCl pH 6.96, 0.28 M NaCl and 1.5 mM Na2HPO4. The resulting mixture was vortexed for 1 min, allowed to stand at room temperature for 20 min and then added drop-wise to the cultures. The cells were placed in an atmosphere of 2.5% CO2 for 16 h at 37°C, then the medium was aspirated and replaced with fresh medium. The cells were incubated for 36 h at 37°C in an atmosphere of 5% CO2 before being prepared for SDS–PAGE.
Indirect immunofluorescence microscopy
Cells fixed in methanol:acetone or sperm pronuclei fixed with EGS were processed for indirect immunofluorescence microscopy as follows. Primary antibody (20 µl), diluted in PBS–NCS, was pipetted over each coverslip. Working dilutions for primary antibodies were as follows: anti-PEP328, 1/10 in PBS–NCS; mAb 414, 1/1000 in PBS–NCS; mAb L6 5D5, 1/500 in PBS–NCS. The coverslips were incubated for 1 h at room temperature or overnight at 4°C in a humidified chamber. The coverslips were washed in five changes of PBS, drained and replaced in the humidified chamber. Twenty microlitres of secondary antibody [fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse and/or tetramethylrhodamine isothiocyanate (TRITC)-conjugated rabbit anti-sheep], diluted in PBS–NCS according to the manufacturer’s instructions, were pipetted onto each coverslip and incubated for a further 1 h at room temperature. The coverslips were washed as described above, drained and placed face down in 5 µl of MOWIOL containing 1 µg/ml DAPI on glass microscope slides. Samples prepared for indirect immunofluorescence were viewed on a Zeiss Axiovert 10 equipped for epifluorescence and fitted with ×40/1.3 N/A PlanNeofluor and ×63/1.4 N/A PlanApochromat lenses. Images were captured with a Digital Pixel CCD camera using IPLab software. To standardize image acquisition, exposure times were as follows: DAPI, 50–150 ms; FITC, 1000 ms; TRITC, 1500 ms. Alternatively, images in each fluorescent channel were collected sequentially using a Zeiss LCM400 laser scanning confocal microscope.
Acknowledgments
Acknowledgements
The authors wish to thank Dr Brian Burke (University of Calgary) for the generous gift of mAb QE5 and rat Nup153 cDNA, Dr Reimer Stick (University of Göttingen) for the generous gift of mAb L6 5D5 and lamin B3 cDNA, Professor Glenn Morris (NE Wales Insititute of Technology) for the gift of emerin cDNA and MANEM3, Professor Gideon Dreyfuss and Dr Sarah Nakielny (Howard Hughes Medical Institute, University of Pennsylvania Medical School) for GST–Nup153 fusion constructs, Professor David Lane (University of Dundee) for the generous gift of mAb PC10 and Dr Douglass Forbes (UCSD) for anti-Nup93 antibodies. We are also grateful to Dr Carol Lyon (University of Dundee) for help with confocal microscopy and to Dr Clare Hall-Jackson (University of Dundee) for help with HEK293 cell transfections and critical reading of the manuscript. This work was funded by a grant from the Cancer Research Campaign, a Wellcome Trust Research Leave Fellowship to C.J.H. and Medical Research Council Unit support to C.S.
References
- Akey C.W. and Radermacher,M. (1993) Architecture of the Xenopus nuclear pore complex revealed by three-dimensional cryo-electron microscopy. J. Cell Biol., 122, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alessi D.R., Andjelkovic,M., Caudwell,B., Cron,P., Morrice,N., Cohen,P. and Hemmings,B.A. (1996) Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J., 15, 6541–6551. [PMC free article] [PubMed] [Google Scholar]
- Aris J.P. and Blobel,G. (1989) Yeast nuclear envelope proteins cross react with an antibody against mammalian pore complex proteins. J. Cell Biol., 108, 2059–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos R., Lin,A., Enarson,M. and Burke,B. (1996) Targeting and function in mRNA export of nuclear pore complex protein Nup153. J. Cell Biol., 134, 1141–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow J.J. and Laskey,R.A. (1986) Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell, 47, 577–587. [DOI] [PubMed] [Google Scholar]
- Bodoor K., Shaikh,S., Salina,D., Raharjo,W.H., Bastos,R., Lohka,M. and Burke,B. (1999) Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. J. Cell Sci., 112, 2253–2264. [DOI] [PubMed] [Google Scholar]
- Cordes V.C., Reidenbach,S., Kohler,A., Stuurman,N., van Driel,R. and Franke,W.W. (1993) Intranuclear filaments containing a nuclear pore complex protein. J. Cell Biol., 123, 1333–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes V.C., Reidenbach,S., Racjwitz,H.R. and Franke,W.W. (1997) Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J. Cell Biol., 136, 515–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delphin C., Guan,T., Melchior,F. and Gerace,L. (1997) RanGTP targets p97 to RanBP2, a filamentous protein localized at the cytoplasmic periphery of the nuclear pore complex. Mol. Biol. Cell, 8, 2379–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond S., Ferrigno,P., Lyon,C., Murphy,J., Goldberg,M., Allen,T., Smythe,C. and Hutchison,C.J. (1999) Temporal differences in the appearance of NEP-B78 and an LBR-like protein during Xenopus nuclear envelope reassembly reflects the ordered recruitment of functionally discrete vesicle types. J. Cell Biol., 144, 225–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer N. and Blobel,G. (1976) A modified procedure for the isolation of a pore complex-lamina fraction from rat liver nuclei. J. Cell Biol., 70, 581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer J.A., Kill,I.R., Pugh,G., Quinlan,R., Lane,E.B. and Hutchison,C.J. (1997) Cell-cycle changes in A-type lamin associations detected with monoclonal antibodies in human dermal fibroblasts. Chromosome Res., 5, 383–394. [DOI] [PubMed] [Google Scholar]
- Ellis D., Jenkins,H., Whitfield,W. and Hutchison,C.J. (1997) GST–lamin fusion proteins act as dominant negative mutants in Xenopus egg extract and reveal the function of the lamina in DNA replication. J. Cell Sci., 110, 2507–2518. [DOI] [PubMed] [Google Scholar]
- Ellis J.A., Craxton,M., Yates,J.R.W. and Kendrick-Jones,J. (1998) Aberrant intranuclear targeting and cell cycle-dependent phosphorylation of emerin contribute to the Emery–Dreifuss muscular dystrophy phenotype. J. Cell Sci., 111, 781–792. [DOI] [PubMed] [Google Scholar]
- Enarson P., Enarson,M., Bastos,R. and Burke,B. (1998) Amino-terminal sequences that direct nucleoporin nup153 to the inner surface of the nuclear envelope. Chromosoma, 107, 228–236. [DOI] [PubMed] [Google Scholar]
- Favreau C., Worman,H.J., Wozniak,R.W., Frappier,T. and Courvalin,J.C. (1996) Cell cycle-dependent phosphorylation of nucleoporins and nuclear pore membrane protein Gp210. Biochemistry, 35, 8035–8044. [DOI] [PubMed] [Google Scholar]
- Firmbach-Kraft I. and Stick,R. (1995) Analysis of nuclear lamin isoprenylation in Xenopus oocytes: isoprenylation of lamin B3 precedes its uptake into the nucleus. J. Cell Biol., 129, 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg M.W. and Allen,T.D. (1996) The nuclear pore complex and lamina: three-dimensional structures and interactions determined by field emission in-lens scanning electron microscopy. J. Mol. Biol., 257, 848–865. [DOI] [PubMed] [Google Scholar]
- Goldberg M., Jenkins,H., Allen,T., Whitfield,W.G. and Hutchison,C.J. (1995) Xenopus lamin B3 has a direct role in the assembly of a replication competent nucleus: evidence from cell-free egg extracts. J. Cell Sci., 108, 3451–3461. [DOI] [PubMed] [Google Scholar]
- Goldberg M.W., Wiese,C., Allen,T.D. and Wilson,K.L. (1997) Dimples, pores, star-rings and thin rings on growing nuclear envelopes: evidence for structural intermediates in nuclear pore complex assembly. J. Cell Sci., 110, 409–420. [DOI] [PubMed] [Google Scholar]
- Grandi P., Dang,T., Panté,N., Shevchenko,A., Mann,M., Forbes,D. and Hurt,E. (1997) Nup93, a vertebrate homolog of yeast Nic96, forms a complex with a novel 205 kD protein and is required for correct nuclear pore assembly. Mol. Biol. Cell, 8, 2017–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi T. et al. (2000) Live fluorescence imaging reveals early recruitment of emerin, LBR, RanBP2 and Nup153 to reforming nuclear envelopes J. Cell Sci., 113, 779–794. [DOI] [PubMed] [Google Scholar]
- Harlow E. and Lane,D. (1988) Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Hutchison C.J. (1994) The use of cell-free extracts of Xenopus eggs for studying DNA replication in vitro. In Fantes,P. and Brookes,R. (eds), The Cell Cycle: A Practical Approach. IRL Press at Oxford University Press, Oxford, UK, pp. 177–195. [Google Scholar]
- Hutchison C.J., Cox,R., Drepaul,R.S., Gomperts,M. and Ford,C.C. (1987) Periodic DNA synthesis in cell-free extracts of Xenopus eggs. EMBO J., 6, 2003–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison C.J., Cox,R. and Ford,C.C. (1988) The control of DNA replication in a cell-free extract that recapitulates a basic cell cycle in vitro. Development, 103, 553–566. [DOI] [PubMed] [Google Scholar]
- Jenkins H., Holman,T., Lyon,C., Lane,B., Stick,R. and Hutchison,C.J. (1993) Nuclei that lack a lamina accumulate karyophilic proteins and assemble a nuclear matrix. J. Cell Sci., 106, 275–285. [DOI] [PubMed] [Google Scholar]
- Jones C. and Smythe,C. (1996) Activation of the Xenopus degradation machinery by full length cyclin A. J. Cell Sci., 109, 1071–1079. [DOI] [PubMed] [Google Scholar]
- Kill I.R., Bridger,J.M., Campbell,K.H., Moldonado-Codina,G. and Hutchison,C.J. (1991) The timing of the formation and usage of replicase clusters in S-phase nuclei of human diploid fibroblasts. J. Cell Sci., 100, 869–876. [DOI] [PubMed] [Google Scholar]
- Kiseleva E., Goldberg,M.W., Daneholt,B. and Allen,T.D. (1996) RNP export is mediated by structural reorganization of the nuclear pore basket. J. Mol. Biol., 260, 304–311. [DOI] [PubMed] [Google Scholar]
- Kiseleva E., Goldberg,M.W., Allen,T.D. and Akey,C.W. (1998) Active nuclear pore complexes in Chironomus: visualization of transporter configurations related to mRNP export. J. Cell Sci., 111, 223–236. [DOI] [PubMed] [Google Scholar]
- Meier J., Campbell,K.H., Ford,C.C., Stick,R. and Hutchison,C.J. (1991) The role of lamin LIII in nuclear assembly and DNA replication, in cell-free extracts of Xenopus eggs. J. Cell Sci., 98, 271–279. [DOI] [PubMed] [Google Scholar]
- Melchior F. and Gerace,L. (1995) Mechanisms of nuclear protein import. Curr. Opin. Cell Biol., 7, 310–318. [DOI] [PubMed] [Google Scholar]
- Moir R., Donaldson,A. and Stewart,M. (1991) Expression in E.coli of human lamins A and C: influence of head and tail domains on assembly properties and paracrystal formation. J. Cell Sci., 99, 363–372. [DOI] [PubMed] [Google Scholar]
- Nakielny S., Shaikh,S., Burke,B. and Dreyfuss,G. (1999) Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J., 18, 1982–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J.W., Wilson,K.L. and Dunphy,W.G. (1990) A lamin-independent pathway for nuclear envelope assembly. J. Cell Biol., 111, 2247–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panté N. and Aebi,U. (1993) The nuclear pore complex. J. Cell Biol., 122, 977–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panté N. and Aebi,U. (1994) Toward the molecular details of the nuclear pore complex. J. Struct. Biol., 113, 179–189. [DOI] [PubMed] [Google Scholar]
- Panté N. and Aebi,U. (1996) Sequential binding of import ligands to distinct nucleopore regions during their nuclear import. Science, 273, 1729–1732. [DOI] [PubMed] [Google Scholar]
- Panté N., Bastos,R., McMorrow,I., Burke,B. and Aebi,U. (1994) Interactions and three-dimensional localization of a group of nuclear pore complex proteins. J. Cell Biol., 126, 603–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu A., Blobel,G. and Moore,M.S. (1995) Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc. Natl Acad. Sci. USA, 92, 1769–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford S.A., Goldberg,M.W. and Allen,T.D. (1997) Three-dimensional visualization of the route of protein import: the role of nuclear pore complex substructures. Exp. Cell Res., 232, 146–160. [DOI] [PubMed] [Google Scholar]
- Shah S., Tugendreich,S. and Forbes,D. (1998) Major binding sites for the nuclear import receptor are the internal nucleoporin Nup153 and the adjacent nuclear filament protein Tpr. J. Cell Biol., 141, 31–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan M.A., Mills,A.D., Sleeman,A.M., Laskey,R.A. and Blow,J.J. (1988) Steps in the assembly of replication-competent nuclei in a cell-free system from Xenopus eggs. J. Cell Biol., 106, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stick R. (1988) cDNA cloning of the developmentally regulated lamin LIII of Xenopus laevis. EMBO J., 7, 3189–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stick R. and Hausen,P. (1985) Changes in the nuclear lamina composition during early development of Xenopus laevis. Cell, 41, 191–200. [DOI] [PubMed] [Google Scholar]
- Sukegawa J. and Blobel,G. (1993) A nuclear pore complex protein that contains zinc finger motifs, binds DNA and faces the nucleoplasm. Cell, 72, 29–38. [DOI] [PubMed] [Google Scholar]
- Theodoropoulos P.A., Polioudaki,H., Koulentaki,M., Kouroumalis,E. and Georgatos,S.D. (1999) PBC68: a nuclear pore complex protein that associates reversibly with the mitotic spindle. J. Cell Sci., 112, 3049–3060. [DOI] [PubMed] [Google Scholar]
- Ullman K.S., Shah,S., Powers,M.A. and Forbes,D. (1999) The nucleoporin nup153 plays a critical role in multiple types of nuclear export. Mol. Biol. Cell, 10, 649–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorburger K., Kitten,G.T. and Nigg,E.A. (1989) Modification of nuclear lamin proteins by a mevalonic acid derivative occurs in reticulocyte lysates and requires the cysteine residue of the C-terminal CXXM motif. EMBO J., 8, 4007–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waseem N.H. and Lane,D.P. (1990) Monoclonal antibody analysis of the proliferating cell nuclear antigen (PCNA). Structural conservation and the detection of a nucleolar form. J. Cell Sci., 96, 121–129. [DOI] [PubMed] [Google Scholar]
- Wiese C., Goldberg,M.W., Allen,T.D. and Wilson,K.L. (1997) Nuclear envelope assembly in Xenopus extracts visualized by scanning EM reveals a transport-dependent ‘envelope smoothing’ event. J. Cell Sci., 110, 1489–1502. [DOI] [PubMed] [Google Scholar]
- Wu J., Matunis,M.J., Kraemer,D., Blobel,G. and Coutavas,E. (1995) Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain and a leucine-rich region. J. Biol. Chem., 270, 14209–14213. [DOI] [PubMed] [Google Scholar]
- Yang L., Guan,T. and Gerace,L. (1997) Lamin-binding fragments of LAP2 inhibits increase in nuclear volume during the cell cycle and progression into S phase. J. Cell Biol., 139, 1077–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Jenkins,H., Goldberg,M.W., Allen,T.D. and Hutchison,C.J. (1996) Nuclear lamina and nuclear matrix organization in sperm pronuclei assembled in Xenopus egg extract. J. Cell Sci., 109, 2275–2286. [DOI] [PubMed] [Google Scholar]