Abstract
Hepatic hydroxylation is an essential step in the metabolism and excretion of bile acids and is necessary to avoid pathologic conditions such as cholestasis and liver damage. In this report, we demonstrate that the human xenobiotic receptor SXR (steroid and xenobiotic receptor) and its rodent homolog PXR (pregnane X receptor) serve as functional bile acid receptors in both cultured cells and animals. In particular, the secondary bile acid derivative lithocholic acid (LCA) is highly hepatotoxic and, as we show here, a metabolic substrate for CYP3A hydroxylation. By using combinations of knockout and transgenic animals, we show that activation of SXR/PXR is necessary and sufficient to both induce CYP3A enzymes and confer resistance to toxicity by LCA, as well as other xenotoxicants such as tribromoethanol and zoxazolamine. Therefore, we establish SXR and PXR as bile acid receptors and a role for the xenobiotic response in the detoxification of bile acids.
In addition to a myriad of hormones, animals confront numerous toxic or potentially toxic endogenous and foreign chemicals (xenobiotics) whose efficient detoxification is essential to the survival of all organisms. The cytochrome P450 (CYP) enzymes, which often catalyze the initial step (e.g., hydroxylation) in such detoxification pathways, are crucial for the detoxification of most xenobiotics, including prescription drugs (1). CYP3A enzymes are of particular significance from a medical perspective because they are involved in the metabolism of a large number of clinically used drugs (2). The concept of General Adaptation Syndrome (GAS; ref. 3), also known as the Adaptive Hepatic Response, clearly implicated the liver as the primary organ responsible for such detoxification as a consequence of the induction of hepatic CYP enzymes in response to xenobiotic inducers (1). The human steroid and xenobiotic receptor (SXR) and its rodent homolog pregnane X receptor (PXR) were isolated as candidate xeno-sensors postulated to regulate CYP3A genes by a feedback mechanism (4–9). Recently, we established unequivocally that SXR and PXR function as xeno-sensors in vivo by demonstrating that targeted disruption of the mouse PXR gene abolishes the xenobiotic response of CYP3A genes. In contrast, expression of an activated SXR transgene results in constitutive up-regulation of CYP3A gene expression and enhanced protection against xenotoxicants (10).
Bile acids are the major products of cholesterol catabolism in the liver. Growing evidence suggests that bile acids, in addition to their physiological roles in the formation of bile and solublizing biliary lipids and promoting their absorption, can regulate the expression of a number of cellular proteins. For example, when bound to bile acids including lithocholic acid (LCA), farnesoid X receptor (FXR; ref. 11) represses transcription of cholesterol 7α-hydroxylase (CYP7A), the rate-limiting enzyme of bile acid synthesis, thereby repressing the conversion of cholesterol to bile acids (12–14). This repression has recently been shown to be mediated by liver receptor homolog-1 (LRH-1) and small heterodimer partner (SHP) (15–17).
Despite their beneficial function in cells, excessive bile acids are potentially toxic when accumulated in the body. For example, the secondary bile acid LCA is a potent cholestatic agent and can cause histologic liver damage and other pathological changes unless it is efficiently eliminated (18, 19). As an average human releases 600 ml of bile a day, the potential for disrupting bile flow (cholestasis) and the resultant accumulation of toxic by-products is significant. Despite such significance and previous effort (e.g., ref. 18), the mechanisms for elimination and detoxification of bile acids remain poorly understood and a role for the xenobiotic response in this process is unknown. The antibiotic rifampicin (RIF), a ligand for SXR and a prototypic CYP3A inducer, has been anecdotally shown to increase urinary output of hydroxylated bile acids in humans (20). This observation, together with the hydroxylation capacity of the CYP3A enzymes, suggests that the xenobiotic response might contribute to the elimination of bile acids. However, it is not known whether bile acids serve as SXR/PXR ligands to induce CYP3A, and, if so, whether such activation could contribute to bile acid homeostasis.
In this report, we identified bile acids as functional ligands for SXR and PXR to induce CYP3A gene. We also showed that a sustained induction of CYP3A is sufficient for the hydroxylation and detoxification of cholestatic bile acids, and thus, establishes the significance of xenobiotic response in the elimination of bile acids.
Materials and Methods
Plasmid Constructs and Transient Transfection.
The reporter plasmids tk-3A4-Luc, tk-USA-Luc (4), and tk-EcRE-Luc (11), and the expression vectors for SXR, PXR, Gal-SXR/LBD (4), and FXR (11) have been described. CV-1 cell transfections using 48-well-plate and DOTAP (N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate) transfection reagent (Boehringer) were carried out as described (4). When necessary, cell were treated with bile acids [chenodeoxycholic acid (CDCA), deoxycholic acid (DCA), and LCA; 100 μM each], RIF, and pregnane-16α-carbonitrile (PCN) (10 μM each). All compounds were purchased from Sigma.
Animals, Drug Treatment, and Histologic Evaluation.
The generation of Alb-VPSXR transgenic and PXR-null mice was described (10) and maintained ad libitum. Mice were given daily treatments of LCA (8 mg/day) or vehicle via gavage, and killed 24 h after the last treatment. For PCN protection, mice were first given a single i.p. injection of PCN (40 mg/kg) before 4 days of treatment with both LCA (8 mg/day) and PCN (13 mg/kg) via gavage. The tribromoethanol anesthesia tests were performed as described (10). For histology evaluation, tissues were fixed in 4% formaldehyde, embedded in paraffin, sectioned at 5 μm, and stained for hematoxylin and eosin.
Northern Blot Analysis.
Total RNA was prepared from tissues by using the TRIZOL Reagent (GIBCO/BRL). Northern hybridization was carried out as described (10). The quantitation was performed with National Institutes of Health image software.
LCA Hydroxylation and Antibody Inhibition Assay.
Recombinant CYP3A4 and CYP3A5 were cloned and expressed by using a vaccinia virus expression system. Human liver samples were obtained through the Organ Procurement Program, University of Rochester (Rochester, NY) and University of Groningen (Groningen, The Netherlands). Preparation of microsomal and cell membrane fractions and bile acid hydroxylase assays were performed as described (21). Bile acid hydroxylase assays were carried out with 50 μM [carboxyl-14C]lithocholic acid (59 mCi/mmol; 1 Ci = 37 GBq) in 50 mM Hepes–NaOH, 0.055 mM EDTA, 2 mM NADPH, 0.625 mg microsomal protein/ml, 75 mM NaCl, 10 mM potassium phosphate, pH 7.4 in a final volume of 80 μl for 10 min at 37°C; products were separated by TLC as described previously (22). For antibody inhibition assay, polyclonal anti-CYP3A IgG was incubated at various concentrations with complete human liver microsomal reaction mixtures minus NADPH, for 40 min at room temperature. Reaction mixtures were warmed to 37°C and LCA hydroxylation was initiated by the addition of NADPH (22).
Results
Bile Acids Are SXR/PXR Activators and CYP3A Inducers.
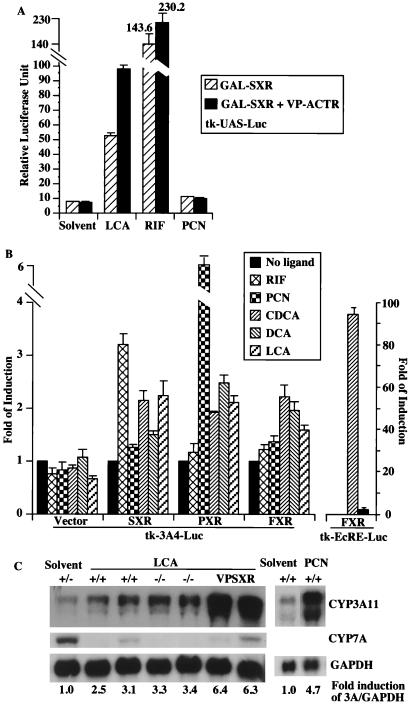
As an initial effort to examine whether bile acids are SXR/PXR ligands, we used a chimeric receptor system in which the ligand-binding domain (LBD) of the human receptor SXR is fused to the DNA binding domain of the yeast transcription factor GAL4. The activity of SXR is determined by using a GAL4 response element-based reporter gene tk-UAS-Luc. As shown in Fig. 1A, LCA transactivated GAL-SXR 5-fold. Two other bile acids, chenodeoxycholic acid (CDCA) and deoxycholic acid (DCA), have similar effect (data not shown). Therefore, bile acids activate the reporter gene activity via DNA-bound SXR-LBD. Moreover, LCA promoted the interaction of SXR with the nuclear receptor coactivator ACTR (23), resulting in a further increase of the reporter activity. RIF and PCN, specific ligands for SXR and PXR, respectively, were also analyzed to verify the responsiveness and specificity of this SXR-based transfection system (Fig. 1A). SXR and PXR are known to regulate CYP3A genes (4–6, 10). To examine whether bile acids can bind and activate SXR/PXR to induce CYP3A gene, we performed an independent ligand activation assay using the full-length SXR or PXR receptor and the CYP3A reporter gene tk-3A4-Luc containing three copies of the IR-6 type of SXR/PXR response element derived from the CYP3A4 gene (4). In SXR- or PXR-transfected cells, this reporter was modestly but consistently activated 2- to 2.5-fold by CDCA, DCA (except for SXR), and LCA (Fig. 1B), comparing to a 3.2- and 6.7-fold activation by RIF and PCN, the prototypic activators for SXR and PXR, respectively (4, 10). It appears that there is no significant species difference between SXR and PXR in their response to bile acids. Interestingly, FXR, the bile acid receptor, also modestly activated this CYP3A4 reporter gene in the presence of CDCA and DCA (Fig. 1B). This is in contrast to a strong induction of the FXR reporter tk-EcRE-Luc (11) by CDCA as expected (Fig. 1B and refs. 12–14). Taken together, these results demonstrate that SXR and PXR can indeed mediate the activation of CYP3A gene by both primary and secondary bile acids.
Figure 1.
Bile acids are SXR/PXR activators and CYP3A inducers. (A) Bile acids activate a reporter gene activity via DNA-bound SXR/LBD. The tk-UAS-Luc reporter was transfected into CV-1 cells together with a chimeric receptor GAL-SXR/LBD alone or in conjunction with VP-ACTR (comprising the receptor interaction domain of ACTR with an amino terminal fusion of the VP16 activation domain). The transfected cells were subsequently treated with indicated compounds. Results represent the average and standard error from triplicate assays. (B) SXR and PXR-mediated activation of CYP3A4 promoter element by bile acids. The SXR/PXR-responsive reporter tk-3A4-Luc or the FXR-responsive reporter tk-EcRE-Luc constructs were transfected into CV-1 cells in the presence of the empty vector or the expression vectors for SXR, PXR, or FXR. The transfected cells were subsequently mock treated or treated with indicated compounds. Results are shown as fold induction over solvent controls and represent the average and standard error from triplicate assays. (C) Induction of CYP3A by LCA in vivo. Total liver RNA isolated from mice of indicated genotypes and treated with LCA (8 mg/day for 4 days) or solvent control were subjected to Northern blot analysis and probed for CYP3A11, CYP7A, and GAPDH mRNA. Similar CYP3A induction by LCA was also seen in mice 24 h after a single dose of 8 mg of LCA, and LCA treatment does not further increase the induction of CYP3A in VPSXR mice (data not shown). Notably, the solvent control is a PXR+/− mouse; we showed before that the basal expression of CYP3A11 gene remains unchanged independent of PXR genotypes (10).
The ability of bile acids to induce CYP3A gene expression was further confirmed by an in vivo activation assay. Treatment of wild-type mice with LCA resulted in a modest induction of hepatic CYP3A11 mRNA as revealed by Northern blot analysis (Fig. 1C). The level of induction is consistent with that in the transient transfection assay where the full-length PXR or SXR and a CYP3A reporter gene were used (Fig. 1B). Having known that bile acids can induce CYP3A gene in wild-type mice, the availability of PXR-null mice (10) enabled us to examine whether PXR is necessary for CYP3A induction by bile acids. Surprisingly, PXR-null mice are remain responsive to LCA to induce CYP3A (Fig. 1C). However, the levels of CYP3A induction in LCA-treated wild-type or PXR-null mice were not as profound as in the Alb-VPSXR transgenic mice or in PCN-treated wild-type animals (Fig. 1C). The Alb-VPSXR transgenic mice express an activated form of SXR under the control of the liver-specific albumin promoter, and exhibit constitutive up-regulation of CYP3A (10). In the same LCA-treated livers, the expression of CYP7A was suppressed by LCA as expected (12–14).
Xenoresponse to Cholestatic LCA in PXR-Null and Alb-VPSXR Transgenic Mice.
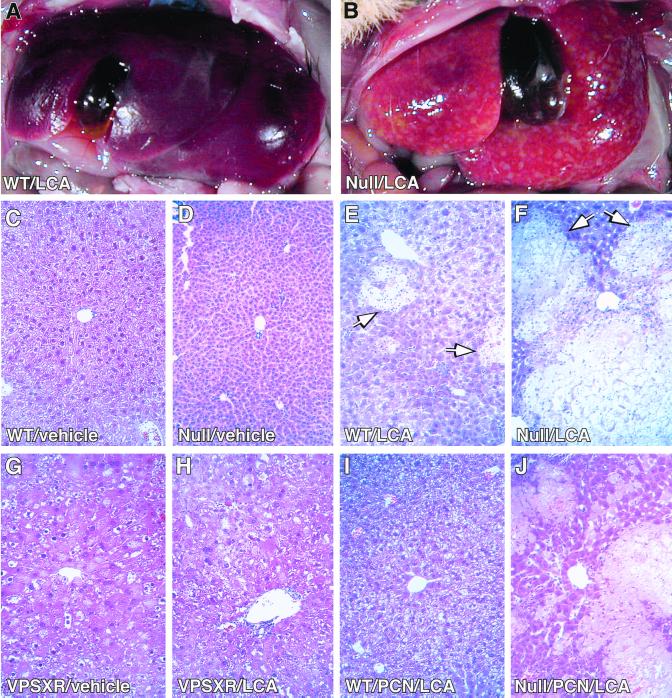
LCA has been shown to cause cholestasis and associated hepatotoxicity (24). The ability of LCA to induce CYP3A gene expression prompted us to speculate that induction of the CYP3A gene might prevent or alleviate cholestatic toxicity by activation of the xenobiotic response. Such an effect has not been described, but seems quite plausible based on our previous observation that genetic activation of SXR and the resultant induction of CYP3A in Alb-VPSXR transgenic mice is sufficient to confer a resistance to xenotoxicants (10). To test this idea, mature wild-type, PXR-null, or Alb-VPSXR transgenic animals were dosed with vehicle solvent or LCA (24). After 4 days of treatment, 58% of wild-type mice (7 of 12) and 100% of PXR-null mice (7 of 7) exhibited areas of liver infarct and/or necrosis when examined at the histological level (Table 1). Shown in the top portion of Fig. 2 are livers from representative LCA-treated animals. The wild-type liver (Fig. 2A) showed resistance to LCA, whereas the livers from a PXR-null mouse (Fig. 2B) exhibited profound subcapsular foci of yellow discoloration. This is most likely corresponding with the areas of saponification/coagulative necrosis that we saw histologically (see below). Unchallenged PXR-null mice exhibited no apparent liver histological abnormalities (compare wild-type in Fig. 2C and PXR-null in Fig. 2D). However, in response to LCA treatment, compared with wild-type mice (Fig. 2E), the histologic damage in the livers of PXR-null mice tended to be more massive (Fig. 2F) with extensive inflammatory and neutrophil infiltration prominent in the necrotic lesions and surrounding areas. The unchallenged livers of Alb-VPSXR transgenic mice (Fig. 2G) exhibited evident, but well tolerated histologic changes (compare Fig. 2G and vehicle control wild-type in Fig. 2C; detailed histology of the transgenic mice will be described elsewhere). In sharp contrast to wild-type and PXR-null mice, the livers of the transgenic animals showed virtually no histologic changes upon LCA treatment (Fig. 2H), compared with its vehicle-treated counterpart in Fig. 2G; also see Table 1). The absence of induced pathology in Alb-VPSXR mice demonstrates that sustained activation of SXR is sufficient to prevent LCA-mediated histologic liver damage.
Table 1.
Loss of PCN-mediated protection from LCA-induced hepatotoxicity in PXR-null mice
| −PCN
|
+PCN
|
||||
|---|---|---|---|---|---|
| WT | PXR-null | VPSXR | WT | PXR-null | |
| LCA-induced histologic liver damage | 7/12 (58.3%) | 7/7 (100%) | 0/5 (0%) | 2/9 (22.2%) | 5/5 (100%) |
WT, wild type.
Figure 2.
LCA-mediated liver damage in wild-type, PXR-null, and transgenic mice. Wild-type (A, C, and E), PXR-null (B, D, and F), or Alb-VPSXR transgenic mice (G and H) were given daily treatments of LCA (A, B, E, F, and H) or vehicle (C, D, and G) via gavage for 4 days (24). Wild-type (I) or PXR-null (J) mice were treated with a single i.p. injection of PCN (40 mg/kg) before 4 days of treatment with both LCA (8 mg/day) and PCN (13 mg/kg). (A and B) Photographs of representative livers. (C–J) Liver paraffin sections stained with hematoxylin and eosin. Regions of liver necrosis are marked by arrows in E and F. (×200.)
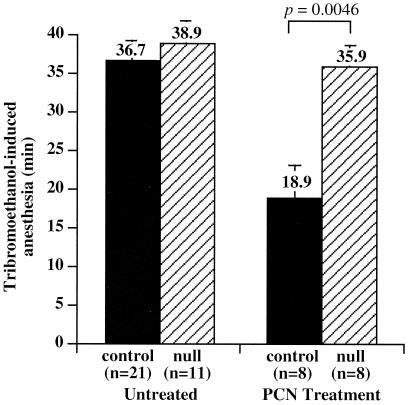
We demonstrated previously that a sustained activation of SXR and the associated induction of CYP3A is sufficient to confer xenoprotection in transgenic mice (10). We next performed in vivo LCA toxicity assays in mice treated with the catatoxic steroid PCN to ascertain whether PXR-mediated induction of CYP3A is necessary for xenoprotection against LCA hepatotoxicity. PCN treatment has been shown to alleviate liver toxicity of a variety of xenotoxicants in animals via its ability to induce CYP enzymes such as CYP3A (Fig. 1C and ref. 3). In principal, such a protective effect should be absent in PXR-null mice because PCN can no longer activate CYP3A gene expression (10). Table 1 shows that, indeed, following PCN treatment of wild-type mice the incidence of LCA-induced histologic liver damage was decreased by about 60% (58% of unprimed mice showing liver toxicity vs. 22% incidence of liver toxicity in PCN-primed mice). In sharp contrast, PCN-treated PXR-null mice remained sensitive to LCA. PCN-induced hepatoprotection was also reflected at the histologic level insofar as liver damage was less severe in PCN-primed (Fig. 2I) compared with the unprimed (Fig. 2E) wild-type animals. By contrast, much less relief was seen in the PXR-null mice (PCN-primed in Fig. 2J; compare with the unprimed control in Fig. 2F). In addition, a significant loss of PCN-mediated xenoprotection in PXR-null mice was also seen when the animals were challenged with two other prototypic xenotoxicants, the anesthetic tribromoethanol (Fig. 3) and the muscle relaxant zoxazolamine (data not shown). Tribromoethanol induced anesthesia equally well in both control (including both PXR+/+ and PXR+/− mice) and PXR-null mice, consistent with the observation that disruption of the mouse PXR gene does not alter the basal expression of mouse CYP3A11 (10). On PCN treatment, control animals were anesthetized for half as long as their untreated counterparts, reflecting the anticipated xenoprotection. In sharp contrast, PXR-null mice remained fully sensitive after PCN treatment (Fig. 3). Therefore, PXR-null mice exhibited a loss of xenoprotection not only to LCA, but also to xenotoxicants in general. These results, together with our previous report of enhanced xenoprotection in Alb-VPSXR mice, provide compelling evidence that SXR/PXR signaling is both necessary and sufficient for xenoprotection.
Figure 3.
Loss of PCN-mediated protection against xenotoxicants in PXR-null mice. Tribromoethanol anesthesia tests were first administered in the absence of PCN as described (10). After a recovery period of 3 days, the same groups of mice were treated with PCN (40 mg/kg) by daily i.p. injections for two days and the anesthesia tests then repeated 24 h after the last PCN injection. Results represent the averages and standard error for the indicated numbers of mice. Controls include both PXR+/+ and PXR +/− mice. The statistical analysis was performed by using INSTAT 2.03.
LCA as a Substrate for CYP3A Hydroxylation.
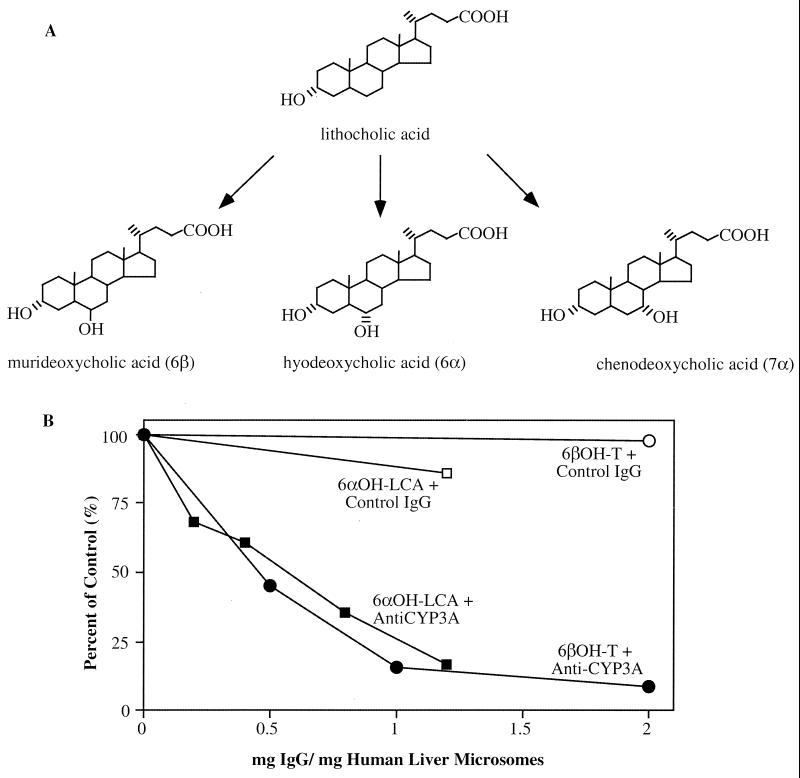
To establish the relevance of the xenobiotic response in protection from LCA toxicity in humans, we set up in vitro enzymatic assays to examine whether LCA is a substrate for hydroxylation by human CYP3A. Fig. 4A shows the P450-dependent pathways of LCA metabolism. LCA was incubated with recombinant human CYP3A4 and 3A5, as well as human liver microsomes (hLM) prepared from four individuals. The products of hydroxylation of LCA were identified as hyodeoxycholic acid (HDCA; 6α-OH), murideoxycholic acid (MDCA; 6β-OH), and CDCA (7α-OH) (21). As shown in Table 2, recombinant CYP3A4 efficiently hydroxylated LCA, whereas the recombinant CYP3A5 enzyme was substantially less active. In each case, the major metabolite for LCA was the 6α-hydroxylated product, followed by 6β-OH and 7α-OH products. Human liver microsomes also hydroxylated LCA in a similar manner, although individual variation in the relative activities is evident. These results are consistent with a recent report by Araya and Wikvall (25). We went on further to perform antibody inhibition studies to examine whether the hydroxylation by liver microsomes is CYP3A-specific. As shown in Fig. 4B, an anti-CYP3A antibody fully inhibited LCA 6α-hydroxylation catalyzed by human liver microsomes (data obtained using human liver sample hLM7; see Table 2). Thus, LCA hydroxylation catalyzed by human liver microsomes is primarily mediated by CYP3A enzymes.
Figure 4.
P450-dependent LCA metabolism. (A) Pathways of LCA hydroxylation. (B) Specific inhibition of human liver microsome-catalyzed LCA 6α-hydroxylation and testosterone 6β-hydroxylation by an anti-CYP3A antibody (22). The latter reaction is an established CYP3A-dependent metabolic reaction in human liver microsomes.
Table 2.
Hydroxylation of LCA by recombinant CYP3A4 and liver microsomes
| Lithocholic acid metabolites | LCA hydroxylation, pmol/mg⋅min
|
|||||
|---|---|---|---|---|---|---|
| CYP3A4 | CYP3A5 | hLM2 | hLM7 | hLM9 | hLM13 | |
| 6α | 8.0 | 0.04 | 9.9 | 38.2 | 9.1 | 6.3 |
| 6β | 2.1 | 0.0 | 7.3 | 8.8 | 3.4 | 4.4 |
| 7α | 0.1 | 0.0 | 2.8 | 5.0 | 0.8 | 3.0 |
Discussion
In this report, using both transient transfection and transgenic models, we identify bile acids as activators for xenobiotic receptors SXR and PXR, and establish the importance of xenobiotic response in the hydroxylation and detoxification of cholestatic bile acids, such as LCA.
The similar responsiveness of wild-type and PXR-null mice to LCA suggests that bile acid induction of CYP3A can be mediated by an alternative and/or compensatory cellular factor(s) other than PXR. Specifically, it is possible that the intact LCA effect to induce CYP3A in PXR-null mice may result from continued expression and signaling of other xenobiotic-regulating nuclear receptors, such as FXR and constitutive androstane receptor (CAR). Indeed, FXR also exhibited a modest activation of a CYP3A promoter element in response to bile acids (Fig. 1B). Moreover, we and others have recently shown that CAR can regulate CYP3A via its ability to adaptively recognize the SXR/PXR response elements (ref. 26, and references therein). However, LCA has no effect on the activity of CYP3A reporter genes in the presence of CAR (data not shown). Nevertheless, our current study provides another example of the proposed fail-safe pathways in xenobiotic regulation (26). However, we cannot exclude the possibility that an additional nuclear receptor(s) or other transcriptional regulators might be involved.
Oral administration of bile acids such as CDCA to patients with cholesterol gallstones can reduce the cholesterol saturation of bile, resulting in a partial to complete dissolution of the gallstones (27). However, the potential clinical benefit of this treatment is compromised because CDCA is partially dehydroxylated by intestinal bacteria to yield the toxic LCA (24). The mechanism by which LCA causes liver toxicity is currently unknown. The enhanced protection against the hepatotoxic effects of LCA described here in Alb-VPSXR mice and in PCN-primed wild type mice suggests that the activation of SXR/PXR provides a simple and natural mechanism for CYP3A metabolism-dependent protection from toxic bile acids. However, the levels of CYP3A induction achieved in LCA-treated wild type or PXR-null mice appear not to be sufficient to confer a complete resistance.
RIF has been shown to relieve pruritus in cholestatic liver disease. The role of bile acids in producing pruritus is still obscure and controversial. It has been anecdotally reported that RIF can stimulate 6α-hydroxylation of bile acids in humans, which in turn facilitates glucuronidation by the UDP-glucuronosyltransferases (UGTs) at the 6α-hydroxy position, followed by renal excretion and a reduction of pruritus (20, 21, 28). This proposed mechanism is consistent with the observation that LCA hydroxylation predominates during recovery from LCA-induced intrahepatic cholestasis (29). Moreover, this observation is in excellent agreement with the identification of RIF as a human-specific SXR activator and CYP3A inducer, supporting our proposal that SXR/PXR plays an important role in xenobiotic response and in bile acid homeostasis.
In summary, bile acids serve as ligands for nuclear receptors. Elegant studies (17, 30) demonstrated that FXR is critical as an endogenous bile acid receptor and a major regulator of bile acid homeostasis. Our present studies focus on bile acid toxicity and the participation of a SXR/PXR-mediated xenobiotic response in bile acid detoxification, and thereby demonstrate an important metabolic link between SXR/PXR, toxic bile acids, and drug metabolizing enzymes, such as CYP3A. These data not only provide a molecular mechanism for the relief of cholestasis-associated pruritus by RIF, but also illustrate a potential therapeutic strategy for the design of drugs targeting cholestasis and other hepatic diseases.
Acknowledgments
We thank Drs. Brent Neuschwander-Tetri and Elizabeth Brunt for their invaluable comments on liver histology; Drs. Phil Guzelian, Bruce Blumberg, and Richard Lin for their critical reading of the manuscript; Alexis Pierce and Henry Juguilon for technical assistance; and Elaine Stevens and Lita Ong for administrative assistance. W.X. and Y.S. are supported by the Susan G. Komen Breast Cancer Foundation. E.S.O. is a research intern from the University of California at Berkeley. R.M.E. is an Investigator of the Howard Hughes Medical Institute at the Salk Institute for Biological Studies and March of Dimes Chair in Molecular and Developmental Biology. This work was supported in part by Mathers Foundation (to R.M.E.), the Howard Hughes Medical Institute (to R.M.E.), and the Superfund Basic Research Center at Boston University (National Institutes of Health Grant ES07381 to D.J.W.).
Abbreviations
- CDCA
chenodeoxycholic acid
- CYP
cytochrome P450 enzyme
- DCA
deoxycholic acid
- FXR
farnesoid X receptor
- LCA
lithocholic acid
- PCN
pregnane-16α-carbonitrile
- PXR
pregnane X receptor
- RIF
rifampicin
- SXR
steroid and xenobiotic receptor
References
- 1.Denison M S, Whitlock J P., Jr J Biol Chem. 1995;270:18175–18178. doi: 10.1074/jbc.270.31.18175. [DOI] [PubMed] [Google Scholar]
- 2.Maurel P. In: Cytochrome P450: Metabolic and Toxicological Aspects. Ioannides C, editor. Boca Raton, FL: CRC; 1996. pp. 241–270. [Google Scholar]
- 3.Selye H. J Pharm Sci. 1971;60:1–28. doi: 10.1002/jps.2600600102. [DOI] [PubMed] [Google Scholar]
- 4.Blumberg B, Sabbagh W, Juguilon H, Bolado J, Jr, Ong E S, Evans R M. Genes Dev. 1998;12:3195–3205. doi: 10.1101/gad.12.20.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kliewer S A, Moore J T, Wade L, Staudinger J L, Jones M A, McKee D D, Oliver B M, Willson T M, Zetterstrom R H, Perlmann T, et al. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 6.Bertilsson G, Heidrich J, Svensson K, Asman M, Jendeberg L, Sydow-Backman M, Ohlsson R, Postlind H, Blomquist P, Berkenstam A. Proc Natl Acad Sci USA. 1998;95:12208–12213. doi: 10.1073/pnas.95.21.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blumberg B, Evans R M. Genes Dev. 1998;12:3149–3155. doi: 10.1101/gad.12.20.3149. [DOI] [PubMed] [Google Scholar]
- 8.Savas U, Griffin K J, Johnson E F. Mol Pharmacol. 1999;56:851–857. doi: 10.1124/mol.56.5.851. [DOI] [PubMed] [Google Scholar]
- 9.Waxman D J. Arch Biochem Biophys. 1999;369:11–23. doi: 10.1006/abbi.1999.1351. [DOI] [PubMed] [Google Scholar]
- 10.Xie W, Barwick J L, Downes M, Blumberg B, Simon C M, Nelson M C, Neuschwander Tetri B A, Brunt E M, Guzelian P S, Evans R M. Nature (London) 2000;406:435–439. doi: 10.1038/35019116. [DOI] [PubMed] [Google Scholar]
- 11.Forman B M, Goode E, Chen J, Oro A E, Bradley D J, Perlmann T, Noonan D J, Burka L T, McMorris T, Lamph W W, et al. Cell. 1995;81:687–693. doi: 10.1016/0092-8674(95)90530-8. [DOI] [PubMed] [Google Scholar]
- 12.Wang H, Chen J, Hollister K, Sowers L C, Forman B M. Mol Cell. 1999;3:543–553. doi: 10.1016/s1097-2765(00)80348-2. [DOI] [PubMed] [Google Scholar]
- 13.Parks D J, Blanchard S G, Bledsoe R K, Chandra G, Consler T G, Kliewer S A, Stimmel J B, Willson T M, Zavacki A M, Moore D D, et al. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- 14.Makishima M, Okamoto A Y, Repa J J, Tu H, Learned R M, Luk A, Hull M V, Lustig K D, Mangelsdorf D J, Shan B. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- 15.Lu T T, Makishima M, Repa J J, Schoonjans K, Kerr T A, Auwerx J, Mangelsdorf D J. Mol Cell. 2000;6:507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- 16.Goodwin B, Jones S A, Price R R, Watson M A, McKee D D, Moore L B, Galardi C, Wilson J G, Lewis M C, Roth M E, et al. Mol Cell. 2000;6:517–526. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 17.Chawla A, Saez E, Evans R M. Cell. 2000;103:1–4. doi: 10.1016/s0092-8674(00)00097-0. [DOI] [PubMed] [Google Scholar]
- 18.Radominska A, Treat S, Little J. Semin Liver Dis. 1993;13:219–234. doi: 10.1055/s-2007-1007351. [DOI] [PubMed] [Google Scholar]
- 19.Erlinger S. In: Bile Flow. Aries I M, Boyer J L, editors. New York: Raven; 1982. pp. 407–427. [Google Scholar]
- 20.Wietholtz H, Marschall H U, Sjovall J, Matern S. J Hepatol. 1996;24:713–718. doi: 10.1016/s0168-8278(96)80268-6. [DOI] [PubMed] [Google Scholar]
- 21.Radominska-Pyrek A, Zimniak P, Irshaid Y M, Lester R, Tephly T R, St. Pyrek J. J Clin Invest. 1987;80:234–241. doi: 10.1172/JCI113053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zimniak P, Radominska A, Zimniak M, Lester R. J Lipid Res. 1988;29:183–190. [PubMed] [Google Scholar]
- 23.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 24.Taylor W, Lesna M. Br J Pharmacol. 1997;61:133–134. [PMC free article] [PubMed] [Google Scholar]
- 25.Araya Z, Wikvall K. Biochim Biophys Acta. 1999;1438:47–54. doi: 10.1016/s1388-1981(99)00031-1. [DOI] [PubMed] [Google Scholar]
- 26.Xie W, Barwick J L, Simon C M, Pierce A M, Safe S, Blumberg B, Guzelian P S, Evans R M. Genes Dev. 2000;14:3014–3023. doi: 10.1101/gad.846800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Danzinger R R, Hofmann A F, Schoenfield L J, Thistle J L. N Engl J Med. 1972;286:1–8. doi: 10.1056/NEJM197201062860101. [DOI] [PubMed] [Google Scholar]
- 28.Pillot T, Ouzzine M, Fournel-Gigleux S, Lafaurie C, Radominska A, Burchell B, Siest G, Magdalou J. J Biol Chem. 1993;268:25636–25642. [PubMed] [Google Scholar]
- 29.Vu D D, Tuchweber B, Plaa G L, Yousef I M. Biochim Biophys Acta. 1992;1126:53–59. doi: 10.1016/0005-2760(92)90216-i. [DOI] [PubMed] [Google Scholar]
- 30.Sinal C J, Tohkin M, Miyata M, Ward J M, Lambert G, Gonzalez F J. Cell. 2000;102:731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]