Abstract
The 26S proteasome is a multisubunit protein- destroying machinery that degrades ubiquitin-tagged proteins. To date only a single species of Rpn10, which possibly functions as a multiubiquitin chain-binding subunit, has been identified in various organisms. Here we report that mouse Rpn10 mRNAs occur in at least five distinct forms, named Rpn10a to Rpn10e, and that they are generated from a single gene by developmentally regulated, alternative splicing. Rpn10a is ubiquitously expressed, whereas Rpn10e is expressed only in embryos, with the highest levels of expression in the brain. Both forms of Rpn10 are components of the 26S proteasome, with an apparently similar affinity for multiubiquitylated [125I]lysozyme in vitro. However, they exert markedly divergent effects on the destruction of B-type cyclin in Xenopus egg extracts. Thus, the 26S proteasome occurs in at least two functionally distinct forms: one containing a ubiquitously expressed Rpn10a and the other a newly identified, embryo-specific Rpn10e. While the former is thought to perform proteolysis constitutively in a wide variety of cells, the latter may play a specialized role in early embryonic development.
Keywords: alternative splicing/development/proteasome/proteolysis/Rpn10
Introduction
In eukaryotic cells, selective breakdown of most cellular proteins is ensured by two distinct, concerted pathways that both require metabolic energy (reviewed in Coux et al., 1996; Hershko and Ciechanover, 1998 and references therein): first, the process of marking appropriate proteins by attaching ubiquitin (Ub), an evolutionarily conserved 8.6 kDa polypeptide, to form the degradation signal; and secondly, the proteolytic attack of multiubiquitylated proteins by the 26S proteasome. There are growing lines of evidence that the Ub–proteasome pathway plays a critical role in various biologically important processes, including cell cycle, apoptosis, signal transduction, development and the immune response (Hershko and Ciechanover, 1998; Tanaka and Kasahara, 1998; Schwartz and Ciechanover, 1999).
Protein ubiquitylation is catalyzed by a multi-enzymatic system consisting of E1 (Ub-activating), E2 (Ub- conjugating) and E3 (Ub-ligating) enzymes. Ub is linked covalently to target proteins via an isopeptide linkage between the C-terminal Gly of Ub and the ε-NH2 group of a Lys residue of an acceptor substrate. In this protein-modifying system, Ub is first activated by E1 in an ATP-dependent manner, being linked to E1 by a high-energy thioester bond. Following activation, Ub forms a thioester bond with E2. E2 catalyzes the formation of the isopeptide bond between Ub and the substrate. In some cases, Ub is transferred directly to target proteins, but the reaction usually requires another enzyme termed E3. In successive reactions, a multi-Ub chain is synthesized by progressive transfer of Ub moieties to Lys48 of the previously attached Ub molecule.
The 26S proteasome, a eukaryotic ATP-dependent protease complex, is composed of a core proteinase, known as the 20S proteasome, and a pair of symmetrically disposed PA700 regulatory particles (also known as the 19S complex) (Coux et al., 1996; Rechsteiner, 1998; Voges et al., 1999; Ferrell et al., 2000). PA700 is attached to both ends of the central 20S proteasome in opposite orientations to form the enzymatically active 26S proteasome in an ATP-dependent manner (Rechsteiner, 1998; DeMartino and Slaughter, 1999). The 26S proteasome with a molecular mass of ∼2500 kDa acts as a highly organized apparatus leading to efficient protein death. Hence, it can be regarded as a protein-destroying machinery (Coux et al., 1996; Voges et al., 1999).
The 20S proteasome, which functions as the proteolytic core of the 26S proteasome, is a protease complex with a molecular mass of 700–750 kDa composed of 28 subunits (Tanaka, 1998). It is a barrel-like particle formed by the axial stacking of four rings made up of two outer α-rings and two inner β-rings, being associated in the order αββα (reviewed in Bochtler et al., 1999). Three β-type subunits of each inner ring have catalytically active threonine residues at their N-termini, and these active sites reside in a chamber formed by the centers of the abutting β-rings.
The regulator, PA700, was discovered as an activator of the 20S proteasome. It is a 700 kDa protein complex composed of ∼20 subunits with sizes of 25–110 kDa (Tanaka, 1998; DeMartino and Slaughter, 1999). PA700 consists of two subcomplexes, known as ‘base’ and ‘lid’, which, in the 26S proteasome, correspond to the portions of PA700 proximal and distal, respectively, to the 20S proteasome (Glickman et al., 1998). The base is made up of six ATPases and the two largest regulatory components named Rpn1 and Rpn2, while the lid contains multiple non-ATPase subunits. The base complex, thought to bind to the outer α-ring of the central 20S proteasome ATP-dependently, is likely to be involved in opening the gate of the α-ring for entry of the protein substrate. The metabolic energy liberated by ATP consumption is probably utilized for assembly of the base complex with the 20S proteasome, although it may also be used for unfolding target proteins so that they can penetrate the channel of the α- and β-rings of the 20S proteasome (Braun et al., 1999). On the other hand, the lid complex is thought to be required for recognition of target proteins.
It is important to understand how the 26S proteasome specifically recognizes ubiquitylated proteins, because this recognition is a key process in the selective degradation of ubiquitylated proteins (Pickart, 1998; Thrower et al., 2000). Previous studies showed that a 50 kDa subunit of human PA700, originally called S5a, could bind to multi-Ub conjugates in vitro, and hence could possibly function as a multi-Ub chain-binding subunit (Deveraux et al., 1994). So far, S5a homologs have been identified in various eukaryotes such as Saccharomyces cerevisiae (van Nocker et al., 1996a; Kominami et al., 1997), Schizosaccharomyces pombe (Wilkinson et al., 2000), Arabidopsis thaliana (van Nocker et al., 1996b), Drosophila melanogaster (Haracska and Udvardy, 1995), Physcomitrella patens (Girod et al., 1999), Oriza sativa (Yanagawa et al., 1998), mouse (Pusch et al., 1998) and human (Ferrell et al., 1996), and given various names such as Mbp1, Mcb1, Sun1, Pus1, µ-54 and Rpn10. Here we call S5a and its homologs Rpn10 in accordance with a recent proposal (Finley et al., 1998).
In the present study, we show that a novel family of Rpn10 is expressed at specific stages of mouse development, forming embryo- and tissue-specific species of 26S proteasomes. Analysis of the mouse Rpn10 gene indicates that all members of the Rpn10 family can be generated from a single gene by alternative splicing. Interestingly, the 26S proteasome with an embryo-specific Rpn10 and that with a previously identified, ubiquitously expressed Rpn10 exert markedly divergent effects on the destruction of B-type cyclin in Xenopus egg extracts.
Results
Multiple forms of Rpn10 are generated by alternative splicing from a single gene
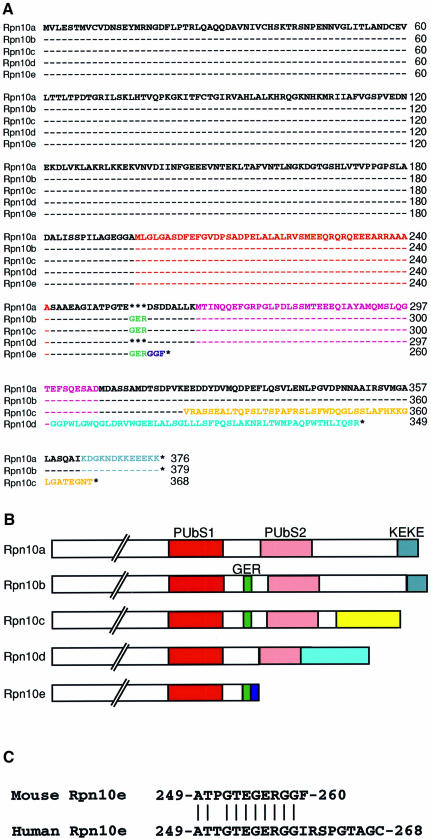
While performing PCR-based differential screening of cDNA libraries from adult and embryonic mouse tissues, we noticed the occurrence of multiple cDNAs for Rpn10. PCR-assisted cloning enabled us to isolate five distinct full-length cDNAs encoding a family of Rpn10 proteins designated Rpn10a to Rpn10e (Figure 1A). The Rpn10a isolated from an adult mouse testis cDNA library is most similar to human S5a (Ferrell et al., 1996), displaying 95% amino acid identity. Thus, Rpn10a is most likely the mouse counterpart of human S5a. The remaining four cDNAs, Rpn10b to Rpn10e, were isolated from mouse cDNA libraries made from embryonic stem cells or day 8.5 or day 16 whole embryos. Sequence alignment of the five Rpn10 proteins (Figure 1A) shows that they all have the same sequence over their N-terminal half regions, whereas their C-terminal halves are highly divergent (for schematic representation see Figure 1B). Human S5a has two segments of ∼30 amino acids, termed PUbS1 and PUbS2, which have been suggested to be involved in the binding to a multi-Ub chain in a cooperative way (Young et al., 1998), although only one segment appears sufficient for the multi-Ub chain-binding activity in other organisms (Fu et al., 1998; Girod et al., 1999). While both segments are present in Rpn10a to Rpn10d, Rpn10e is truncated at its C-terminal and has only PUbS1. We also identified the human counterpart of Rpn10e from a human fetal brain cDNA library, confirming that the occurrence of this form is not a peculiarity of mouse Rpn10 (Figure 1C). Rpn10e structurally resembles the multi-Ub-binding subunit of the budding yeast 26S proteasome, known as Rpn10, Mcb1 or Sun1 (van Nocker et al., 1996a; Kominami et al., 1997), and its fission yeast counterpart Pus1 (Wilkinson et al., 2000), in that they all have approximately the same relative molecular mass (∼29 kDa) and lack PUbS2 (van Nocker et al., 1996a; Kominami et al., 1997; Wilkinson et al., 2000). Intriguingly, Rpn10b is identical to Rpn10a, except that the former contains an insertion of nine nucleotides coding for Gly-Glu-Arg between the PUbS1 and PUbS2 segments. This insertion is also present in Rpn10c and Rpn10e. The C-terminal regions of Rpn10a and Rpn10b contain the KEKE domain, enriched in alternating Lys and Glu, which has been proposed to participate in protein–protein interactions (Realini et al., 1994). In contrast, Rpn10c and Rpn10d have distinct C-terminal extension sequences consisting of 45 and 51 amino acid residues, respectively. Thus, each Rpn10 form has a unique structural feature that distinguishes one from another.
Fig. 1. A family of mouse Rpn10 proteins. (A) Sequence alignment of the Rpn10 proteins deduced from the cDNA sequences. The multi-Ub-binding domains [PUbS1 (red) and PUbS2 (pink)], the Gly-Glu-Arg (GER) sequence (green), the KEKE domain (gray) and the specific C-terminal sequences in Rpn10c (yellow), Rpn10d (pale blue) and Rpn10e (dark blue) are indicated by colored letters. Dashes indicate identical amino acid residues, and asterisks indicate absence of residues. (B) Schematic representation of the C-terminal half structures of multiple Rpn10 proteins. The amino acid sequences written in color in (A) are shown as boxes of the same color. (C) C-terminal sequence alignment of the mouse and human Rpn10e deduced from the cDNA sequences. Vertical bars indicate identical amino acids. Numbers on the right indicate the residue numbers of the C-terminal amino acids. Note that the regions not shown are identical between the mouse and human Rpn10e.
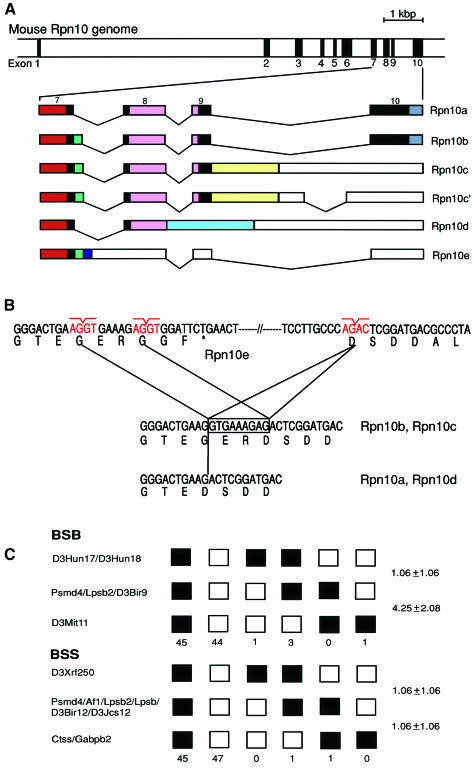
To understand how the multiple Rpn10 cDNAs are produced, we isolated genomic clones for Rpn10 from a mouse λ phage library. Only one Rpn10 gene, designated Psmd4, was identified. Comparison of the genomic and cDNA sequences revealed that alternative splicing of pre-mRNA generates all distinct forms of Rpn10 cDNAs (Figure 2A). The nine nucleotides coding for Gly-Glu-Arg are inserted into or deleted from the cDNAs because of differential usage of the splice donor sites in intron 7 (Figure 2B). The Rpn10d and Rpn10c cDNAs are generated because of the failure to splice out introns 8 and 9, respectively. The cDNA for Rpn10e is generated because intron 7 is not excised. This creates a stop codon 19 bp downstream from the 3′-end of exon 7, resulting in a smaller protein truncated downstream from the PUbS1 segment (Figure 2B).
Fig. 2. Genomic organization and chromosomal localization of the mouse Rpn10 (Psmd4) gene. (A) Physical map of the Rpn10 gene. Exons are indicated by filled boxes and numbered from 1 to 10 (upper panel). The structures of multiple Rpn10 mRNAs generated by alternative splicing are shown schematically. Solid and open boxes indicate the coding region and the 3′-untranslated region, respectively (lower panel). Colored boxes correspond to those of the same color in Figure 1B. Rpn10c and Rpn10c′ code for an identical protein. (B) Differential usage of the splice donor sites in intron 7 accounts for the presence or absence of the Gly-Glu-Arg sequence. An asterisk in Rpn10e indicates a termination codon. (C) Mapping of the Rpn10 gene to chromosome 3. The sequences of the primers used for PCR were 5′-GGGAACAGAGTTTAGCCAAGAATCG-3′ and 5′-TGACGTCATAGTCATCCTCCTC-3′. In BSB, open boxes represent the C57BL/6J-M.spretus heterozygote pattern, and closed boxes the homozygous C57BL/6J pattern. In BSS, open boxes represent the homozygous SPRET/Ei pattern, and closed boxes the C57BL/6Ei-SPRET/Ei heterozygote pattern. The number of mice carrying each type of chromosome is shown below the boxes. For each panel, 94 animals were typed. Recombination frequencies between pairs of loci (percentage recombination with standard errors) are given at the right side. The loci not separated by recombination are indicated by ‘/’.
Chromosomal localization of the Psmd4 gene was determined using two panels of interspecific backcross mice generated at The Jackson Laboratory (Rowe et al., 1994) (Figure 2C). The primer pair described in the figure legends amplified DNA fragments of 590 and 569 bp in C57BL/6J and SPRET/Ei mice, respectively. We used this length variation for typing interspecific backcross mice. Analysis of the BSB panel [(C57BL/6J × M.spretus) F1 × C57BL/6J] showed that Psmd4 maps to the vicinity of Lpsb2 (the gene coding for the liver protein, selenium binding, 56 kDa) on chromosome 3. This localization was confirmed by analysis of the BSS panel [(C57BL/6Ei × SPRET/Ei)F1 × SPRET/Ei]. Here, Psmd4 was found to map to the vicinity of Af1 (the gene coding for the antisecretory factor 1), Lpsb2 and Lpsb (the gene coding for the liver protein, selenium binding, 56 kDa). No recombination was observed among Psmd4, Af1, Lpsb2 and Lpsb, indicating that they lie within 3.8 cM at the 95% confidence level.
Expression of multiple forms of Rpn10 is developmentally regulated
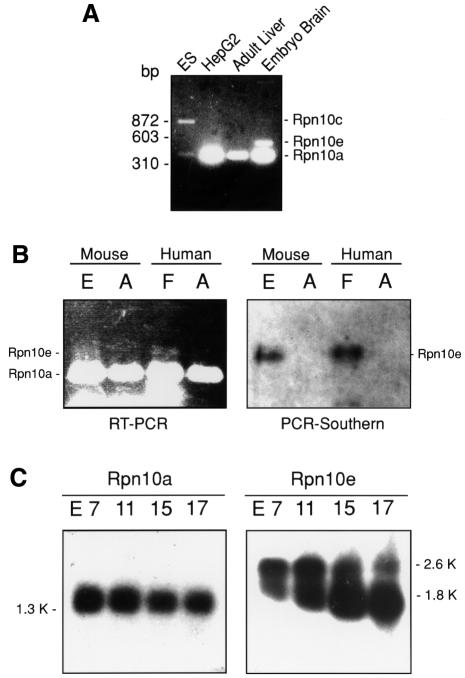
Expression profiles of different Rpn10 mRNAs were analyzed by PCR using a set of primers corresponding to the conserved sequences of the Psmd4 gene. The fragment of ∼370 bp was amplified from all cDNA libraries irrespective of tissues and developmental stages (Figure 3A). However, the fragments of ∼520 and ∼870 bp were amplified only from embryonic brain and embryonic stem cell cDNA libraries, respectively. Partial sequence analysis of these fragments showed that the ∼870, ∼520 and ∼370 bp bands correspond to the Rpn10c, Rpn10e and Rpn10a mRNAs, respectively. We then investigated whether expression of the Rpn10e mRNA is truly embryo-specific by comparing its expression between fetal and adult brains. Semi-quantitative RT–PCR amplified a faint band of ∼520 bp only in mouse embryonic and human fetal brains, while the ∼370 bp band corresponding to the Rpn10a mRNA was amplified in both adult and embryonic (fetal) brains (Figure 3B, left panel). The identity of the ∼520 bp band was confirmed by Southern blot analysis with an Rpn10e-specific probe (Figure 3B, right panel) and by direct sequencing. Northern blot analysis showed that the Rpn10a transcript is expressed constitutively during embryonic development from 7 to 17 days post coitum (d.p.c.) as a single prominent band of 1.35 kb (Figure 3C, left panel). This transcript was also found abundantly in all adult tissues examined (data not shown), indicating ubiquitous expression of the Rpn10a mRNA. In contrast, northern blot analysis with the probe specific for Rpn10e detected two hybridizing bands of ∼1.8 and ∼2.6 kb in embryonic tissues (Figure 3C, right panel), but no bands in various adult tissues (data not shown). Intriguingly, the intensity of the ∼1.8 kb, but not the ∼2.6 kb band increased dramatically from 7 to 17 d.p.c., peaking at 15 d.p.c. The expression levels of the Rpn10e protein as detected by western blot analysis increased roughly in proportion to those of the ∼1.8 kb band (Figure 4B and C), suggesting that this band corresponds to the major form of the translatable Rpn10e mRNA. The exact identity of the ∼2.6 kb band is not clear.
Fig. 3. Expression of Rpn10e mRNA is developmentally regulated. (A) PCR-based expression analysis with cDNA libraries. Degenerate PCR primers were designed for the conserved sequence motifs of S5a: 218LALRVS223 and 336QSVLEN341. PCR was performed using the cDNA libraries as templates. Note that the ∼520 bp (Rpn10e) and ∼870 bp (Rpn10c) bands were amplified only from the embryonic stem cell (ES) and embryonic brain cDNA libraries. (B) Expression of Rpn10a and Rpn10e mRNAs in embryonic, fetal and adult brains. Semi-quantitative RT–PCR (left panel) was performed using the primer pair used in (A). Southern blot analysis of PCR products (right panel) was carried out using an Rpn10e-specific cDNA probe. The sources of the mRNAs were as follows: E, day 13 mouse embryo; F, human fetal brain; A, mouse and human adult brains. (C) Northern blot analysis of Rpn10a and Rpn10e mRNAs during mouse embryonic development. Numbers above the panels stand for days post coitum (d.p.c.).
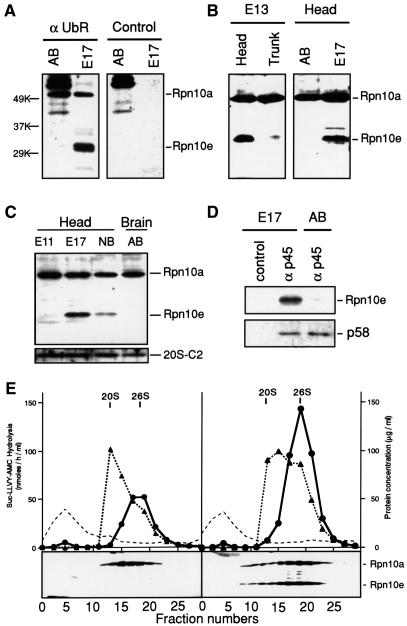
Fig. 4. Identification of embryo-specific 26S proteasomes that contain Rpn10e as a subunit. (A–C) Western blot analysis using the antibody against the N-terminal fragment of human S5a (equivalent to Rpn10a). In (A), non-immune IgG was used as a control (right panel). Samples of soluble proteins were prepared from the following sources: 11 d.p.c. embryo head (E11), 13 d.p.c. embryo head and trunk (E13), 17 d.p.c. embryo head (E17), newborn brain (NB) and adult brain (AB). In (C), western blot analysis was performed also with the antibody specific for the C2 subunit of the human 20S proteasome (lower panel). (D) Rpn10e is co-precipitated with the p45 subunit of the PA700 regulatory complex. Extracts of 17 d.p.c. embryonic heads or adult brains were immunoprecipitated with the antibody specific for p45 (αp45) or non-immune IgG (control). The resulting precipitates were subjected to western blot analysis with the antibody specific for S5a or p58. (E) Sedimentation velocity analysis. Aliquots (10 mg) of adult brain (left panel) and 17 d.p.c. embryonic head (right panel) extracts were fractionated by glycerol density-gradient centrifugation (10–40% glycerol). Aliquots (20 µl) of individual fractions were assayed for proteasomal activity using Suc-LLVY-AMC as the substrate in the presence (filled triangles) or absence (filled circles) of 0.05% SDS. The dotted line represents UV absorbance (total protein). In lower panels, proteins in each fraction (200 µl) were precipitated with acetone, subjected to SDS–PAGE, transferred to a PVDF membrane, and stained with the antibody specific for S5a. The vertical lines indicate the positions of 20S and 26S proteasomes.
The Rpn10e subunit is the component of embryo-specific 26S proteasomes
Because Rpn10a and Rpn10e differ markedly in terms of expression pattern and structures, we decided to perform detailed biochemical and functional analysis of these isoforms. To this end, we first examined whether Rpn10a and Rpn10e are expressed at the protein level. Western blot analysis with an antibody raised against the N-terminal region of human S5a detected two bands of 50 and 32 kDa, corresponding to Rpn10a and Rpn10e, respectively, in 17 d.p.c. mouse whole embryos (Figure 4A). In contrast, the same antibody detected only the 50 kDa band in adult brains. When 13 d.p.c. embryos were divided into head and trunk, the 32 kDa band was detected exclusively in the head (Figure 4B, left panel). As expected, the 50 kDa band was found both in embryonic heads and trunks. The 32 kDa band was detected only scarcely in the head of 11 d.p.c. embryos, but its intensity increased dramatically in the heads of 13 and 17 d.p.c. embryos; this band became less intense in the head of newborn mice (Figure 4C). The expression levels of the 50 kDa band, like those of the C2 subunit of the 20S proteasome, did not change during embryonic development (Figure 4C).
We then examined whether Rpn10a and Rpn10e occur as components of the 26S proteasome. First, we performed co-precipitation experiments with an antibody against the p45 ATPase, a subunit of the PA700 regulatory complex of the 26S proteasome (Hendil et al., 1998) (Figure 4D). While this antibody specifically co-precipitated Rpn10e as well as the PA700 subunit p58 from 17 d.p.c. embryonic head extracts, no 32 kDa band was co-precipitated from adult brain extracts. Consistent with this observation, sedimentation velocity analysis with glycerol gradient showed that both 50 and 32 kDa proteins co-migrated with the 26S proteasome in the 17 d.p.c. embryonic head extracts (Figure 4E, right panel), suggesting that both Rpn10a and Rpn10e are integral components of the 26S proteasome in mouse embryos. Neither Rpn10a nor Rpn10e was detectable in top fractions in adult or embryo tissues (Figure 4E, right and left panels). Thus, unlike the yeast multi-Ub-binding receptor Rpn10, which exists in both free and 26S proteasome-integrated forms (van Nocker et al., 1996a; Wilkinson et al., 2000), no free form of Rpn10a or Rpn10e was detected in mouse tissues, indicating that they both fulfill their roles solely as subunits of 26S proteasomes. Thus, we conclude that Rpn10e is a novel embryo-specific component of the 26S proteasome, which forms the embryo-specific 26S proteasome. It is noteworthy that 26S proteasome activities in the embryo head extracts are ∼2-fold higher than those in the adult brain extracts (Figure 4E, left panel). It is, however, not clear whether this elevation is attributable to the presence of Rpn10e-containing 26S proteasomes.
Rpn10a and Rpn10e exert markedly divergent effects on B-type cyclin destruction in Xenopus egg extracts
We next examined whether there is any functional difference between Rpn10a and Rpn10e. To this end, we prepared recombinant Rpn10 proteins expressed in Escherichia coli. Far-western blot analysis, performed as reported previously (Deveraux et al., 1994; Haracska and Udvardy, 1997; Fu et al., 1998; Young et al., 1998), showed that recombinant Rpn10a and Rpn10e were capable of binding to in vitro prepared multiubiquitylated [125I]lysozyme, but not to free [125I]lysozyme (Figure 5A), indicating that they all function as a multi-Ub-binding receptor for this model substrate. A previous study (Deveraux et al., 1995) showed that free S5a could block the ability of the 26S proteasome to degrade multiubiquitylated [125I]lysozyme in a competitive manner. Therefore, we examined whether Rpn10a and Rpn10e exert similar effects (Figure 5B). The two Rpn10 isoforms inhibited degradation of multiubiquitylated [125I]lysozyme with similar efficiency. Furthermore, proteasomal degradation of high-molecular-mass forms of multiubiquitylated [125I]lysozyme was markedly suppressed by addition of Rpn10a or Rpn10e (Figure 5C). These results indicate that Rpn10e is functionally equivalent to Rpn10a in trapping the multi-Ub chain of this model substrate for subsequent degradation.
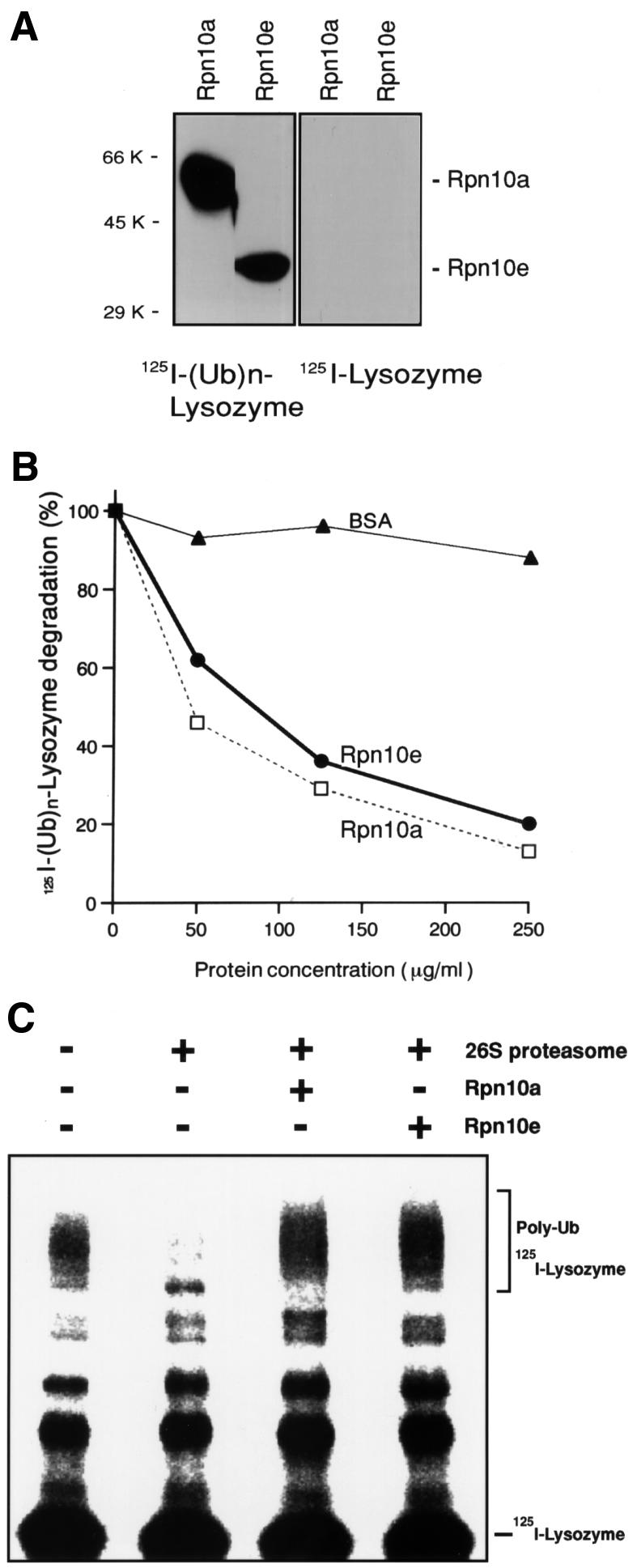
Fig. 5. Both Rpn10a and Rpn10e can bind to multiubiquitylated lysozyme. (A) Far-western blot analysis of multi-Ub chain-binding activities. Multiubiquitylated and free forms of 125I-labeled lysozyme (left and right panels, respectively) were overlaid on recombinant Rpn10a and Rpn10e proteins (4 µg) that had been transferred to a nylon membrane after SDS–PAGE. After extensive washing, the membrane was subjected to autoradiography. Note that, like S5a (Ferrell et al., 1996; Young et al., 1998), recombinant Rpn10a migrated to a position estimated to be ∼50 kDa on SDS–PAGE, although their true molecular masses deduced from the amino acid compositions are ∼40 kDa. (B and C) Effects of recombinant Rpn10a and Rpn10e on the degradation of multiubiquitylated [125I]lysozyme by the purified 26S proteasome. In (B), conversion of [125I]lysozyme to acid-soluble fragments was measured in the presence of Rpn10a, Rpn10e or bovine serum albumin (BSA) at the concentrations indicated. Degradation of [125I]lysozyme–Ub conjugates is expressed as a percentage of the value obtained in control experiments without any additives. The spontaneous degradation rate in the absence of any additive was ∼30%/h. In (C), electrophoretic analysis was performed in the presence or absence of Rpn10a or Rpn10e.
It has been reported that free S5a can block the degradation of cyclin B in Xenopus egg extracts (Deveraux et al., 1995). Therefore, we examined whether Rpn10a and Rpn10e have such activities (Figure 6A). When we triggered cell-cycle progression by Ca2+, cyclin B2 was rapidly degraded as a function of time. Addition of recombinant Rpn10a inhibited this process almost completely. However, Rpn10e had no significant effects, even if the recombinant protein was added at very high concentrations, implying that Rpn10e is much less potent in interacting with ubiquitylated cyclin B2 than Rpn10a. In contrast, both Rpn10a and Rpn10e almost completely suppressed activation of the Cdc2 kinase as well as chromosome condensation before entry into mitosis in cycling extracts of Xenopus eggs (Figure 6B). These observations suggest that Rpn10a and Rpn10e presumably have different roles in the regulation of cell cycles in embryos.
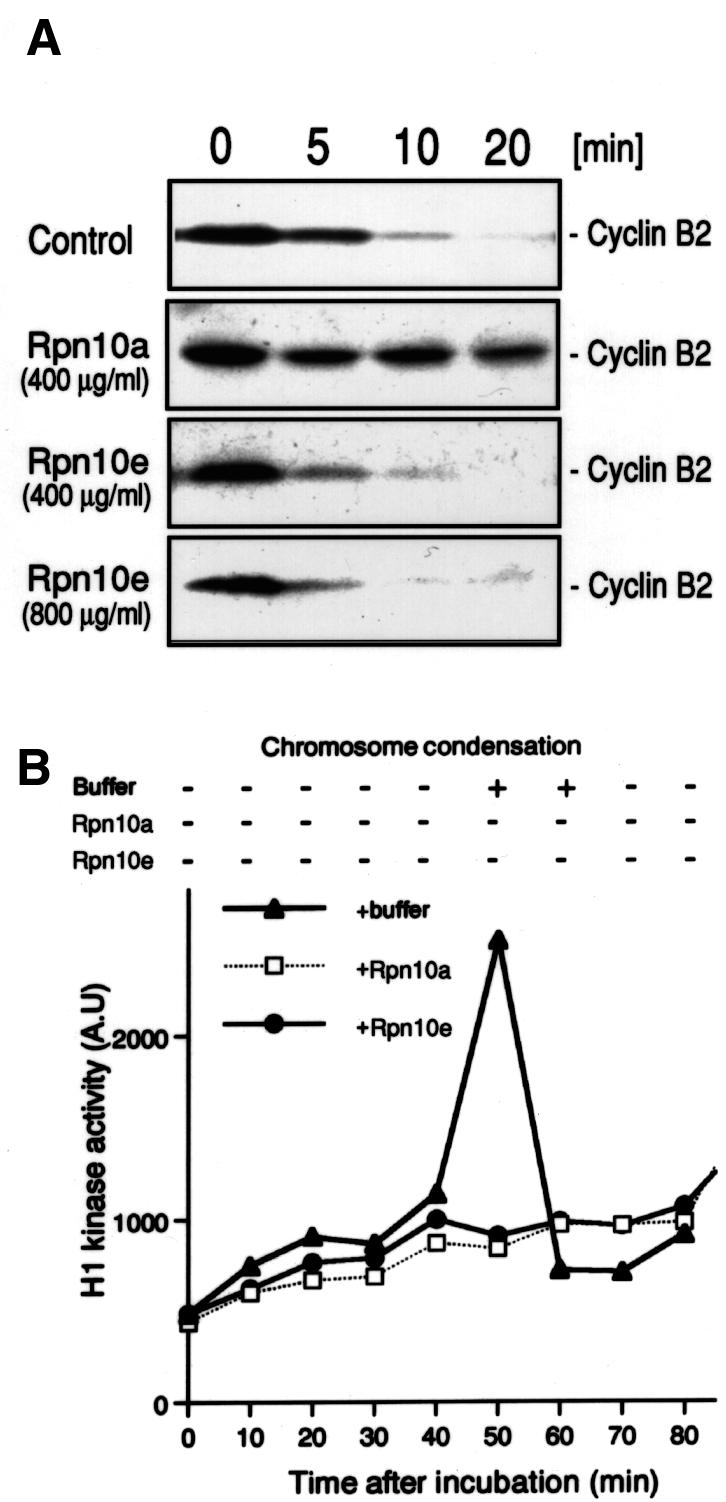
Fig. 6. Effects of recombinant Rpn10a and Rpn10e on cell-cycle progression in Xenopus cell-free extracts. (A) Recombinant Rpn10 proteins expressed in E.coli were added to CSF (cytostatic factor)-arrested extracts prepared from unfertilized Xenopus eggs. Cyclin B2 destruction was then measured as a function of time after triggering cell-cycle progression by the addition of 2 mM Ca2+. CSF extracts were periodically harvested and subjected to western blot analysis with an anti-cyclin B2 antibody. (B) Recombinant Rpn10 proteins (500 µg/ml) were added to the cell-cycle extracts prepared from activated Xenopus eggs. The extracts were periodically harvested after incubation at 23°C, and Cdc2 (histone H1) kinase activity was measured (lower panel). Chromosome condensation was monitored simultaneously by Hoechst staining (upper panel).
Discussion
Until now, only a single evolutionarily conserved species of Rpn10 has been identified in various eukaryotic cells (Haracska and Udvardy, 1995; Ferrell et al., 1996; van Nocker et al., 1996a,b; Kominami et al., 1997; Pusch et al., 1998; Yanagawa et al., 1998; Girod et al., 1999; Wilkinson et al., 2000). In the present study, we obtained evidence for the existence of multiple forms of mouse Rpn10 that are expressed differentially during development. Analysis of the mouse Rpn10 gene indicates that all members of the Rpn10 family can be generated from a single gene by alternative splicing (Figures 1 and 2). This type of genetic mechanism has been shown to confer functional diversity upon a variety of genes including those coding for transcription factors and polypeptide receptors (reviewed in Lopez, 1998).
The most interesting observation made in this study is that some forms of Rpn10 are expressed only at certain developmental stages (Figure 3). In particular, Rpn10e is expressed only in embryos, with the highest levels of expression in the brain. Because it is an integral component of the 26S proteasome (Figure 4), embryonic brains contain a specialized form of the 26S proteasome with an embryo-specific Rpn10. Biochemical analysis showed that Rpn10e and Rpn10a had similar affinities for multiubiquitylated [125I]lysozyme in vitro and that they could inhibit the degradation of the model substrate by the 26S proteasome with similar efficiency (Figure 5). However, unlike Rpn10a, Rpn10e was unable to affect the destruction of cyclin B2 in Xenopus egg extracts (Figure 6A), suggesting that the two forms of Rpn10 have distinct biological activities. This suggestion is reinforced by our recent observation that Rpn10e and Rpn10a affect cell-cycle progression in Xenopus oocytes differently in vivo (data not shown). Thus, the embryonic brain-specific 26S proteasome that contains Rpn10e might play a unique role in re-modeling of cellular proteins during early embryonic development. Figure 4E shows that both Rpn10a and Rpn10e are expressed in embryonic brains. It is, however, not known whether they are co-expressed in a single cell. Also unknown is whether the two forms of Rpn10 are incorporated into the 26S proteasome at equal efficiency in the assembly process.
Recognition of multiubiquitylated substrates by the 26S proteasome is a key step leading to the selective degradation of various cellular proteins. It is, therefore, surprising that disruption of the gene orthologous to Rpn10a (rpn10) did not cause any growth defect in budding (van Nocker et al., 1996a; Kominami et al., 1997) and fission (Wilkinson et al., 2000) yeast. Recent work using a plant, P.patens, indicates that deletion of the Rpn10a ortholog causes developmental arrest and loss of the ability to form buds (Girod et al., 1999). However, complementation analysis in the mutant P.patens strain showed that the multi-Ub chain-binding site was not essential for the wild-type phenotype (Girod et al., 1999). This is consistent with the previous report that the residues near the N-terminus, thus not the multi-Ub chain-binding site, are most critical for Rpn10 function in vivo in yeast (Fu et al., 1998). Therefore, the 26S proteasome may have an Rpn10-independent mechanism for trapping ubiquitylated substrate, although there is no experimental evidence to support such speculations. Interestingly, mice deficient in the Rpn10 gene are embryonic lethal (S.Murata, H.Kawahara, M.Kasahara, K.Yamamoto, K.Tanaka and T.Chiba, manuscript in preparation). Thus, unlike the situation in yeast, Rpn10 appears to play an essential role for embryonic development in mammals.
Like the budding yeast Rpn10 (van Nocker et al., 1996a; Kominami et al., 1997) and the fission yeast Pus1 (Wilkinson et al., 2000), Rpn10e has only one multi-Ub-binding site corresponding to PUbS1 of human S5a (Young et al., 1998). Other eukaryotic homologs have a second possible multi-Ub-binding site, termed PUbS2, in their C-termini (Haracska and Udvardy, 1997; Fu et al., 1998; Young et al., 1998; Girod et al., 1999). Young et al. (1998) reported that PUbS1 and PUbS2 bind multi-Ub chains in a cooperative manner. However, Rpn10e and Rpn10a were equally effective in binding multiubiquitylated [125I]lysozyme (Figure 5A) and also in suppressing the degradation of the model substrate by the purified 26S proteasome (Figure 5B and C). This is consistent with the finding that PUbS2 is dispensable for multi-Ub chain binding in the Arabidopsis Mbp1 protein (Fu et al., 1998). In considering different effects of Rpn10a and Rpn10e on the degradation of cyclin B2 in Xenopus egg extracts (Figure 6A) however, the exact role of PUbS2 in higher eukaryotic Rpn10 remains to be uncovered.
The 26S proteasome has a highly conserved structure in a variety of species ranging from yeast to human (Tanaka, 1998). However, recent studies have uncovered that the structures of the proteasome are not identical in mammals and yeast. For example, γ-interferon (IFN-γ) induces replacement of three constitutive β-type subunits of the mammalian 20S proteasome by IFN-γ-inducible subunits, thereby producing ‘immunoproteasomes’ (reviewed in Tanaka and Kasahara, 1998). Furthermore, in mammals, IFN-γ induces the proteasome activator known as PA28, REG or the 11S regulator, consisting of two related subunits designated α and β (Tanaka and Kasahara, 1998; DeMartino and Slaughter, 1999; Rechsteiner et al., 2000), leading to the formation of ‘hybrid proteasomes’ containing PA28 at one end and PA700 at the other end of the 20S proteasome (Hendil et al., 1998; Tanahashi et al., 2000). These IFN-γ-inducible subunits are absent in yeast and appear to have emerged close to the origin of vertebrates to accommodate the need to produce antigenic peptides capable of binding to major histocompatibility complex class I molecules efficiently (Tanaka and Kasahara, 1998). The existence of multiple forms of Rpn10 in mammals, and its apparent absence in yeast may represent yet another example of adaptive changes.
Clearly, there are many unsolved questions regarding the biological functions of Rpn10 and the significance of the multiplicity of Rpn10. However, the existence of multiple Rpn10 isoforms with distinct biological activities suggests that multi-Ub chains may not always be recognized by the 26S proteasome through the same mechanism, but rather multiple pathways may operate for targeting of substrates to the 26S proteasome. We favor the hypothesis that substrate discrimination occurs also at the level of the 26S proteasome by temporally and spatially regulated expression of the Rpn10 family proteins. Accordingly, it is of obvious interest to examine whether the remaining three isoforms of Rpn10—Rpn10b, Rpn10c and Rpn10d—have specialized functions in early embryonic development.
Materials and methods
Cloning of mouse and human Rpn10 family cDNAs
Full-length cDNAs were amplified by PCR from the mouse cDNA libraries made from the following sources: adult testis, embryonic stem cells, and 8.5 and 16 day whole embryos. Human Rpn10e cDNA was isolated from a fetal brain cDNA library.
Cloning and mapping of the mouse Rpn10 (Psmd4) gene
The Psmd4 gene was isolated by screening a λ FIX II genomic library of 129/SvJ mice (Stratagene) using the full-length Rpn10a cDNA as a probe. Gene mapping by PCR was performed as described previously (Noguchi et al., 1995) using the [(C57BL/6J × M.spretus)F1 × C57BL/6J] (abbreviated as BSB) and [(C57BL/6Ei × SPRET/Ei)F1 × SPRET/Ei] (abbreviated as BSS) interspecific backcross DNA panels obtained from The Jackson Laboratory, Bar Harbor, ME.
Biochemical analyses
Northern blot hybridization was performed with a probe specific for Rpn10a or Rpn10e using the standard protocol. Hydrolysis of the synthetic peptide succinyl-Leu-Leu-Val-Tyr-7-amino-4-methylcoumarin (Suc-LLVY-AMC) was measured as described previously (Kanayama et al., 1992). For sedimentation velocity analysis, samples (10 mg) of tissue extracts were subjected to glycerol density-gradient centrifugation with 10–40% glycerol. After centrifugation at 83 000 g for 22 h, the gradient was separated into 30 fractions of 1 ml each (Kanayama et al., 1992). Western blot analysis was performed using an anti-peptide antibody specific for S5a or p58 and a monoclonal antibody, mAb2-17, specific for the C2 subunit of the human 20S proteasome (Kumatori et al., 1990). The anti-peptide antibodies were raised by immunizing rabbits with 37CHSKTRSNPENNVGLITLAN56 for S5a and 514CREQQDLEF AKEMAED529 for p58. Immunoprecipitation of the 26S proteasome was conducted using an antibody specific for the p45 ATPase subunit of the human 26S proteasome (Hendil et al., 1998) and protein A–Sepharose 4B. The binding assay of multiubiquitylated 125I-labeled lysozyme was carried out with recombinant His-tagged Rpn10 proteins purified by a Ni2+-agarose column as previously reported (Deveraux et al., 1994; Young et al., 1998). Proteins separated by SDS–PAGE were electrophoretically transferred to a membrane, which was then subjected to far-western blot analysis with multiubiquitylated or free 125I-labeled lysozyme. Purification of the rat 26S proteasome and an assay of degradation of multiubiquitylated [125I]lysozyme were performed as described previously (Kanayama et al., 1992).
Assay of cell-cycle progression in Xenopus egg extracts
Unfertilized Xenopus eggs were crushed with EGTA at the second meiotic metaphase to prepare cytostatic factor (CSF)-arrested extracts (Lorca et al., 1993). Initiation of mitosis was triggered with Ca2+ using the extracts supplemented with or without recombinant Rpn10 proteins (Lorca et al., 1993). Cyclin B2 degradation was monitored by western blot analysis with an antibody specific for cyclin B2 (Hartley et al., 1996). Cell-cycle extracts were prepared from Ca2+ ionophore-activated eggs at 0°C. The extracts, with or without recombinant Rpn10 proteins, were incubated at 23°C. After incubation, activation of the Cdc2 kinase and chromosome condensation were monitored as previously described (Hartley et al., 1996).
The nucleotide sequence data reported in this paper will appear in the DDBJ/EMBL/GenBank Nucleotide Sequence Databases with the following accession Nos: Rpn10a cDNA (AB029142), Rpn10b cDNA (AB029143), Rpn10c cDNA (AB029144), Rpn10d cDNA (AB029145), Rpn10e cDNA (AB029146), human Rpn10e cDNA (AB033605) and the mouse Rpn10 (Psmd4) gene (AB029090).
Acknowledgments
Acknowledgements
This work was supported in part by Grants-in-Aid for Scientific Research on Priority Areas (Intracellular Proteolysis) from the Ministry of Education, Science, Sports and Culture of Japan.
References
- Bochtler M., Ditzel,L., Groll,M., Hartmann,C. and Huber,H. (1999) The proteasome. Annu. Rev. Biophys. Biomol. Struct., 28, 295–317. [DOI] [PubMed] [Google Scholar]
- Braun B.C., Glickman,M., Kraft,R., Dahlmann,B., Kloetzel,P.M., Finley,D. and Schmidt,M. (1999) The base of the proteasome regulatory particle exhibits chaperone-like activity. Nature Cell Biol., 1, 221–226. [DOI] [PubMed] [Google Scholar]
- Coux O., Tanaka,K. and Goldberg,A.L. (1996) Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem., 65, 801–847. [DOI] [PubMed] [Google Scholar]
- DeMartino G.N. and Slaughter,C.A. (1999) The proteasome, a novel protease regulated by multiple mechanisms. J. Biol. Chem., 274, 22123–22126. [DOI] [PubMed] [Google Scholar]
- Deveraux Q., Ustrell,V., Pickart,C. and Rechsteiner,M. (1994) A 26S protease subunit that binds ubiquitin conjugates. J. Biol. Chem., 269, 7059–7061. [PubMed] [Google Scholar]
- Deveraux Q., van Nocker,S., Mahaffey,D., Vierstra,R. and Rechsteiner,M. (1995) Inhibition of ubiquitin-mediated proteolysis by the Arabidopsis 26S protease subunit S5a. J. Biol. Chem., 270, 29660–29663. [DOI] [PubMed] [Google Scholar]
- Ferrell K., Deveraux,Q., van Nocker,S. and Rechsteiner,M. (1996) Molecular cloning and expression of a multiubiquitin chain binding subunit of the human 26S protease. FEBS Lett., 381, 143–148. [DOI] [PubMed] [Google Scholar]
- Ferrell K., Wilkinson,C.R.M., Dubiel,W. and Gordon,C. (2000) Regulatory subunit interactions of the 26S proteasome, a complex problem. Trends Biochem. Sci., 25, 83–88. [DOI] [PubMed] [Google Scholar]
- Finley D. et al. (1998) Unified nomenclature for subunits of the Saccharomyces cerevisiae proteasome regulatory particle. Trends Biochem. Sci., 23, 244–245. [DOI] [PubMed] [Google Scholar]
- Fu H., Sadis,S., Rubin,D.M., Glickman,M., van Nocker,S., Finley,D. and Vierstra,R.D. (1998) Multiubiquitin chain binding and protein degradation are mediated by distinct domains within the 26S proteasome subunit Mcb1. J. Biol. Chem., 273, 1970–1981. [DOI] [PubMed] [Google Scholar]
- Girod P.A., Fu,H., Zryd,J.P. and Vierstra,R.D. (1999) Multiubiquitin chain binding subunit MCB1 (RPN10) of the 26S proteasome is essential for developmental progression in Physcomitrella patens. Plant Cell, 11, 1457–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman M.H., Rubim,D.M., Coux,O., Wefes,I., Pfeifer,G., Cjeka,Z., Baumeister,W., Fried,V.A. and Finley,D. (1998) A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9 signalosome and elF3. Cell, 94, 615–623. [DOI] [PubMed] [Google Scholar]
- Haracska L. and Udvardy,A. (1995) Cloning and sequencing a non-ATPase subunit of the regulatory complex of the Drosophila 26S protease. Eur. J. Biochem., 231, 720–725. [DOI] [PubMed] [Google Scholar]
- Haracska L. and Udvardy,A. (1997) Mapping the ubiquitin-binding domains in the p54 regulatory complex subunit of the Drosophila 26S protease. FEBS Lett., 412, 331–336. [DOI] [PubMed] [Google Scholar]
- Hartley R.S., Rempel,R.E. and Maller,J.L. (1996) In vivo regulation of the early embryonic cell cycle in Xenopus. Dev. Biol., 173, 408–419. [DOI] [PubMed] [Google Scholar]
- Hendil K.B., Khan,S. and Tanaka,K. (1998) Simultaneous binding of PA28 and PA700 activators to 20S proteasomes. Biochem. J., 332, 749–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A. and Ciechanover,A. (1998) The ubiquitin system. Annu. Rev. Biochem., 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Kanayama H., Tamura,T., Ugai,S., Kagawa,S., Tanahashi,N., Yoshimura,T., Tanaka,K. and Ichihara,A. (1992) Demonstration that a human 26S proteolytic complex consists of a proteasome and multiple associated protein components and hydrolyzes ATP and ubiquitin-ligated proteins by a closely linked mechanism. Eur. J. Biochem., 206, 567–578. [DOI] [PubMed] [Google Scholar]
- Kominami K. et al. (1997) Yeast counterparts of subunits S5a and p58 (S3) of the human 26S proteasome are encoded by two multicopy suppressors of Nin1-1. Mol. Biol. Cell, 8, 171–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumatori A., Tanaka,K., Inamura,N., Sone,S., Ogura,T., Matsumoto,T., Tachikawa,T., Shin,S. and Ichihara, A. (1990) Abnormally high expression of proteasomes in human leukemic cells. Proc. Natl Acad. Sci. USA. 87, 7071–7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez A.J. (1998) Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet., 32, 279–305. [DOI] [PubMed] [Google Scholar]
- Lorca T., Cruzalegui,F.H., Fesquet,D., Cavadore,J.C., Mery,J., Means,A. and Doree,M. (1993) Calmodulin-dependent protein kinase II mediates inactivation of MPF and CSF upon fertilization of Xenopus eggs. Nature, 366, 270–273. [DOI] [PubMed] [Google Scholar]
- Noguchi M., Kitabatake,A., Ishibashi,T. and Kasahara,M. (1995) The MHC class I-like Zn-α2-glycoprotein gene maps to mouse chromosome 5. Immunogenetics, 42, 72–74. [DOI] [PubMed] [Google Scholar]
- Pickart C.M. (1998) Targeting of substrates to the 26S proteasome. FASEB J., 11, 1055–1066. [DOI] [PubMed] [Google Scholar]
- Pusch W., Jahner,D. and Ivell,R. (1998) Molecular cloning and testicular expression of the gene transcript encoding the murine multiubiquitin-chain-binding protein (Mcb1). Gene, 207, 19–24. [DOI] [PubMed] [Google Scholar]
- Realini C., Rogers,S.W. and Rechsteiner,M. (1994) KEKE motif. FEBS Lett., 348, 109–113. [DOI] [PubMed] [Google Scholar]
- Rechsteiner M. (1998) The 26S proteasome. In Peters,J.-M., Harris,J.R. and Finley,D. (eds), Ubiquitin and the Biology of the Cell. Plenum Press, New York, NY, pp. 147–189. [Google Scholar]
- Rechsteiner M., Realini,C. and Ustrell,V. (2000) The proteasome activator 11S REG (PA28) and class I antigen presentation. Biochem. J., 345, 1–15. [PMC free article] [PubMed] [Google Scholar]
- Rowe L.B., Nadeau,J.H., Turner,R., Frankel,W.N., Letts,V.A., Eppig,J.T., Ko,M.S., Thurston,S.J. and Birkenmeier,E.H. (1994) Maps from two interspecific backcross DNA panels available as a community genetic mapping resource. Mamm. Genome, 5, 253–274. [DOI] [PubMed] [Google Scholar]
- Schwartz A.L. and Ciechanover,A. (1999) The ubiquitin–proteasome pathway and pathogenesis of human diseases. Annu. Rev. Med., 50, 57–74. [DOI] [PubMed] [Google Scholar]
- Tanahashi N., Murakami,Y., Minami,Y., Shimbara,N., Hendil,K.B. and Tanaka,K. (2000) Hybrid proteasomes: induction by interferon-γ and contribution to the ATP-dependent proteolysis. J. Biol. Chem., 275, 14336–14345. [DOI] [PubMed] [Google Scholar]
- Tanaka K. (1998) Molecular biology of proteasomes. Biochem. Biophys. Res. Commun., 247, 537–541. [DOI] [PubMed] [Google Scholar]
- Tanaka K. and Kasahara,M. (1998) The MHC class I ligand generating system: roles of immunoproteasomes and INF-γ-inducible PA28. Immunol. Rev., 163, 161–176. [DOI] [PubMed] [Google Scholar]
- Thrower J.S., Hoffman,L., Rechsteiner,M. and Pickart,C.M. (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J., 19, 94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nocker S., Sadis,S., Rubin,D.M., Glickman,M., Fu,H., Coux,O., Wefes,I., Finley,D. and Vierstra,R.D. (1996a) The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol. Cell. Biol., 16, 6020–6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nocker S., Deveraux,Q., Rechsteiner,M. and Vierstra,R.D. (1996b) Arabidopsis MBP1 gene encodes a conserved ubiquitin recognition component of the 26S proteasome. Proc. Natl Acad. Sci. USA, 93, 856–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voges D., Zwickl,P. and Baumeister,W. (1999) The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem., 68, 1015–1068. [DOI] [PubMed] [Google Scholar]
- Wilkinson C.R.M., Ferrell,K., Penney,M., Wallace,M., Dubiel,W. and Gordon,C. (2000) Analysis of a gene encoding Rpn10 of the fission yeast proteasome reveals that the polyubiquitin-binding site of this subunit is essential when Rpn12/Mts3 activity is compromised. J. Biol. Chem., 275, 15182–15192. [DOI] [PubMed] [Google Scholar]
- Yanagawa Y., Ueda,T., Yamamato,K., Sasaki,T., Tanaka,K., Hashimoto,J., Sato,T. and Nakagawa,H. (1998) Cloning and sequencing of cDNA encoding a non-ATPase subunit homolog of Arabidopsis thaliana MBP1 from the Oriza sativa 26S proteasome. Plant Biotechnol., 15, 147–150. [Google Scholar]
- Young P., Deveraux,Q., Beal,R.E., Pickart,C.M. and Rechsteiner,M. (1998) Characterization of two polyubiquitin binding sites in the 26S protease subunit 5a. J. Biol. Chem., 273, 5461–5467. [DOI] [PubMed] [Google Scholar]