Abstract
The Wnt/β-catenin signaling pathway is frequently activated in hepatocellular carcinoma (HCC). Downstream signaling events involving the Wnt/β-catenin cascade occur through T-cell factor (TCF) proteins. The human TCF-4 gene is composed of 17 exons with multiple alternative splicing sites. However, the role of different TCF-4 isoforms in the pathogenesis of HCC is unknown. The purpose of this study was to identify and characterize TCF-4 isoforms in HCC. We identified 14 novel TCF-4 isoforms from four HCC cell lines. Functional analysis following transfection and expression in HCC cells revealed distinct effects on the phenotype. The TCF-4J isoform expression produced striking features of malignant transformation characterized by high cell proliferation rate, migration and colony formation even though its transcriptional activity was low. In contrast, the TCF-4K isoform displayed low TCF transcriptional activity; cell proliferation rate and colony formation were reduced as well. Interestingly, TCF-4J and TCF-4K differed by only five amino acids (the SxxSS motif). Thus, these studies suggest that conserved splicing motifs may have a major influence on the transcriptional activity and functional properties of TCF-4 isoforms and alter the characteristics of the malignant phenotype.
Keywords: TCF-4 isoform, HCC, Wnt
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors worldwide and the third most common cause of cancer mortality [1, 2]. Although the major etiologies of HCC are now well-defined and include chronic viral hepatitis B (HBV) and C (HCV), toxins and drugs, and metabolic liver diseases, the molecular mechanisms that contribute to tumor initiation or progression of HCC are poorly understood. Past studies revealed the presence of genetic alterations in the Wnt pathway as well as mutations in tumor suppressor genes such as p53, and retinoblastoma that are believed to be important in hepatic oncogenesis. However, there is increasing evidence that aberrantly activated Wnt signaling due to over-expression of components of this pathway such as Frizzled (FZD) receptors and Wnt ligands is a common early event in the molecular pathogenesis of this disease [3, 4].
Canonical Wnt signaling is mediated by cell surface FZD and lipoprotein receptor-related protein (LRP) receptors and their interaction promote Wnt signals [5]. In the presence of active Wnt signaling, β-catenin accumulates in the cytoplasm, translocates into the nucleus, and binds to T-cell factor/lymphoid enhancer factor (TCF/LEF) proteins to activate Wnt responsive target genes [6]. The TCF/LEF family in mammals is comprised of four different proteins: LEF-1, TCF-1, TCF-3, and TCF-4 [7, 8]. Several functional domains have been mapped on the TCF family of proteins as depicted as Fig. 1A. The high mobility group (HMG) DNA binding domain is the most highly conserved region of the TCF proteins. The second most highly conserved motif is a β-catenin binding domain at the N-terminus. Binding of β-catenin to a N-terminal domain of TCFs facilitates assembly of multimeric complexes containing transcriptional co-activators, such as CBP/p300 [9] and BCL9/Legless [10] and Pygopus [11–13], which can activate downstream target genes.
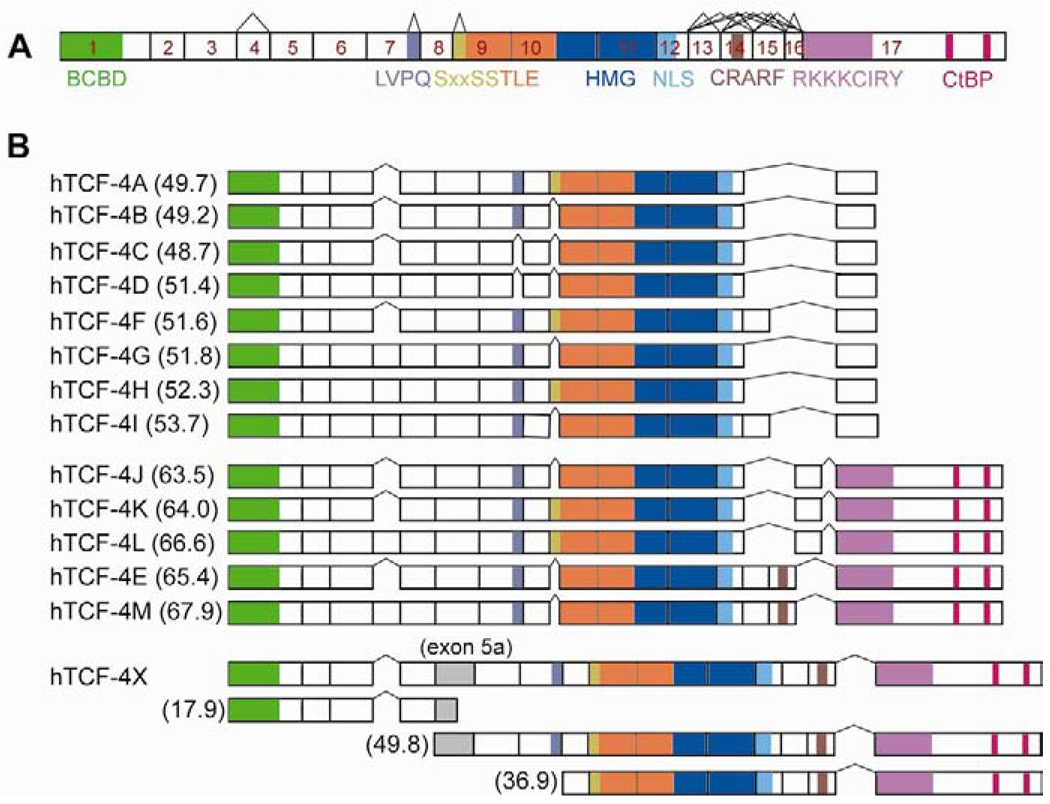
Fig. 1.
Identification of novel TCF-4 splicing variants in HCC cell lines. (A) A schematic representation of human TCF-4 gene (TCF7L2) comprising 17 exons based on reported cDNA sequences [16]. BCBD, N-terminal β-catenin binding domain; TLE, Groucho/TLE co-repressor binding site; HMG, high mobility group; NLS, nuclear localization signal; CtBP, C-terminal binding protein binding site. Alternatively splicing sites are exon 4, LVPQ and SxxSS motifs, and exon13–16. (B) Summary of the organizational structure of 14 TCF-4 isoforms. In HCC cell lines, 14 different TCF-4 mRNA splicing variants were identified by cloning and DNA sequencing. hTCF-4B and E were designated as short and long isoforms, respectively. The remaining 12 were named alphabetically based on their calculated molecular weight represented in parentheses (kDa). Note that hTCF-4X included an extra exon 5a, which may produce three different proteins.
Several investigations suggest that TCF-4 was highly expressed among the TCF family of proteins in HCC. Over-expression of TCF-4 mRNA was found in human HCC [14] and it has been reported that TCF-4/β-catenin complex formation was increased in transgenic mice that develop HCC [15]. The human TCF-4 gene (TCF7L2) is composed of 17 exons with several alternatively splicing sites including a C-terminal tail (exon13–17) and exon 4 [16]. In addition, alternative splicing in the central domain of TCF-4 is also important [17], and generates isoforms with or without the highly conserved LVPQ and SxxSS motifs, which are located at the end of exon 7 and beginning of exon 9, respectively [16–18].
Even though β-catenin is a main effector of Wnt signaling, the success of relaying signals to the nucleus depends on the presence of TCF-4 transcription factors. Thus, TCF-4 is equally important in mediating the final transcriptional program, and such activity may be regulated according to isoform diversity and abundance. Despite the fact that TCF proteins are bi-functional context-dependent regulators that bind DNA and act as either transcriptional activators or repressors (depending on their association with other proteins), the function of TCF transcription factors have been generally proposed to have activating properties that either upregulate proto-oncogenes or inhibit apoptosis [19–21]. Moreover, the role of different TCF-4 splicing variants in this process is unknown and there is no information on their function in HCC, which may be critical to understand the oncogenic process.
We suspect that TCF-4 isoform expression may be associated with different biologic properties of tumor cells depending on the isoforms generated by splicing events during hepatic transformation and they will influence development of the malignant phenotype. In the present study, we identified and cloned 14 different TCF-4 isoforms and characterized their biologic functions in HCC cell lines. These studies provide evidence that certain TCF-4 isoforms may function as either transcriptional activators or repressors, to produce distinct cellular phenotypes during hepatocarcinogenesis.
Materials and methods
Cloning of TCF-4 isoforms
Total cellular RNA was extracted from four human HCC cell lines (FOCUS, Huh7, Hep3B, and HepG2) using TRIzol reagent (Invitrogen, Carlsbad, CA). The quality of the RNA samples was determined by ethidium bromide-stained agarose gel electrophoresis to verify intact 18S and 28S rRNA bands. To obtain first-strand complementary DNA (cDNA), 500 ng of total RNA was treated with DNase-I, RNase inhibitor, and reverse transcribed with oligo-dT primer and AMV reverse transcriptase (Roche Diagnostics, Indianapolis, IN). For identifying different TCF-4 isoforms, PCR was carried out with a primer pair 1F (forward, 5’-CCGCTCGAGCGGATGCCGCAGCTGAACGGCGG-3’) and 17R (reverse, 5’-CGCGGATCCGCGCTATTCTAAAGACTTGGTGACGAGCGACAGCGGCT-3’) that span from the beginning of exon 1 to the end of exon 17. The PCR products from all four HCC cell lines were cloned into the pCR2.1 vector using a TOPO TA cloning kit (Invitrogen) and the individual clones were DNA sequenced. To verify sequences of the TCF-4 isoforms, a ClustalW2 program (http://www.ebi.ac.uk/Tools/clustalw2/index.html) was used to align sequences of all isoforms against the reference sequences as following: 1) The total genomic sequence of TCF-4 gene was obtained from the NCBI website under the name of TCF7L2 (transcription factor 7-like 2). 2) The cDNA sequences of TCF-4 mRNA were also acquired from the NCBI.
To generate TCF-4-myc mammalian expression plasmid, TCF-4 isoform cDNAs were subcloned into a pcDNA3.1/myc-HisB vector plasmid. All plasmids constructed were verified by sequencing. Dominant-negative form (dn) of TCF-4 and β-catenin expression plasmids were obtained from Millipore/Upstate (Billerica, MA) and OriGene (Rockville, MD), respectively.
Semi-quantitative reverse transcription (RT)-PCR
PCR was performed using eight different sets of splicing site-specific primers (P1-F/P1-R, P1-F/P2-R, P2-F/P1-R, P2-F/P2-R, P1-F/P3-R, P2-F/P3-R, P1-F/P4-R, P2-F/P4-R) shown in Figure 2A and Table 1. The PCR products generated by three primers sets producing multiple isoforms (P1-F/P1-R, P2-F/P1-R, P1-F/P3-R), were separated on a 3.5% NuSieve 3:1 agarose gel (Lonza Rockland, Inc., Rockland, ME). This analytic technique is capable of resolving DNA fragments from 10 to 1500 bp. Isoforms E and X generated from primers P1-F and P4-R were isolated on 2% agarose gel due to the size difference of their PCR products. Single PCR products were analyzed on 2% agarose gel. Each TCF-4 isoform plasmid (100 fmol) was amplified to generate PCR product and used as an internal calibrator for determining the expression level of TCF-4 isoform. As positive controls for such experiments, each TCF-4 isoform plasmid was transfected into Huh7 cells followed by RT-PCR.
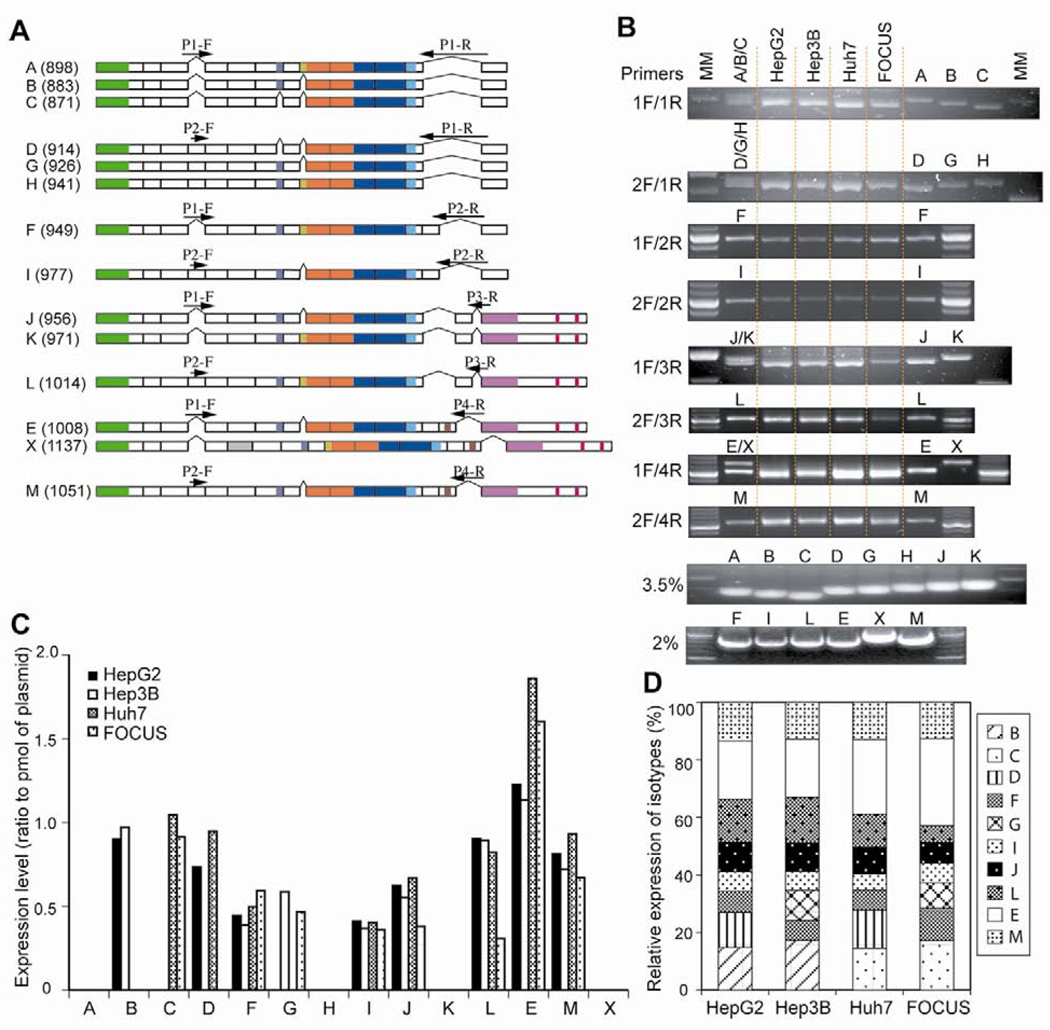
Fig. 2.
Expression of TCF-4 mRNA splicing variants in HCC cell lines. (A) Schematic diagram illustrating the positions of primer pairs for RT-PCR. Numbers in parentheses represent the size of PCR products (bp). (B) Agarose gel electrophoresis of PCR products stained by ethidium bromide. Each TCF-4 isoform plasmid was transfected into Huh7 cells and used as a positive control for RT-PCR. For multiple PCR products, positive controls were applied to lanes as both multiple (A/B/C) and single (A, B, and C) species. All of positive controls revealed the expected size of the PCR product. PCR products using 3 sets of primers which generated multiple isoforms (P1F/P1R, P2F/P1R, P1F/P3R), were separated on a 3.5% NuSieve 3:1 agarose gel, which is capable of resolving DNA fragments from 10 to 1500 bp. Isoforms E and X generated by primers P1F and P4R was isolated on 2% agarose gel due to the size difference of their PCR products. Single PCR products were also analyzed on 2% agarose gels. A defined amount (100 fmol) of each TCF-4 isoform plasmid DNA was subjected to PCR and separated on 3.5 or 2% agarose gels. The intensity of the PCR product was used as an internal calibrator for the expression level of each isoform. (C) Expression level of 14 TCF-4 isoforms in HCC cell lines. TCF-4 expression in each sample is represented as a ratio related to the TCF-4 plasmid DNA (pmol). (D) Relative expression pattern of these isoforms in 4 different HCC cell lines. TCF-4E (long form) was abundant in all of HCC cell lines.
Table 1.
Primers sequences used for RT-PCR
| Primer | Sequence (5’-3’) |
|---|---|
| P1-F | CCCGAACCTATCTCCAGATG |
| P1-R | TTTTCTCCATTGGTCTCTCC |
| P2-F | AGCACACATTACTCTGCGTA |
| P2-R | TTTTCTCCTGTAATCGGA |
| P3-R | TTTTCTCCTGCACGGTTTG |
| P4-R | TTTTTCTCCTGCAAGGGC |
| GAPDH-F | CTTAGCACCCCCTGGCCAAG |
| GAPDH-R | GATGTTCTGGAGAGCCCCG |
Cell culture and transfection studies
Human HCC cell lines (FOCUS, Huh7, Hep3B, and HepG2) were propagated in DMEM with 10% fetal bovine serum (FBS). For transient transfection, cells were plated in either 6-well or 12-well plates and plasmids were transfected by a TransIT-LT1 transfection reagent (Mirus Bio Co., Madison, WI). To establish Huh7 HCC cells stably expressing TCF-4 isoforms, we transfected TCF-4B, J, and K, or empty vector as a control, followed by selection of colonies using 400 µg/mL of geneticin (G418, Invitrogen) and expanded for further characterization.
For analysis of TCF transcriptional activity, we employed a TOPFlash/FOPFlash reporter gene assay as previously described [4]. In brief, each TCF-4 expressing plasmid was co-transfected with TOPFlash or FOPFlash, and β-galactosidase in the presence or absence of a β-catenin expression construct into HEK293 and Huh7 HCC cells. The β-galactosidase activity was used for normalization of transfection efficiency. At 48 h post-transfection, the luciferase activity was measured and the relative transcriptional activity was determined by the ratio of TOP value to FOP basal activity.
Cyclin D1 promoter activity was measured with cyclin D1 promoter reporter plasmids (−163CD1Luc, −163ΔLefCD1Luc) [22], which were kindly provided by Dr. Pestell (Thomas Jefferson University). Briefly, −163CD1LUC or −163ΔLefCD1LUC (a negative control) was co-transfected with β-galactosidase in the presence or absence of a β-catenin expression plasmid into Huh7 cells stably expressing TCF-4B, J or K isoform. At 48 h after transfection, the luciferase activity was measured and the relative activity determined by the ratio of −163CD1Luc to −163ΔLefCD1Luc basal value.
To examine protein expression of TCF-4 isoforms and β-catenin, Western blot analysis was performed as previously described [23] using anti-TCF-4 (Upstate/Millipore, Billerica, MA), anti-Myc-tag, anti-β-catenin (Cell Signaling, Danvers, MA), anti-HA (BD Transduction Lab), and anti-actin antibodies (Santa Cruz).
Cell proliferation assay
Cell growth rate was measured by using the CellTiter 96 AQeous Non-Radioactive Cell Proliferation Assay (Promega, Madison, WI). Briefly, Huh7 cells were co-transfected with each of the TCF-4 isoform expression plasmid or empty vector and β-catenin expressing plasmid. Twenty-four h after transfection, the cells were plated in 96-well plates at a density of 2000 cells/well. Cell proliferation was measured with a combined MTS/PMS solution (Promega), a colorimetric method for determining viable proliferating cells. Results are presented as the average absorbance of six wells in one experiment and reported as means of three independent assays.
Wound healing and transwell cell motility assays
Cell migration was assessed using a scratch wound assay [4]. In brief, a scratch was made using a sterile micropipette tip and cells were washed to remove floating cells and debris. The wound closure, as an index of cell migration, was photographed at the indicated times from the same area. Phase-contrast images were acquired with a MicroFire® Microscope Digital CCD Camera (Optronics, Goleta, CA) under an Olympus IX70 fluorescence microscope (DSC Optical Services, Newton Centre, MA) using Picture Frame software (Optronics). The mean wound area was calculated using Image J software. Results were expressed as a percent of wound closure normalized to wound width at time 0 h. Results were derived from three independent measurements.
Cell motility was evaluated using a luminescence-based assay as described previously with some modifications [24]. In brief, Huh7 cells (1×105) were seeded into the upper chamber of 12-well plate separated by a Transparent PET Membrane (BD Biosciences, San Jose, CA). Migrated cells were harvested with a cotton swab and luminescence was measured after incubation with ATPLite substrate (PerkinElmer, Waltham, MA). The results are expressed as a percent of migrated to total number of cells and the experiments were performed in triplicate.
Colony formation in soft-agar
Anchorage independent colony growth was assessed in soft-agar assay using Cell Transformation Detection Assay kit (Chemicon/Millipore, Billerica, MA) according to the manufacture’s instruction. In brief, cells (2.5 × 104) were suspended in 0.4% top agar over a bottom layer of 0.8% base agar in 6-well plates. The solidified soft-agar was overlaid with DMEM media containing 10% FBS. The media was changed every 4–5 days. Colonies were visualized by staining with cell stain solution, and the number and size of colonies analyzed using Image J software.
Statistical Analysis
Statistic for dual comparisons was generated using Student’s t tests assuming unequal variances. A P value less than 0.05 was considered to be statistically significant in all analyses. The results were reported as mean ± SD or SE.
Results
Identification of novel TCF-4 isoforms in HCC cell lines
To identify different TCF-4 mRNA isoforms, we performed RT-PCR using primers specific for exon 1 (forward) and exon 17 (reverse) of the human TCF-4 gene (TCF7L2) in four HCC cell lines (HepG2, Hep3B, Huh7, and FOCUS). The PCR products were cloned into the pCR2.1-TOPO vector using a TA cloning kit and the individual clones were sequenced. DNA sequencing results revealed 14 different TCF-4 isoforms that were expressed in HCC cell lines as depicted in Fig. 1B. Two previously known species designated as short (TCF-4B) and long (TCF-4E) forms [25] were identified among the 14 isoforms. These 12 new TCF-4 isoforms were named in alphabetical order according to the calculated size of their protein product. They showed several distinct features: 1) six isoforms were found to include exon 4, 2) six others include exon 9L, which encodes for a SxxSS motif that has not been previously described in human tumors, 3) three isoforms transcribed exon 15, and 4) hTCF-4X (X for unknown) which includes an extra exon 5a at intron/exon boundaries embedded within intron 5. This hTCF-4X mRNA can generate protein products of various sizes (17.9, 49.8 and 36.9 kDa) due to insertion of exon 5a as demonstrated in Fig. 1B. Although exon 16 has been described in colorectal cancer (CRC) cell lines [16], we did not find any evidence of this isoform in HCC cell lines.
Using the same cloning approach, we also identified and characterized TCF-4 isoforms derived from 5 normal human liver samples. In this regard, all clones sequenced represented the single isoform of TCF-4B.
Expression profiles of TCF-4 isoforms in HCC cell lines
Semi-quantitative RT-PCR was performed to confirm the expression of TCF-4 isoforms in four cell lines. Since these isoforms were generated from multiple splicing sites, it was not possible to detect a single isoform by RT-PCR (such as TCF-4A, B, and C). Thus, we employed RT-PCR using eight different sets of site-specific primers at potential splicing domains as shown in Fig. 2A (P1-F/P1-R, P1-F/P2-R, P2-F/P1-R, P2-F/P2-R, P1-F/P3-R, P2-F/P3-R, P1-F/P4-R, P2-F/P4-R) and analyzed products on agarose gels as described in Materials and methods. For multiple PCR products, positive controls were applied to lanes as multiple (A/B/C) and single (A, B, and C) species. All positive controls revealed the expected size of PCR product. As shown in Fig. 2B and C, we found that expression of 10 isoforms were detected in the 4 different HCC cell lines studied. TCF-4A, H, K and X isoforms were undetectable by RT-PCR, possibly due to low expression levels. The TCF-4E, J, and M isoforms were abundant in all HCC cell lines studied. There was different pattern of expression in four HCC cell lines according to degree of HCC differentiation, i.e. HepG2 > Hep3B > Huh7 > FOCUS [24]. TCF-4B was detectable only in well differentiated HepG2 and Hep3B cell lines, whereas TCF-4C was found in poorly differentiated Huh7 and FOCUS cells.
Analysis of β-catenin dependent-TCF transcriptional activity
To evaluate the functional properties of each TCF-4 isoform in HCC cell line, we generated TCF-4-myc plasmids using pcDNA3.1/myc-His vector; protein expression was confirmed by Western blot analysis after transient transfection into HEK293 and Huh7 cells. The TCF-4X isotype was expressed as one (49.8 kDa) of the predicted proteins, which included exon5a.
Since the TCF-4 isoforms expressed the predicted size proteins, we assessed their transcriptional activity using the TOPFlash reporter assay in HEK293 and Huh7 HCC cells. To exclude the effect of different expression levels of TCF-4 isoforms and/or β-catenin on TCF transcriptional activity, transfection was carried out in duplicate and Western blot analysis and TCF activity assay were performed simultaneously. As shown in Fig. 3, the expression level of 14 TCF-4 isoforms was comparable as detected with both anti-Myc-tag and anti-TCF-4 antibodies. The dominant negative form (dn) of TCF-4 was detected with both anti-HA and anti-TCF-4 antibodies. The expression level of β-catenin was constant in all samples tested including dnTCF-4 and empty vector transfected cells indicating that there was no variation of β-catenin expression levels to effect TCF transcriptional activity. Nevertheless, TCF-4/β-catenin-dependent transcriptional activity exhibited different are interesting features (Fig. 4A) as following: 1) Each TCF-4 isoform exhibited the same pattern of transcriptional activity in two different cell lines (HEK293 and Huh7 HCC cells). 2) The transcriptional activity was increased with β-catenin expression (black bars) as compared to transfections that lack β-catenin expression (white bars). 3) TCF-4B, C, and G isoforms revealed higher transcriptional activity than other isoforms. Interestingly, many but not all of the isoforms displayed lower activity than the empty vector control (EV) and may represent repressive isoforms that inhibit TCF transcriptional activity. These observations support the hypothesis that some of TCF-4 isoforms may have a role as transcriptional repressors in HCC.
Fig. 3.
Ectopic expression of TCF-4 isoforms and β-catenin in HEK293 and Huh7 cells. Each isoform-containing plasmid was co-transfected with TOP, β-galactosidase, and β-catenin plasmids. Representative immunoblot results from HEK293 (A) and Huh7 (B) cell lines. TCF-4 isoforms containing Myc-tag at the C-terminus were detected with anti-Myc-tag or anti-TCF-4 antibodies (*). A dominant negative form (dn) of TCF-4 (HA-tag at the N-terminus) was detected by anti-HA or anti-TCF-4 antibodies (**). Actin serves as the protein loading control.
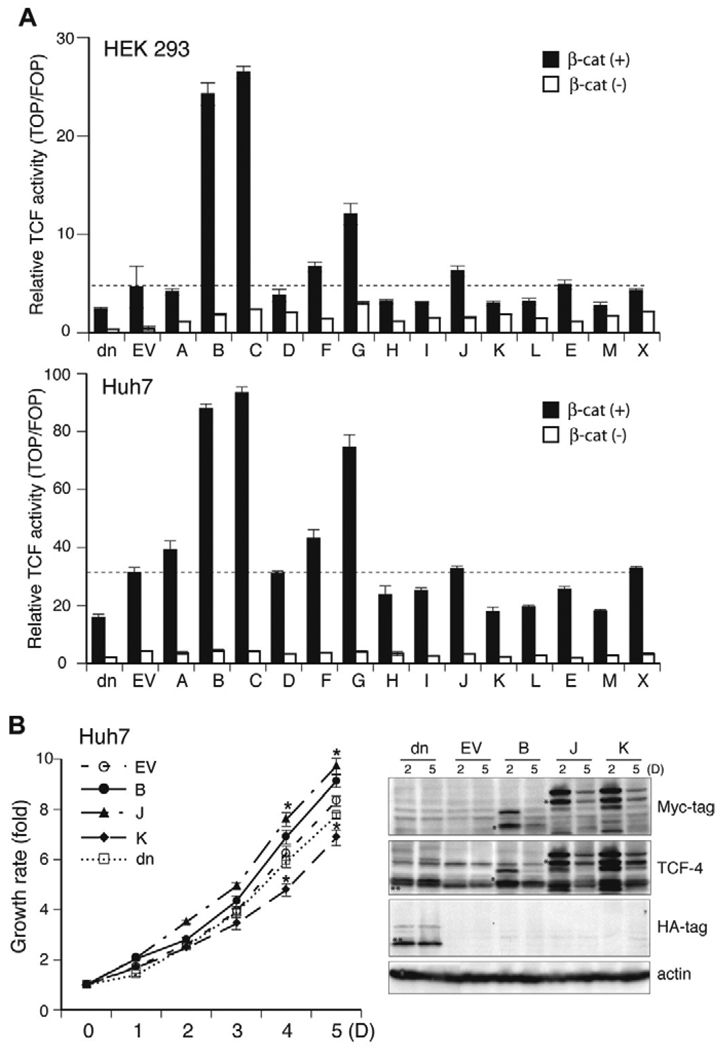
Fig. 4.
Effects of TCF-4 isoforms on TCF transcriptional activity and Huh7 cell proliferation. (A) TCF Transcriptional activities of 14 distinct TCF-4 isoforms after transient transfection in HEK293 and Huh7 HCC cells. Activity was measured using a TOPFlash luciferase reporter gene system. Each isoform-containing plasmid was co-transfected with TOP or FOP, and β-galactosidase, with/without β-catenin plasmids. Luciferase activity was measured using the Luciferase Assay System (Promega), and relative luciferase activity was represented by the ratio of TOP value to FOP after normalization of transfection efficiency with β-galactosidase activity. A dominant negative form (dn) of TCF, which is a N-terminal deletion mutant lacking the β-catenin binding domain was used as a control for assessment of repressed TCF transcriptional activity. (B) Cell proliferation rate of Huh7 HCC cells over-expressing TCF-4 isoforms after transient transfection. Each TCF-4 isoform expressing plasmid was transiently co-transfected with β-catenin into Huh7 cells and growth rate was measured for 5 days using CellTiter 96 AQeous Non-radioactive cell proliferation assay. Empty vector (EV) and dominant negative (dn) TCF-4 expression plasmids were used as controls. Note that TCF-4J isoform produced highest cell growth rate, whereas TCF-4K reduced the cell growth of Huh7 cells even further compared to empty vector (EV) and dnTCF-4 controls. Results shown are the means ± S.D.; * p < 0.05 versus control (EV) (n = 3 independent experiments, assayed in quadruplicate). (Right panel) Expression levels of dnTCF-4, TCF-4B, J and K at days 2 and 5 post-transfection. TCF-4B, J and K isoforms containing Myc-tag at the C-terminus were detected with anti-Myc-tag and anti-TCF antibodies (*). A dominant negative form (dn) of TCF-4 (HA-tag at the N-terminus) was detected by anti-HA and anti-TCF-4 antibodies (**). Actin serves as the protein loading control. Note that protein expression was still detected at 5 day post-transfection.
The effect of TCF-4 splicing isoforms on cell proliferation
Next, the effect of TCF-4 isoforms on HCC cell proliferation was examined in Huh7 cells. Each TCF-4 isoform expression plasmid was transiently co-transfected with β-catenin into Huh7 cells and growth rate was measured over 5 days. Empty vector (EV) and dnTCF-4 expression plasmids were used as controls. As shown by a representative example presented in Fig. 4B, TCF-4B, which has enhanced TCF transcriptional activity (Fig. 4A), increased cell proliferation compared to empty vector control. In contrast, the TCF-4J isoform produced the highest cell growth rate but had lower transcriptional activity than TCF-4B. Moreover, TCF-4K reduced the cell growth of Huh7 cells even further compared to empty vector and dnTCF-4 controls.
Analysis of TCF-4B, J and K isoforms in relation to HCC cell migration and colony formation
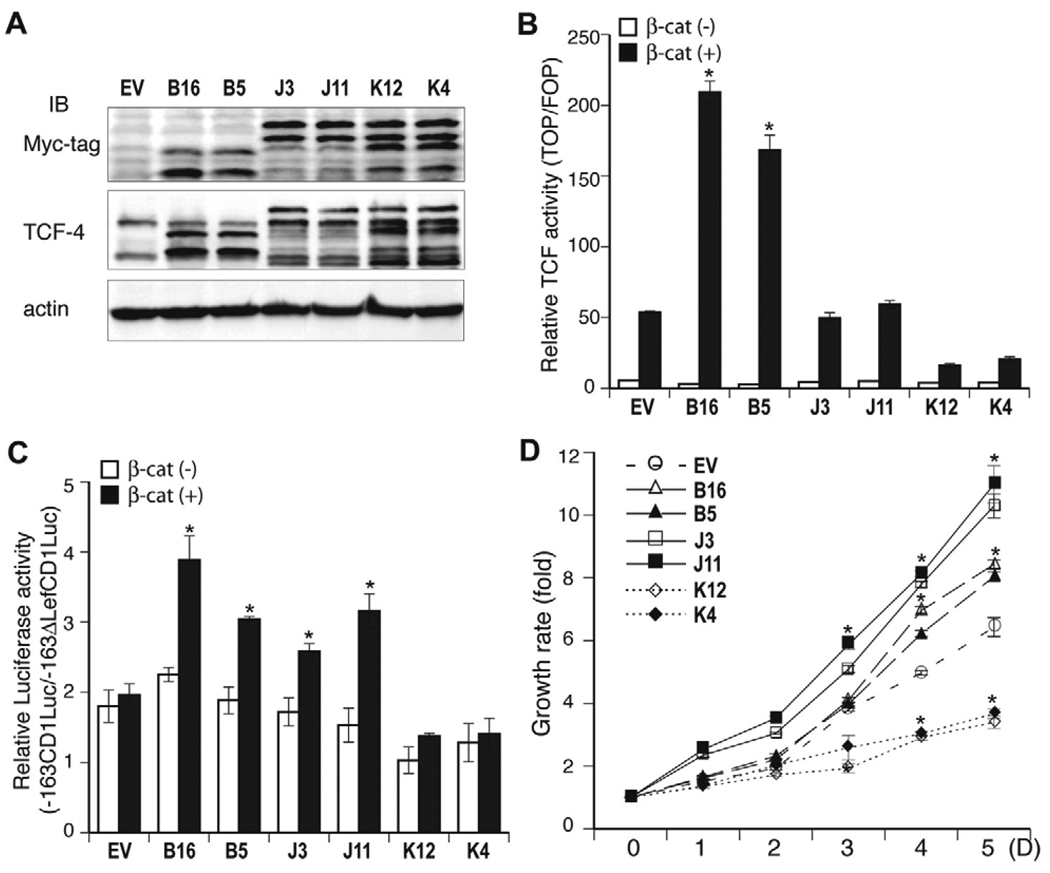
The observation that TCF-4B, J, and K exhibited different transcriptional activities and cell proliferation rates, led us to consider that these isoforms may have different biologic functions and influence the phenotype of HCC cells. To further explore the properties of these isoforms, we generated stable Huh7 clones which over-expressed TCF-4B, J, and K. To exclude clonal artifacts, two different stable clones representative of each isoform were used for analysis. An empty vector (EV) was also stably transfected into Huh7 cells and used as a control. Expression of TCF-4B, J and K isoform was confirmed by Western blot analysis (Fig. 5A). TCF transcriptional activity (Fig. 5B) exhibited by Huh7-stable expression clones was consistent with that of transient transfection experiments. Despite low TCF transcriptional activity, transient expression of TCF-4J resulted in enhanced cell proliferation suggesting that TCF-4J may activate target genes related to cell proliferation. In this regard, we tested whether TCF-4J had effect on cyclin D1 gene promoter activation. As shown in Fig. 5C, TCF-4J-overexpressing Huh7 cells (J3, J11) increased cyclin D1 promoter activity compared to control and cyclin D1 promoter activity was correlated with cell proliferation (Fig. 5D). In contrast, TCF-4K revealed a lower proliferation rate and low TCF and cyclin D1 transcriptional activities. These results raise the possibility that different TCF-4 isoforms may activate different target genes and produce effects on HCC cell proliferation that may not be directly related to the magnitude of TCF transcriptional activity on artificial reporter genes used in transient transfection assays.
Fig. 5.
TCF and cyclin D1 transcriptional activities and cell proliferation of Huh7-stable clones over-expressing TCF-4B, J, and K isoforms. (A) Expression of TCF-4B, J, and K were detected by anti-Myc-tag and anti-TCF-4 antibodies in stable Huh7 clones of TCF-4B (B16, B5), TCF-4J (J3, J11) and TCF-4K (K12, K4). (B) TCF transcriptional activity was measured in Huh7-stable clones over-expressing empty vector (EV), B (B16, B5), J (J3, J11), and K (K12, K4) isoforms. Note that activity of Huh7 cells over-expressing these isoforms was consistent with the findings after transient transfection. Data shown are the means ± S.D.; * p < 0.05 versus control (EV) (n = 3 independent experiments, assayed in triplicate) (C) Cyclin D1 promoter activity was measured in Huh7-stable clones. Cyclin D1 promoter reporter (−163CD1Luc) was co-transfected with β- galactosidase in the presence or absence of β-catenin followed by measurement of luciferase activity. A plasmid (−163ΔLefCD1Luc) was used as a negative control and activity was determined as a ratio of −163CD1LUC to −163ΔLefCD1LUC. Note that expression of TCF-4B and J increased cyclin D1 transcriptional activity. Results represent the mean ± S.D.; * p < 0.05 versus control (EV) (n = 3 independent experiments, assayed in triplicate) (D) Cell proliferation assay results of stable Huh7 clones expressing TCF-4B, J, and K. TCF-4B (B16, B5) and J (J3, J11) isoforms show an enhanced growth rate, while TCF-4K (K12, K4) reveals slower cell growth, which is lower than that found with the control (EV). Results are expressed as the mean ± S.D.; * p < 0.001 versus control (EV) (n = 3 independent experiments, assayed in quadruplicate).
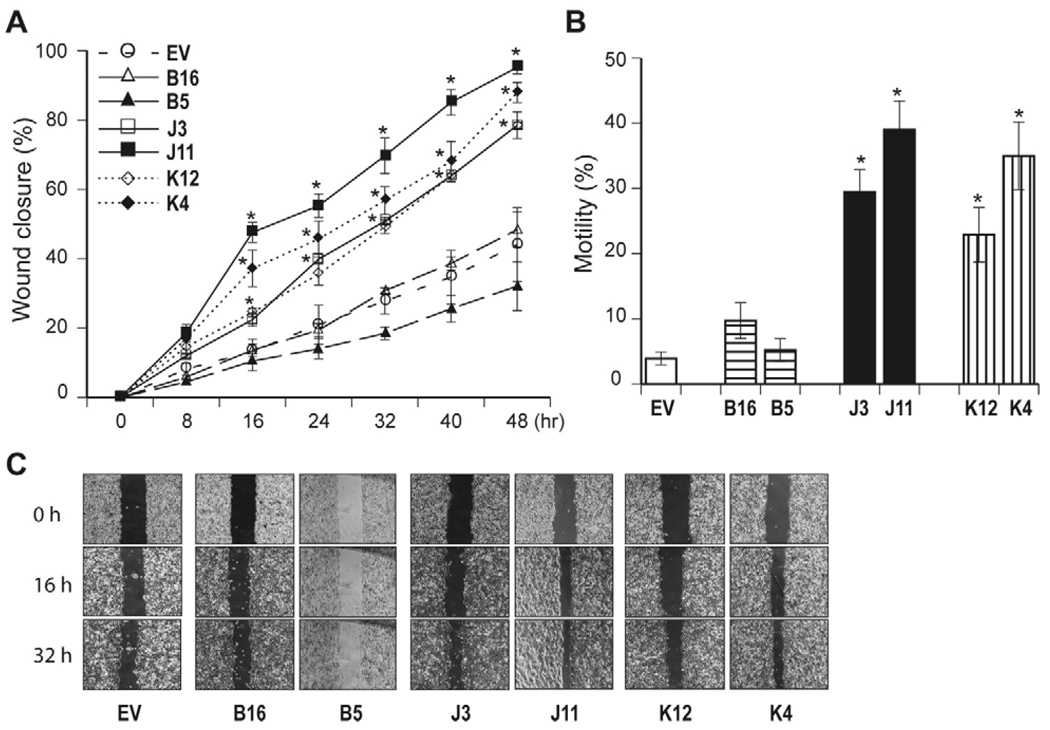
The effects of TCF-4B, J, and K on HCC cell migration and motility were then examined. As shown in Fig. 6A and C, Huh7 stable cell lines over-express both TCF-4J and K isoforms resulted in increased cell migration as assessed by a Wound healing assay. In contrast, TCF-4B revealed no difference of cell migration compared to control. The findings regarding TCF-4B, J and K on cell motility as assessed by a second luminescence-based assay was consistent with the observations made with wound closure assay (Fig. 6B).
Fig. 6.
The effect of TCF-4B, J and K on HCC cell migration and motility. (A) A wound healing assay was performed in Huh7 cells to evaluate migration. Cell migration of stable TCF-4B (B16, B5), J (J3, J11), and K (K12, K4) over-expressing clones were compared to control (EV). The cells were photographed at the identical location as indicated. The graph depicts a percent of wound closure plotted against time. Cell migration of TCF-4B expressing clones (B16, B5) was slower than control, whereas TCF-4J and K over-expressing Huh7 cells exhibited increased cell migration. Data shown are the means ± S.D. * p < 0.01 versus control (EV) (n = 3 independent assays in duplicate). (B) Cell motility assay using a luminescence-based assay. The results are expressed as a percent of migrated to total number of cells, and the experiments were performed in triplicate (n = 3 independent cell preparations). * p < 0.05 versus control (EV) The effect of TCF-4B, J and K on cell motility was consistent with observations made with wound closure. (C) Representative examples of wound-healing assays.
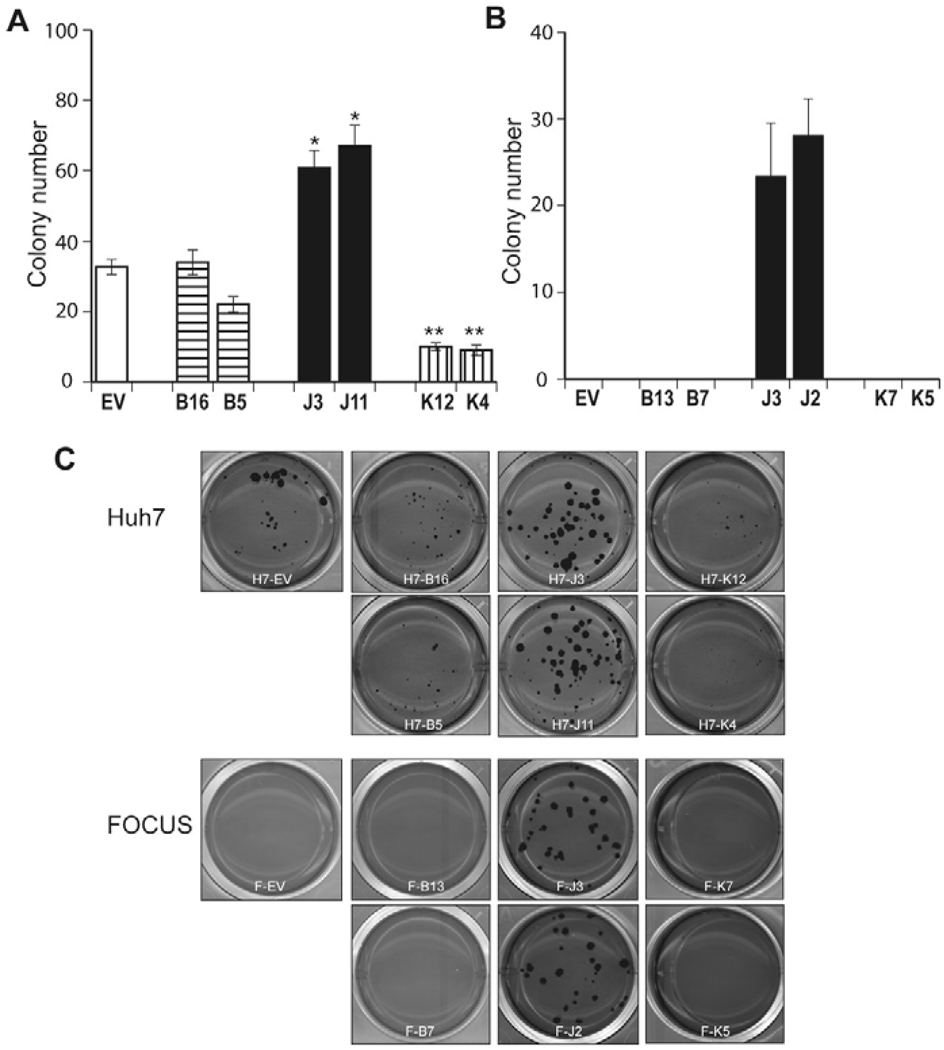
Finally, to determine if TCF-4B, J, and K play a role in anchorage-independent cell growth, a colony-forming assay in soft-agar was performed. This approach is considered to be a reliable test for malignant transformation of cells. As shown in Fig. 7A and C, colony formation was significantly increased in TCF-4J over-expressing Huh7 cells (J3, J11), and was reduced in TCF-4K expressing cells (K12, K4) compared to control. Notably colonies derived from Huh7 cells expressing TCF-4J were not only increased in number, but were also found to be consistently larger compared to control. In FOCUS HCC cells, there was no colony formation in EV, TCF-4B (B13, B7) or K (K7, K5) stable expressing clones. Large size colonies were found only in FOCUS cells over-expressing TCF-4J (J3, J2), which was consistent with the observations made with the Huh7 cells (Fig. 7B, C).
Fig. 7.
Colony formation in soft-agar. Huh7 and FOCUS stable clones over-expressing empty vector (EV), TCF-4B, TCF-4J, and TCF-4K were applied to soft-agar colony formation assay to evaluate anchorage independent colony growth. Twenty-four days after seeding the cells, the colonies were stained and analyzed for the number and size using Image J software. (A) Bar graph shows numbers of colonies in Huh7 stable clones. There was no difference in size or number of colonies between empty vector control and TCF-4B (B16, B5) isoform. In contrast, TCF-4J expressing clones (J3, J11) resulted in increased number as well as size of colonies compared to control. In contrast, TCF-4K (K12, K4), which showed repressed TCF transcriptional activity, cell proliferation, but enhanced cell migration revealed very lower colony formation activity compared to control. Data shown are the means ± S.D.; n = 3 independent experiments, assayed in triplicate. * p < 0.05, ** p < 0.01 versus control (EV) (B) Bar graph shows numbers of colonies in FOCUS stable expressing clones; there was no colony formation in EV, TCF-4B (B13, B7) or K (K7, K5) stable expressing clones. Large size colonies were found only in FOCUS cells over-expressing TCF-4J (J3, J2), which was consistent with observation in Huh7 cells. (C) Representative examples of soft-agar plates illustrating transformed colonies derived from Huh7 and FOCUS stable expressing clones.
Taken together, our studies suggest that an individual TCF-4 isoform may exhort a novel biologic function to produce a different HCC phenotype. Expression of TCF-4B (short form) had the highest TCF and cyclin D1 transcriptional activities and also exhibited increased cell proliferation, but there was no apparent effect on cell migration and colony formation compared to control. In contrast, TCF-4J isoform expression increased cell proliferation, migration as well as colony formation, which may have influenced, in part, by increased cyclin D1 promoter activity even though the overall TCF transcriptional activity was low. In contrast, TCF-4K isoform expression inhibited TCF and cyclin D1 transcriptional activities as well as colony formation, supporting the hypothesis that some TCF-4 isoforms may act as suppressors under certain conditions. Moreover, even though TCF-4J and TCF-4K varied by only five amino acids (the SxxSS motif), they exhibited substantial differences in generating features of the malignant phenotype. It will be of importance to explore the molecular mechanisms by which the SxxSS motif expressed in such TCF-4 isoforms influences HCC malignant behavior in the future.
Discussion
The TCF proteins mediate Wnt signals in the nucleus as bipartite factors that act as either transcriptional activators or repressors according to their association with other proteins. Transcriptional activation is driven, in part, by β-catenin, whereas, repression is mediated by co-repressors that may interact either with β-catenin [26–28] or directly with TCF proteins [29–32]. In the absence of a Wnt signal and nuclear β-catenin, TCFs can associate with several different transcriptional co-repressors such as Groucho/TLE and CtBP, resulting in inhibition of target gene expression. In this regard, definition of TCF transcriptional activity by β-catenin or the co-repressors is essential to produce biological effects, such as an ensuring correct physiological development or preventing oncogenesis. It is likely that TCF-4 has several isoforms that may be important in hepatocyte transformation and their activity influenced by inclusion of functional motifs related to splicing events. In the present study, we hypothesized that different TCF-4 isoforms may exhibit different biologic properties that could participate in hepatic oncogenesis.
To identify various TCF-4 splicing isoforms, we utilized four different human HCC cell lines such as HepG2, Hep3B, Huh7, and FOCUS. The FOCUS and Huh7 cell lines were derived from HBV-associated HCC [33, 34]. The HepG2 cell line was derived from well-differentiated hepatoblastoma, which carries a deletion in the exon 3 of the CTNNB1 gene; this event leads to stabilization and cellular accumulation of β-catenin, and was associated with aberrant activation of canonical Wnt/β-catenin signaling [35]. Several studies have shown deregulation of the Wnt/β-catenin signaling pathway in these cells. In addition, up-regulation of FZD receptors was correlated with HCC cell proliferation and enhanced motility [23, 36]. Furthermore, small molecule antagonists of TCF-4/β-catein complex formation inhibited cell growth of HepG2 and Huh7 cells [37]. These four cell lines have shown different degrees of cellular differentiation (FOCUS < Huh7 < Hep3B < HepG2) with respect to morphology, growth rate, production of liver specific proteins such as albumin, α-anti-trypsin and transferrin, altered an anchorage-independent growth in soft agar and tumor formation in nude mice. Interestingly, different expression levels of these isoforms were found in the four HCC cell lines that was related to the degree of cellular differentiation. In addition, the pattern of expression was also different in these four cell lines (Fig. 2D), suggesting that the TCF transcriptional activity may function in aggregate depending on differential expression of these isoforms in the presence of β-catenin.
We have identified 14 different TCF-4 isoforms by sequencing cDNAs generated from complete mRNA clones derived from four HCC cell lines. Two are previously known as TCF-4B (NM_001146286.1) and TCF-4E (NM_030756.4 or Q9NQB0-8). Isoforms TCF-4G (Q9NQB0-9), TCF-4I (Q9NQB0-6), and TCF-4M (Q9NQB0-1) have been suspected to exist since their sequences have been deduced by overlapping expressed sequence tags (ESTs) and by primer walking between BAC clones or DNA contigs available at UniProt [16]. However, these 14 TCF-4 isoforms were characterized as full-length cDNA clones derived from HCC cell lines to perform functional analysis. The TCF-4X (X for unknown and extra exon) isoform contains a novel sequence, of 114 nucleotides between exons 5 and 6. It is referred to as exon 5A in order to keep the same exon nomenclature as previously proposed [16]. The DNA sequence of TCF-4 gene (TCF7L2) revealed a potential exon with ag/gt dinucleotides at intron/exon boundaries embedded within the intron 5. Therefore, TCF7L2 has the 18th exon in the genome, and the number of exons may increase since TCF7L2 isoform 3 (NM_001146283.1) also includes an additional sequence between exons 5 and 6.
To confirm expression of TCF-4 isoforms in HCC cell lines, we were unable to perform real-time RT-PCR because it was not possible to design a unique RT-PCR primer pair for each isoform. Although a given primer pair may distinguish one isoform from another closely-related one, it will still detect other isoforms. For example, a primer pair, which detects and amplifies the region of the LVPQ and SxxSS motifs, can distinguish TCF-4A from TCF-4B and C. However, this primer pair would also detect isoforms TCF-4F, H, K, L, and X as they have the same motifs with more splicing exons located elsewhere. Therefore, we had to employ a RT-PCR method that combines high-resolution agarose gel electrophoresis to semi-quantify expression of each isoform. In this regard, expression of these isoforms was confirmed using RT-PCR. It is noteworthy that four (TCF-4A, H, K and X) of 14 isoforms were undetectable in HCC cell lines, due to low expression levels. However, we found that all 14 isoforms were expressed in either human HCC tumors, or peritumoral tissues, or normal liver. Consistent with results derived from HCC cell lines, TCF-4A, H, and K isoforms revealed exhibited very low or no expression in human HCC tumors compared to normal and petitumor tissues; expression levels of TCF-4X was low in all HCC tumor tissues studied (unpublished observations).
Assessment of TCF transcriptional activity revealed several interesting features. A potentially important finding was the differential transcriptional activity observed in three isoform pairs namely: TCF-4A and B, TCF-4G and H, and TCF-4J and K. Comparison of their organizational structure revealed identity except for the presence or absence of a conserved SxxSS motif (Fig. 1B). Indeed, the transcriptional activity measured from isoforms TCF-4A, H and K were lower than that observed with TCF-4B, G and J, which contains this motif. Therefore, the SxxSS sequence may be responsible, in part, for the reduced transcriptional activity observed even in the presence of β-catenin activation (Fig. 3A). It is of interest that HCC cell lines tend to express more of the TCF-4 isoforms that do not contain the SxxSS motif such as TCF-4B, C, D, G, J, E and M. Another interesting finding was the differential transcriptional activity observed between the TCF-4C and TCF-4D isoforms. Transcriptional activity of TCF-4D, which has exon 4, is lower than empty vector, whereas TCF-4C lacking exon 4 has the highest transcriptional activity. These results suggest that exon 4 may also have a repressor function and further study will be required.
In summary, this is the initial identification, description, and characterization of 14 different TCF-4 isoforms of 12 novel species. Results reveal the presence of different TCF-4 isoforms with divergent properties that may influence the phenotype of HCC cells and function as transcription activators or repressors. In particular, comparison of TCF-4J and K isoforms suggests that a conserved splicing motif may have a major influence on the transcriptional activity and function and alter the characteristics of the malignant phenotype. How these TCF-4 isoforms are linked to the progression and course of HCC tumors is unknown. It will be important to define the molecular pathways activated, to promote a better understanding the oncogenesis of these devastating tumors. Furthermore, to identify interactions with growth factor signaling cascades may eventually provide novel molecular targets for therapy of HCC.
Acknowledgements
The work was supported in part by grants from National Institutes of Health CA-35711, CA 123544 (JRW), P20 RR015578 and Developmental Research Award from Rhode Island Hospital (MK).
Abbreviations
- CtBP
C-terminal binding protein
- FZD
Frizzled receptor
- HCC
hepatocellular carcinoma
- HMG
high mobility group
- LEF
lymphoid enhancer factor
- RT
reverse transcription
- TCF
T-cell factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers
TCF-4A, GenBank ID: HM352849
TCF-4B, GenBank ID: HM352842
TCF-4C, GenBank ID: HM352843
TCF-4D, GenBank ID: HM352840
TCF-4F, GenBank ID: HM352838
TCF-4G, GenBank ID: HM352846
TCF-4H, GenBank ID: HM352841
TCF-4I, GenBank ID: HM352844
TCF-4J, GenBank ID: HM352847
TCF-4K, GenBank ID: HM352839
TCF-4L, GenBank ID: HM352851
TCF-4E, GenBank ID: HM352848
TCF-4M, GenBank ID: HM352850
TCF-4X, GenBank ID: HM352845
References
- 1.Bosch FX, Ribes J, Cleries R, Diaz M. Epidemiology of hepatocelluar carcinoma. Clin. Liver Dis. 2005;9:191–211. doi: 10.1016/j.cld.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 2.Bruix J, Hessheimer AJ, Forner A, Boix L, Vilana R, Llovet JM. New aspects of diagnosis and therapy of hepatocellular carcinoma. Oncogene. 2006;25:3848–3856. doi: 10.1038/sj.onc.1209548. [DOI] [PubMed] [Google Scholar]
- 3.Bengochea A, de Souza MM, Lefrancois L, Roux EL, Galy O, Chemin I, Kim M, Wands JR, Trepo C, Hainaut P, Scoazec J-Y, Vitvitski L, Merle P. Dysregulation of specific Wnt and Frizzled genes: a striking event in human hepatocellular carcinoma. Br. J. Cancer. 2008;99:143–150. doi: 10.1038/sj.bjc.6604422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim M, Lee HC, Tsedensodnom O, Hartley R, Lim Y-S, Yu E, Merle P, Wands JR. Functional interaction between Wnt3 and Frizzled-7 leads to activation of the Wnt/beta-catnein signaling pathway in hepatocellular carcinoma cells. J. Hepatol. 2008;48:780–791. doi: 10.1016/j.jhep.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 6.Behrens J, von Kries JP, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of β-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 7.Travis A, Amsterdam A, Belanger C, Grosschedl R. LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor alpha enhancer function. Genes Dev. 1991;5:880–894. doi: 10.1101/gad.5.5.880. [DOI] [PubMed] [Google Scholar]
- 8.van de Wetering M, Oosterwegel M, Dooijes D, Clevers H. Identification and cloning of TCF-1, a T lymphocyte-specific transcription factor containing a sequence-specific HMG box. EMBO J. 1991;10:123–132. doi: 10.1002/j.1460-2075.1991.tb07928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bienz M, Clevers H. Armadillo/beta-catenin signals in the nucleus-proof beyond a resonable doubt? Nat. Cell Biol. 2003;5:179–182. doi: 10.1038/ncb0303-179. [DOI] [PubMed] [Google Scholar]
- 10.Kramps T, Peter O, Brunner E, Nellen D, Froesch B, Chatterjee S, Murone M, Zullig S, Basler K. Wnt/Wingless signaling requires BCL9/Legless-mediated recruitement of Pygopus to the nuclear beta-catenin-TCF complex. Cell. 2002;109:47–60. doi: 10.1016/s0092-8674(02)00679-7. [DOI] [PubMed] [Google Scholar]
- 11.Parker DS, Jemison J, Cadigan KM. Pygopus, a nuclear PHD-finger protein required for Wingless signaling in Drosophila. Development. 2002;129:2565–2576. doi: 10.1242/dev.129.11.2565. [DOI] [PubMed] [Google Scholar]
- 12.Belenkaya TY, Han C, Standley HJ, LIn X, Houston DW, Heasman J, Lin X. Pygopus encides a nuclear protein essential for Wingless/Wnt signaling. Development. 2002;129:4089–4101. doi: 10.1242/dev.129.17.4089. [DOI] [PubMed] [Google Scholar]
- 13.Thompson B, Townsley F, Rosin-Arbesfeld R, Musisi H, Bienz M. A new nuclear component of the Wnt signaling pathway. Nat. Cell Biol. 2002;4:367–373. doi: 10.1038/ncb786. [DOI] [PubMed] [Google Scholar]
- 14.Cui J, Zhou X, Liu Y, Tang Z, Romeih M. Alterations of beta-catenin and Tcf-4 instead of GSK-3β contribute to activation of Wnt pathway in hepatocellular carcinoma. Chin. Med. J. 2003;116:1885–1892. [PubMed] [Google Scholar]
- 15.Yanagitani A, Yamada S, Yasui S, Shimomura T, Murai R, Murawaki Y, Hashiguchi K, Kanbe T, Saeki T, Ichiba M, Tanabe Y, Yoshida Y, Morino S, Kurimasa A, Usuda N, Yamazaki H, Kunisada T, Ito H, Murawaki Y, Shiota G. Retinoic acid receptor a dominant negative form causes steatohepatitis and liver tumors in transgenic mice. Hepatology. 2004;40:366–375. doi: 10.1002/hep.20335. [DOI] [PubMed] [Google Scholar]
- 16.Duval A, Rolland S, Tubacher E, Bui H, Thomas G, Hamelin R. The human T-cell transcription factor-4 gene: structure, extensive characterization of alternative splicings, and mutational analysis in colorectal cancer cell lines. Cancer Res. 2000;60:3872–3879. [PubMed] [Google Scholar]
- 17.Pukrop T, Gradl D, Henningfeld K, Knochel W, Wedlich D, Kuhl M. Identification of two regulatory elements within the high mobility group box transcription factor XTCF-4. J. Biol. Chem. 2001;276:8968–8978. doi: 10.1074/jbc.M007533200. [DOI] [PubMed] [Google Scholar]
- 18.Young RM, Reyes AE, Allende ML. Expression and splice varient analysis of the zebrafish tcf4 transcription factor. Mech. Dev. 2002;117:269–273. doi: 10.1016/s0925-4773(02)00180-6. [DOI] [PubMed] [Google Scholar]
- 19.Muncan V, Sansom O, Tertoolen L, Phesse T, Begthel H, Sancho E, Cole A, Gregorieff A, de Alboran I, Clevers H, Clarke A. Rapid loss of intestinal cryps upon conditinal deletion of the Wnt/Tcf-4 target gene c-Myc. Mol. Cell. Biol. 2006;26:8418–8426. doi: 10.1128/MCB.00821-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Ray G, Yoo B, Redogan M, Rosen K. Down-regulation of death-associated protein-kinase-2 is required for β-catenin-induced anoikis resistance of maligant epithelial cells. J. Biol. Chem. 2009;284:2012–2022. doi: 10.1074/jbc.M805612200. [DOI] [PubMed] [Google Scholar]
- 21.Nejak-Bowen K, Zeng G, Tan X, Cieply B, Monga S. β-catenin regulates vitamin C biosynthesis and cell survival in murine liver. J. Biol. Chem. 2009;284:28115–28127. doi: 10.1074/jbc.M109.047258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shtutman M, Zhurinsky J, Simcha I, Albanese C, D'Amico M, Pestell R, Ben-Ze'ev A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad Sci U S A. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merle P, de la Monte S, Kim M, Herrmann M, Tanaka S, Von Dem Bussche A, Kew MC, Trepo C, Wands JR. Functional consequences of frizzled-7 receptor overexpression in human hepatocellular carcinoma. Gastroenterology. 2004;127:1110–1122. doi: 10.1053/j.gastro.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 24.Lee H, Tian B, Sedivy JM, Wands JR, Kim M. Loss of Raf Kinase Inhibitor Protein promotes cell proliferation and migration of human hepatoma cells. Gastroenterology. 2006;131:1208–1217. doi: 10.1053/j.gastro.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korinek V, Barker N, Morin P, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a β-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 26.Bauer A, Chauvet S, Huber O, Usseglio F, Rothbacher U, Aragnol D, Kemler R, Pradel J. Pontin52 and reptin52 function as antagonistic regulators of β-catenin signalling activity. EMBO J. 2000;19:6121–6130. doi: 10.1093/emboj/19.22.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tago K, Nakamura T, Nishita M, Hyodo J, Nagai S, Murata Y, Adachi S, Ohwada S, Morishita Y, Shibuya H, Akiyama T. Inhibition of Wnt signaling by ICAT, a novel β-catenin-interacting protein. Genes Dev. 2000;14:1741–1749. [PMC free article] [PubMed] [Google Scholar]
- 28.Sakamoto I, Kishida S, Fukui A, Kishida M, Yamamoto H, Hino S, Michiue T, Takada S, Asashima M, Kikuchi A. A novel β-catenin-binding protein inhibits β-catenin-dependent Tcf activation and axis formation. J. Biol. Chem. 2000;275:32871–32878. doi: 10.1074/jbc.M004089200. [DOI] [PubMed] [Google Scholar]
- 29.Waltzer L, Bienz M. Drosophila CBP represses the transcription factor TCF to antigonize Wingless signalling. Nature. 1998;395:521–525. doi: 10.1038/26785. [DOI] [PubMed] [Google Scholar]
- 30.Brannon M, Brown JD, Bates R, Kimelman D, Moon RT. XCtBP is a XTcf-3 co-repressor with roles thoughout Xenopus development. Development. 1999;126:3159–3170. doi: 10.1242/dev.126.14.3159. [DOI] [PubMed] [Google Scholar]
- 31.Sampson EM, Haque ZK, Ku M-C, Tevosian SG, Albanese C, Pestell RG, Paulson KE, Yee AS. Negative regulation of the Wnt-β-catenin pathway by the transcriptional repressor HBP1. EMBO J. 2001;20:4500–4511. doi: 10.1093/emboj/20.16.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snider L, Thirlwell H, Miller JR, Moon RT, Groudine M, Tapscott SJ. Inhibition of Tcf3 binding by I-mfa domain proteins. Mol. Cell Biol. 2001;21:1866–1873. doi: 10.1128/MCB.21.5.1866-1873.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- 34.He L, Isselbacher KJ, Wands JR, Goodman HM, Shih C, Quaroni A. Establishment and characterization of a new human hepatocellular carcinoma cell line. In Vitro. 1984;20:493–504. doi: 10.1007/BF02619623. [DOI] [PubMed] [Google Scholar]
- 35.Lopez-Terrada D, Cheung SW, Finegold MJ, Knowles BB. HepG2 is a hepatoblastoma-derived cell line. Hum. Pathol. 2009;40:1512–1515. doi: 10.1016/j.humpath.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 36.Fujimoto T, Tomizawa M, Yokosuka O. SiRNA of frizzled-9 suppresses proliferation and motility of hepatoma cells. Int. J. Oncol. 2009;35:861–866. doi: 10.3892/ijo_00000400. [DOI] [PubMed] [Google Scholar]
- 37.Wei W, Chua MS, Grepper S, So S. Small molecule antagonists of Tcf4/beta-catenin complex inhibit the growth of HCC cells in vitro and in vivo. Int. J. Cancer. 2010;126:2426–2436. doi: 10.1002/ijc.24810. [DOI] [PubMed] [Google Scholar]