Abstract
We examined the role of the microtubule cytoskeleton during vaccinia virus infection. We found that newly assembled virus particles accumulate in the vicinity of the microtubule-organizing centre in a microtubule- and dynein–dynactin complex-dependent fashion. Microtubules are required for efficient intracellular mature virus (IMV) formation and are essential for intracellular enveloped virus (IEV) assembly. As infection proceeds, the microtubule cytoskeleton becomes dramatically reorganized in a fashion reminiscent of overexpression of microtubule-associated proteins (MAPs). Consistent with this, we report that the vaccinia proteins A10L and L4R have MAP-like properties and mediate direct binding of viral cores to microtubules in vitro. In addition, vaccinia infection also results in severe reduction of proteins at the centrosome and loss of centrosomal microtubule nucleation efficiency. This represents the first example of viral-induced disruption of centrosome function. Further studies with vaccinia will provide insights into the role of microtubules during viral pathogenesis and regulation of centrosome function.
Keywords: centrosome/MAPs/microtubules/vaccinia virus
Introduction
Intracellular bacterial and viral pathogens have evolved numerous mechanisms to appropriate and exploit different systems of the host during their life cycles in order to facilitate their spread during entry and exit from the host (Cudmore et al., 1997; Finlay and Cossart, 1997; Dramsi and Cossart, 1998). In the case of viruses, perhaps the best studied example is the exploitation of the actin cytoskeleton by vaccinia virus during its exit from infected cells (Cudmore et al., 1997). Vaccinia virus is a large DNA virus with a genome of ∼191 kb encoding 260 open reading frames (ORFs) that is a close relative of variola virus, the causative agent of smallpox (Johnson et al., 1993; Massung et al., 1993). Vaccinia virus morphogenesis is a complex process which occurs in the cytoplasm of infected cells and results in the formation of the intracellular mature virus (IMV) and the intracellular enveloped virus (IEV). IMV consist of a viral core of DNA and protein enveloped in a membrane cisterna derived from the intermediate compartment (Sodeik et al., 1993). The IMV core contains five major proteins, A3L, A4L, A10L, F17R and L4R (Vanslyke and Hruby, 1994; Jensen et al., 1996a), while 12 proteins, A12L, A13L, A14L, A14.5L, A17L, A27L, D8L, G4L, G7L, H3L, I5L and L1R, are associated with the membranes around the virus particle (Jensen et al., 1996a; Betakova et al., 2000). Depending on the virus strain and cell type, a proportion of IMV can become enwrapped by a membrane cisterna derived from the trans-Golgi apparatus to give rise to IEV particles (Schmelz et al., 1994). To date, six IEV-specific proteins, A33R (Roper et al., 1996), A34R (Duncan and Smith, 1992), A36R (Parkinson and Smith, 1994), A56R (Payne and Norrby, 1976; Shida, 1986), B5R (Engelstad et al., 1992; Isaacs et al., 1992) and F13L (Hirt et al., 1986), have been identified. Studies using recombinant viruses have shown that A33R, A34R, B5R and F13L play an important role in IEV assembly (Blasco and Moss, 1991; Engelstad and Smith, 1993; Wolffe et al., 1993, 1997; Roper et al., 1998; Sanderson et al., 1998a; Röttger et al., 1999). Vaccinia virus is thought to leave the cell by fusion of the outer IEV membrane with the plasma membrane, to give rise to the extracellular enveloped virus (EEV) (Morgan, 1976; Payne, 1980; Blasco and Moss, 1991) or the cell-associated enveloped viruses (CEV) which remain associated with the outer surface of the plasma membrane (Blasco and Moss, 1992).
During the complex vaccinia infection process, the actin cytoskeleton is dramatically reorganized and numerous actin comet-like tails are induced by IEV particles (Cudmore et al., 1995; Röttger et al., 1999). Using actin polymerization as the driving force, IEV particles are propelled on actin tails until they contact the plasma membrane and extend outwards, thereby facilitating infection of neighbouring cells (Cudmore et al., 1995). In addition, vaccinia infection results in stimulation of cell motility, loss of contact inhibition and changes in cell adhesion (Sanderson and Smith, 1998; Sanderson et al., 1998b). Vaccinia virus-induced cell motility can be subdivided further into cell migration and extension of neurite-like projections, the latter of which is dependent on microtubules (Sanderson et al., 1998b). The dependence of neurite-like projection formation on microtubules suggests that the microtubule cytoskeleton may also play a role during the life cycle of vaccinia virus. Indeed, recently, the vaccinia A27L protein and microtubules have been shown to be required for efficient IMV dispersion (Sanderson et al., 2000). Furthermore, in the absence of vaccinia actin-based motility, cell to cell spread still occurs although it is less efficient (Wolffe et al., 1997, 1998; Sanderson et al., 1998a), suggesting that additional transport mechanisms must exist.
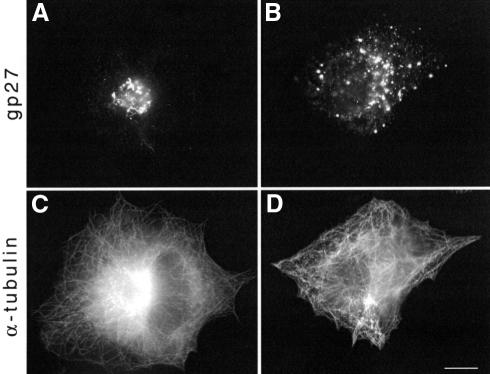
Given these observations, we wondered whether the microtubule cytoskeleton has a function during the life cycle of vaccinia virus. We now report that the microtubule cytoskeleton and the dynein–dynactin complex play an important role during the early stages of vaccinia infection. However, later during the infection cycle, loss of centrosome function and accumulation of viral-encoded microtubule-associated proteins (MAPs) result in a dramatic rearrangement of the microtubule cytoskeleton.
Results
Vaccinia localization in the vicinity of the MTOC depends on microtubules and the dynein–dynactin complex
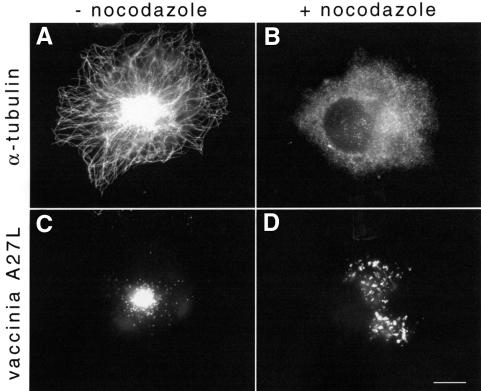
Indirect immunofluorescence labelling shows that by 6 h post-infection the majority of vaccinia virus particles are concentrated in the area coinciding with the centre of the microtubule aster (Figure 1A and C). To examine whether this localization is indeed microtubule dependent, we infected cells pre-treated with nocodazole to depolymerize microtubules. In the absence of microtubules, virus particles were distributed throughout the cytoplasm (Figure 1B and D). The accumulation of virus particles in the area around the centre of the microtubule aster suggested that a microtubule minus end-directed motor may be involved in establishing the position of the virus in this location. To examine this possibility, we infected cells overexpressing p50/dynamitin which acts as a dominant-negative for dynein–dynactin function (Echeverri et al., 1996). We found in cells overexpressing p50/dynamitin that virus particles did not accumulate at the centre of the microtubule aster but rather throughout the cytoplasm, as occurs in the absence of microtubules (compare Figure 2B with Figure 1D).
Fig. 1. Vaccinia virus localization in the vicinity of the MTOC depends on microtubules. HeLa cells infected with vaccinia virus in the absence (A and C) or presence of nocodazole (B and D) fixed 6 h post-infection. Depolymerization of microtubules results in dispersed cytoplasmic viral assembly and loss of localization at the MTOC area (B and D). The microtubule cytoskeleton (A and B) and vaccinia virus (C and D) are visualized with anti-α-tubulin and anti-A27L antibodies respectively. Scale bar = 10 µm.
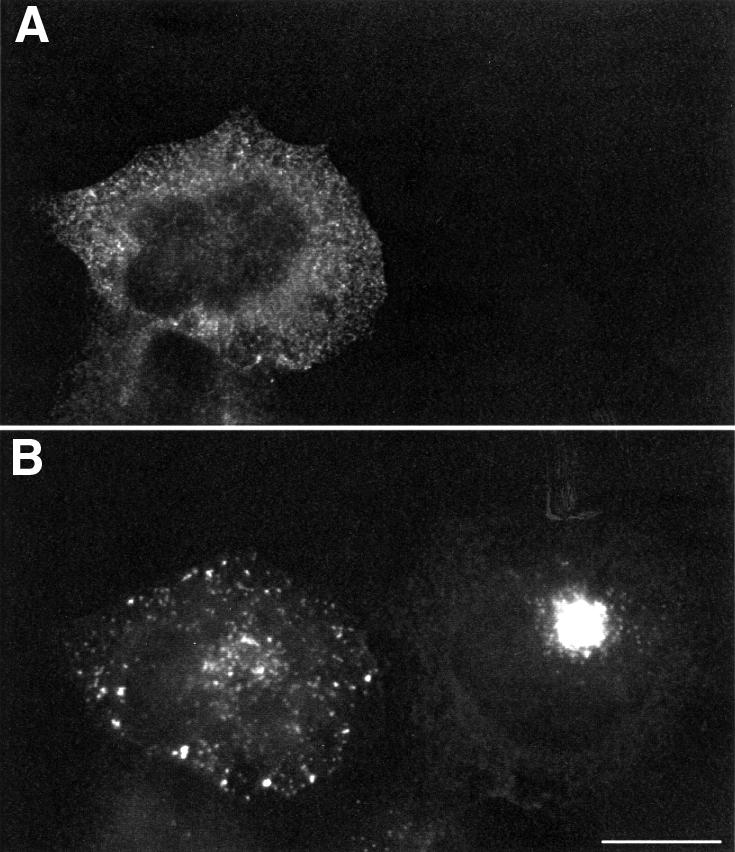
Fig. 2. Disruption of the dynein–dynactin complex results in dispersed cytoplasmic viral localization. p50/dynamitin is overexpressed in the left cell as judged by detection of the myc epitope tag (A) although the anti-A27L antibody labelling shows that both cells are infected (B). Scale bar = 10 µm.
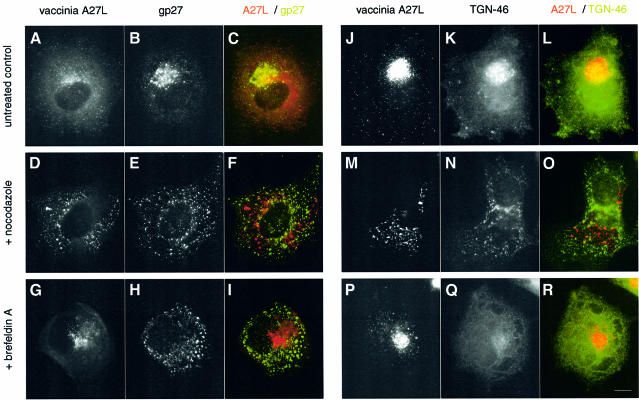
As vaccinia morphogenesis involves wrapping by host membranes, it was possible that the effects of nocodazole and p50/dynamitin on virus localization were in fact due to disruption of the intermediate compartment and Golgi apparatus by these reagents (Burkhardt et al., 1997). However, two independent experiments showed that this is not the case. First, in cells infected in the absence of microtubules, the Golgi apparatus as well as vaccinia virus particles are dispersed throughout the cytoplasm but do not co-localize (Figure 3F and O). Secondly, vaccinia particles remain in the vicinity of the microtubule-organizing centre (MTOC) when the Golgi but not the microtubules was disrupted by treatment with brefeldin A (Figure 3G and P). Similar results were obtained using other markers: A17L for vaccinia, galactosyltransferase for the Golgi or ERGIC53 for the intermediate compartment (data not shown). Taken together, our data indicate that the microtubule cytoskeleton is required for the localization of newly assembled virus particles in the vicinity of the MTOC during vaccinia infection.
Fig. 3. Vaccinia virions do not co-distribute with disrupted Golgi markers. Golgi and vaccinia localization in control (A–C and J–L), nocodazole- (D–F and M–O) and brefeldin A- (G–I and P–R) treated cells. Vaccinia virus particles are visualized with the anti-A27L antibody, while the cis-Golgi and trans-Golgi network are labelled with the anti-gp27 and anti-TGN-46 antibodies, respectively. Scale bar = 10 µm.
Formation of functional IEV, but not IMV, is microtubule dependent
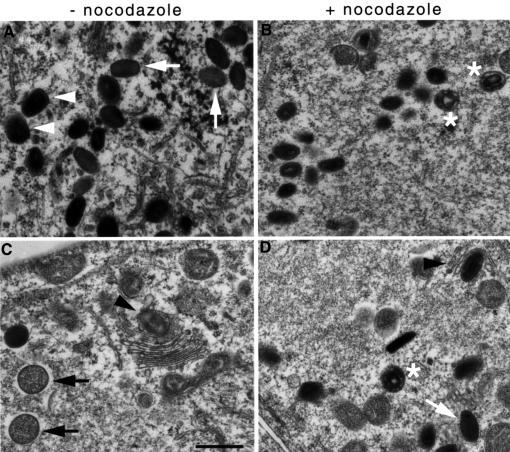
Given the requirement for microtubules in vaccinia localization, we subsequently examined whether this localization has a role in morphogenesis of the two different intracellular forms of vaccinia virus, IMV and IEV. From electron microscopic examination of cells infected in the presence of nocodazole, it became clear that IMV particles which are morphologically indistinguishable from controls are formed (Figure 4). Although IMV particles are assembled in the absence of microtubules, we wondered whether their number is reduced and whether those that are formed are infectious, since the integrity of the intermediate compartment depends on microtubules (Burkhardt et al., 1997). To address this question, three independent virus stocks were prepared in the presence or absence of nocodazole. To simplify the interpretation of the data, we used the recombinant vaccinia virus mutant ΔF13L, which is unable to form IEV (Blasco and Moss, 1991). The final concentration of virus particles produced, as determined by the method of Joklik (1962), was 30.2 ± 5.2 × 1010 particles/ml in the presence of microtubules and 9.0 ± 6.7 × 1010 particles/ml in the absence of microtubules. Although there is a 3-fold decrease in the number of virus particles formed in the absence of microtubules, the particles that are formed are infectious (data not shown).
Fig. 4. IMV but not IEV particles assemble in the absence of microtubules. Thin section electron microscope micrographs of HeLa cells infected with vaccinia virus in the absence (A and C) or presence of nocodazole (B and D) fixed 8 h post-infection. White arrows point to IMV particles, white arrowheads to IEVs, black arrows to IV particles, black arrowheads to IMVs in the process of wrapping with the trans-Golgi network to become IEVs. Asterisks indicate aberrant virus particles. Scale bar = 500 nm.
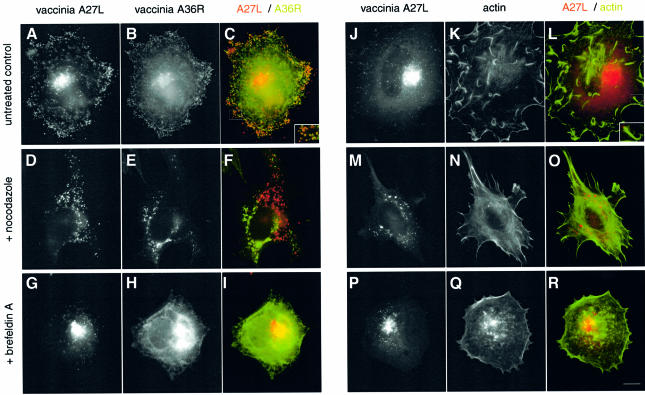
While infectious IMV are formed in the absence of microtubules, we found no evidence for IEV formation, based on electron microscope examination of cells infected in the presence of nocodazole (Figure 4). We did, however, observe IMV particles partially wrapped in trans-Golgi membranes most probably in the process of abortive IEV formation (Figure 4D). Given these data, we examined by indirect immunofluorescence whether low amounts of IEV particles are formed in the absence of microtubules. However, we could find no evidence for co-localization of the IEV protein markers A36R, A34R or A33R with vaccinia particles formed in the presence of nocodazole (Figure 5F). We also found no evidence for IEV formation, based on their ability to nucleate actin tails (Figure 5O). As IEV particle assembly involves wrapping by the Golgi apparatus (Schmelz et al., 1994), we examined the effects of only disrupting this membrane compartment using brefeldin A. We could find no evidence for IEV formation, based on co-localization of IEV protein markers with virus particles and actin tails in cells infected in the presence of brefeldin A (Figure 5G–I and P–R). Indeed, in brefeldin A-treated cells, the IEV membrane proteins required for assembly were observed in the endoplasmic reticulum and not the trans-Golgi (Figure 5H). In summary, our data indicate that the microtubule cytoskeleton is required for efficient IMV assembly and is essential for IEV formation.
Fig. 5. IEV formation is microtubule and Golgi dependent. IEV particles, which are identified by co-labelling with antibodies against A27L and the A36R IEV membrane protein (A–C), are not formed in the presence of nocodazole (D–F) or brefeldin A (G–I). Actin tails normally induced by IEV (J–L) are also absent in nocodazole- (M–O) or brefeldin A- (P–R) treated cells. Scale bar = 10 µm.
Vaccinia infection disrupts microtubule organization
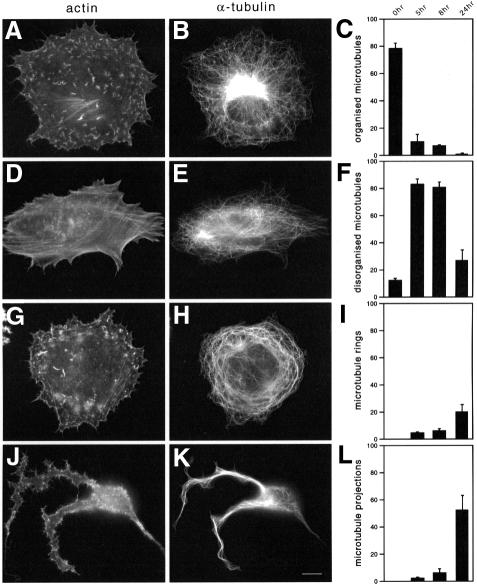
In the course of our experiments, it became obvious that the Golgi apparatus becomes progressively dispersed during infection co-concominantly with disruption of the microtubule network (Figure 6). Further analysis showed that during infection the normal morphology of the microtubule cytoskeleton is replaced by morphologically aberrant microtubule forms, which vary among each other but have in common the absence of a discrete MTOC (Figure 7). These aberrant forms can be broadly classified into three types: (i) cells with a disorganized microtubule network where microtubules seem randomly oriented (Figure 7E); (ii) cells in which microtubules form rings around the nucleus and throughout the cytoplasm (Figure 7H); or (iii) cells with long projections consisting of microtubule bundles (Figure 7K). We quantified the appearance of the different morphological forms in five independent infection experiments, in which 200 cells were counted for each time point for each experiment (Figure 7C, F, I and L). Small compact cells, representing 20.7 ± 2.6, 21.8 ± 12.4 and 29.7 ± 15.2% for 5, 8 and 24 h post-infection, respectively, in which the microtubule cytoskeleton morphology was not evident were not included in the analysis. Already by 5 h post-infection, when virus particle assembly has occurred, the normal aster microtubule configuration has been disrupted and replaced in the majority of cells by microtubules without obvious organization from the MTOC (Figure 7F). Furthermore, ∼10% of cells have microtubule rings and 5% of cells have long projections by this time point (Figure 7I and L). As the infection proceeds, microtubules become progressively more disrupted and bundled (Figure 7I and L).
Fig. 6. Vaccinia infection induces disruption of the microtubule cytoskeleton and the Golgi apparatus. Vaccinia-infected HeLa cells labelled 6 h post-infection with anti-gp27 and anti-α-tubulin antibodies to visualize the Golgi apparatus (A and B) and the microtubule cytoskeleton (C and D), respectively. The Golgi apparatus is dispersed in infected cells whose microtubule cytoskeleton is also disrupted (B, D) but not in cells with normal microtubule morphology (A, C). Scale bar = 10 µm.
Fig. 7. Vaccinia virus infection induces severe changes in microtubule organization. Examples of the four different classes of microtubule cytoskeleton morphologies observed in infected cells are shown (B, E, H and K) together with their corresponding actin cytoskeletons (A, D, G and J). Quantification of the relative amounts of these different microtubule cytoskeleton morphologies at 5, 8 and 12 h post-infection is indicated (C, F, I and L). Scale bar = 10 µm.
From our observations, there seems to be no obvious connection between the disruption and changes in the actin and the microtubule cytoskeletons (Figure 7). Moreover, the same reorganization of the microtubule network occurs in cells infected with the vaccinia deletion mutants ΔF13L and ΔA36R which do not make actin tails (data not shown). The effects of vaccinia virus infection on the reorganization of the microtubule cytoskeleton were also observed in all cell lines we examined (BHK-21, C2C12, PtK2, RK13 and Swiss 3T3) to varying degrees (data not shown). Our data show that vaccinia infection results in severe disruption of the normal morphology of the microtubule cytoskeleton.
The vaccinia virus genome encodes proteins with MAP-like properties
The formation of microtubule bundles and the loss of organization from the MTOC in vaccinia-infected cells is strongly reminiscent of the phenotype observed in cells overexpressing a MAP (Weisshaar et al., 1992; Togel et al., 1998). As overexpression of MAPs stabilizes microtubules, we examined whether the microtubule cytoskeleton in infected cells was more resistant to depolymerization by nocodazole or cold treatment (Figure 8). This was indeed the case, suggesting that the virus genome may encode viral proteins with MAP-like properties. To identify viral proteins which exhibit microtubule-binding properties, we performed microtubule co-sedimentation assays using extracts prepared from uninfected and vaccinia-infected cells (Figure 9). Initial experiments, however, revealed that intact virus particles in the extracts were prone to pellet even in the absence of microtubules, making identification of viral MAPs impossible. To avoid this problem, we prepared extracts from cells infected in the presence of rifampicin, a drug that inhibits vaccinia virus particle assembly but does not affect viral protein expression (Moss et al., 1969; Tan and McAuslan, 1970). The morphological effects of vaccinia infection on the microtubule cytoskeleton were the same in the presence or absence of rifampicin (data not shown). Comparison of the proteins present in pellets from microtubule co-sedimentation assays reveals that a number of additional prominent and minor bands are present in extracts prepared from vaccinia-infected but not from uninfected cells (Figure 9). Co-sedimentation assays performed in the presence of nocodazole or with cold-treated extracts reveal that the majority of these additional bands disappear in the absence of microtubules. To identify the viral proteins co-sedimenting with microtubules, we performed in-gel protease digestion followed by analysis of the resulting peptides by MALDI mass spectrometry. Using this approach, we identified a number of potential vaccinia-encoded MAPs: A10L (a structural protein), I1L and L4R (which are DNA-binding proteins), all of which are associated with viral cores (Vanslyke and Hruby, 1994; Jensen et al., 1996a; Klemperer et al., 1997), and A6L which is conserved in all poxvirus genomes but is of unknown function (Figure 9).
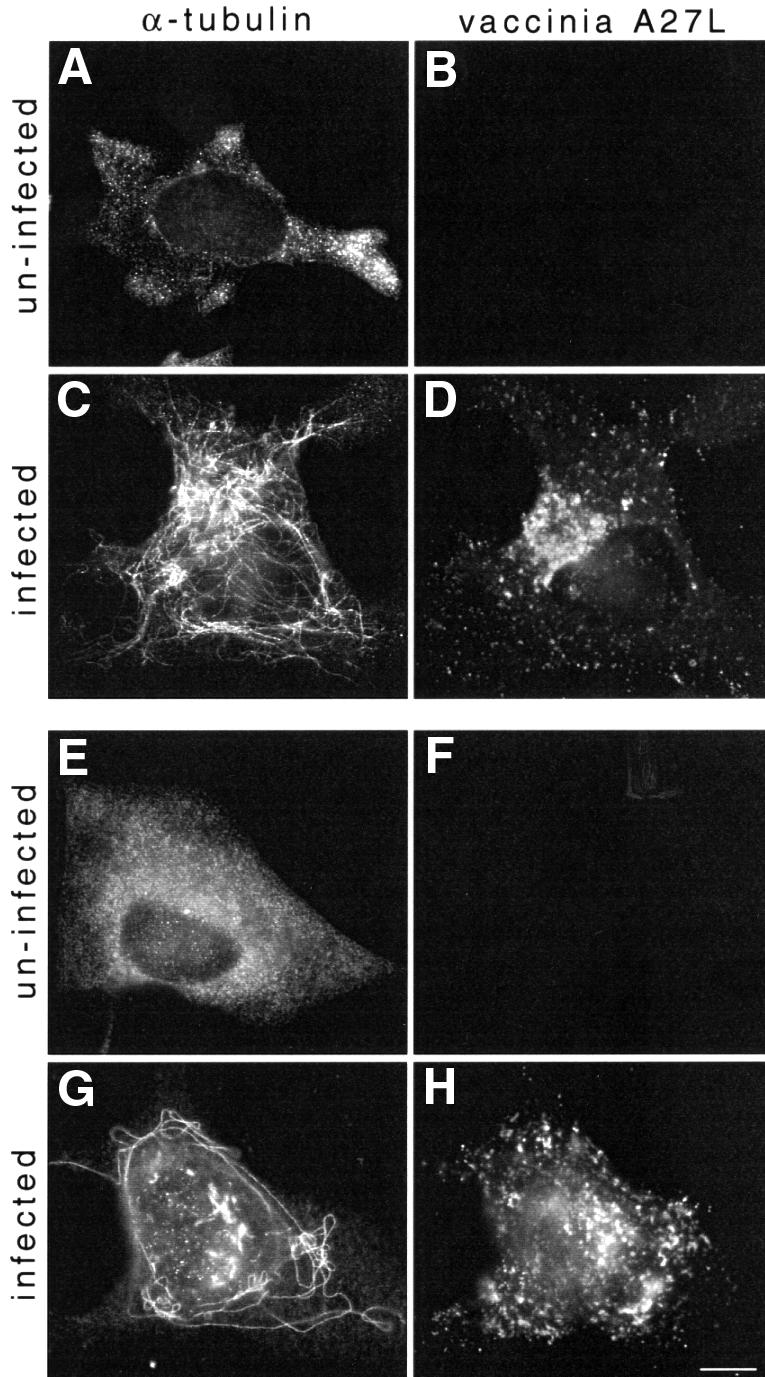
Fig. 8. Vaccinia virus infection stabilizes the microtubule cytoskeleton. In uninfected cells, microtubules are depolymerized by treatment with 10 µM nocodazole for 1 h (A and B) or by cold treatment for 1 h (E and F) while in vaccinia virus-infected cells a subpopulation of microtubules is resistant to nocodazole (C and D) or cold (G and H) depolymerization. Scale bar = 10 µm.
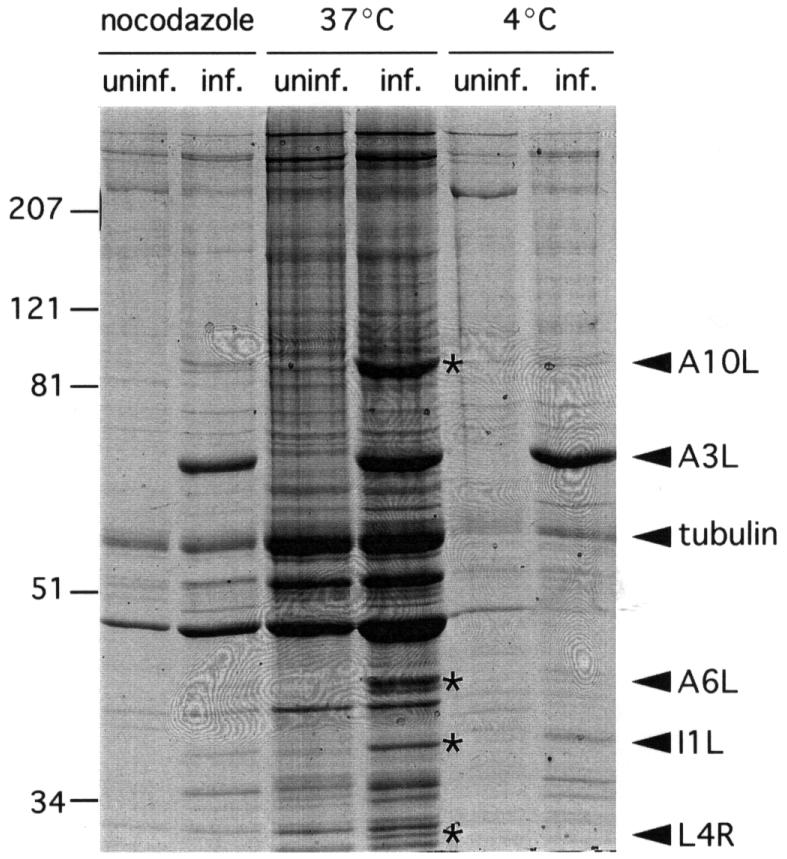
Fig. 9. Vaccinia encodes proteins that co-sediment with microtubules. Analysis of pellets from in vitro microtubule co-sedimentation assays performed with protein extracts from vaccinia-infected (inf.) and uninfected (uninf.) cells. Twice the amount of pellet has been loaded in control assays performed in the absence of microtubules (nocodazole or 4°C). Proteins co-sedimenting with microtubules that were only present in extracts from infected cells are indicated by an asterisk. The identity of proteins determined by in-gel proteolysis MALDI mass spectrometry is indicated (arrowheads).
A10L and L4R associate with microtubules in vivo and mediate binding of viral cores to microtubules in vitro
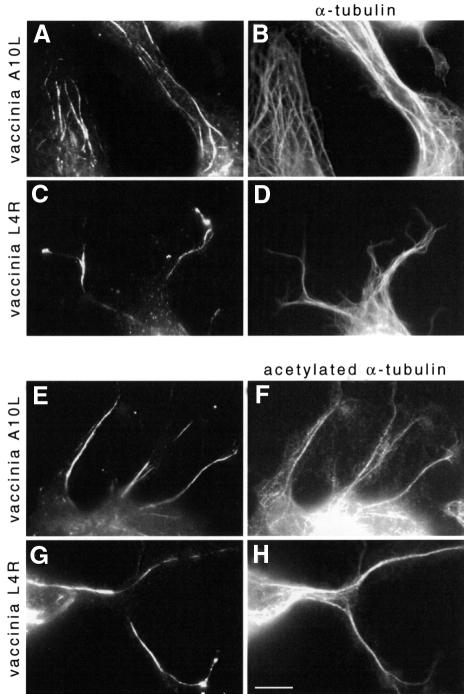
Using available antibodies, we examined the localization of A10L, L4R and I1L in infected cells to see whether they associate with microtubules in vivo, in addition to their essential role in the virus core (Vanslyke and Hruby, 1994; Jensen et al., 1996a). As a negative control, we also examined the localization of the A3L core protein which was identified as the prominent 70 kDa protein pelleting in the absence of microtubules (Figure 9). Indirect immunofluorescence analysis showed that A10L and L4R are associated with microtubules, in both the presence and absence of rifampicin (Figure 10). As expected, A10L and L4R were also associated with viral particles (data not shown). In contrast, I1L and A3L were never observed in association with microtubules, regardless of the fixation conditions, but were localized to viral factories and viral particles, respectively (data not shown). Interestingly, A10L and L4R were not associated with all microtubules but were co-localized with a subset of acetylated microtubules (Figure 10).
Fig. 10. A10L and L4R associate with a subset of microtubules in infected cells. HeLa cells 24 h post-infection with vaccinia virus are labelled with anti-A10L (A and E) or anti-L4R (C and G) and anti-α-tubulin (B and D) or anti-acetylated α-tubulin (F and H). Scale bar = 10 µm.
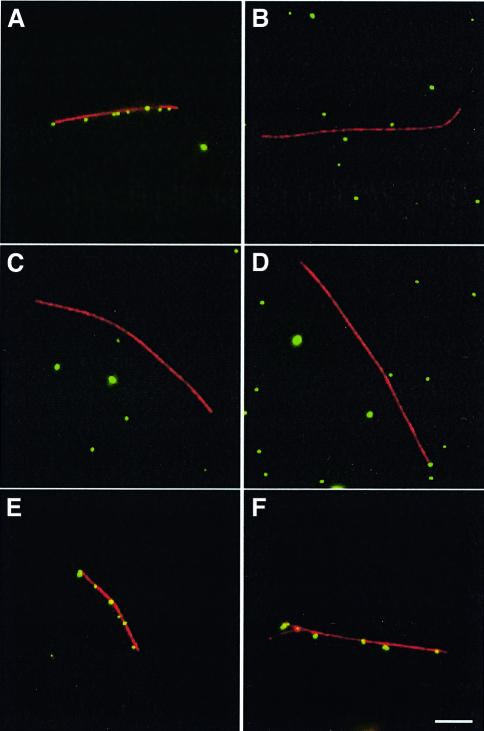
The association of A10L and L4R with virus particles and microtubules raises the question of whether there is a role for this microtubule-binding activity during infection. We wondered whether these two proteins mediate the interaction of incoming viral cores with microtubules at the beginning of infection, as cores and not virus particles are released in the cytoplasm at the start of the infection cycle (Ichihashi, 1996; Vanderpasschen et al., 1998; Pedersen et al., 2000). To examine this possibility, we investigated whether purified viral cores would bind microtubules in vitro. We found that viral cores were able to bind microtubules, while protease-treated cores showed no association (Figure 11A and B). Pre-incubation of purified viral cores with antibodies against A10L and L4R specifically inhibited the interaction of viral cores with microtubules (Figure 11C and D); in contrast, IgG or antibodies against A3L had no inhibitory effect (Figure 11E and F). Taken together, our data suggest that A10L and L4R have MAP-like properties and may play a role in mediating interactions of incoming viral cores with microtubules.
Fig. 11. Vaccinia cores bind directly to microtubules in vitro. Purified viral cores labelled by DAPI (green) bind to rhodamine-labelled micro tubules (red) in the absence of fixation (A). Binding to microtubules is not observed if cores are pre-treated with protease (B) or pre-incubated with antibodies against the A10L (C) or L4R (D) proteins. In contrast, pre-incubation of purified viral cores with control IgG (E) or antibody against the A3L protein (F) does not inhibit their interaction with microtubules. Scale bar = 5 µm.
Vaccinia virus infection disrupts centrosome function
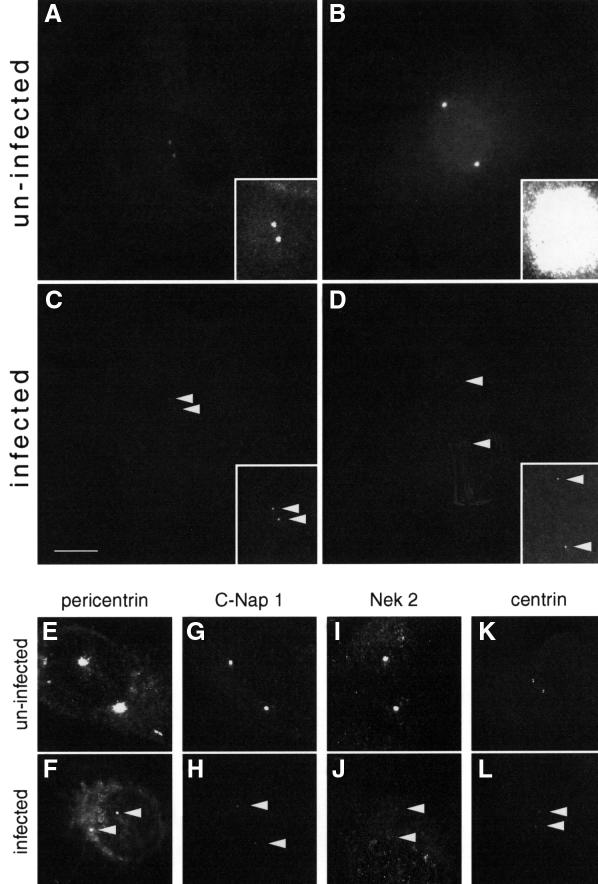
The dramatic rearrangement of the microtubule cytoskeleton which occurs during vaccinia infection is unlikely to be attributed exclusively to the action of A10L and L4R since they only associate with a subset of microtubules (Figure 10). Furthermore, the loss of microtubule organization precedes detectable association of A10L and L4R with microtubules, which occurs from ∼8 h post-infection. We therefore wondered whether vaccinia infection disrupts centrosome function, given the loss of microtubule aster configuration during infection (Figure 7). Since microtubules are nucleated by the centrosome in animal cells, we examined whether vaccinia infection affects γ-tubulin, which is critically required for this process (Stearns and Kirschner, 1994). We observed that γ-tubulin labelling of the centrosome is greatly reduced from as early as 2 h post-infection (Figure 12). The same result was obtained when we infected PtK1 cells stably expressing green fluorescent protein (GFP)-labelled γ-tubulin (Khodjakov and Rieder, 1999). In addition, the centrosomal and centriolar components pericentrin, C-Nap 1, Nek 2 and centrin are reduced by immunofluorescence in the centrosomes/centrioles of vaccinia-infected cells (Figure 12). Furthermore, the reduction of centrosomal markers requires viral protein synthesis as their levels are not affected when cells are infected in the presence of cycloheximide (data not shown).
Fig. 12. Vaccinia infection dramatically reduces levels of centrosomal components. Immunofluorescent γ-tubulin labelling of centrosomes in uninfected control cells (A and B) and in cells 2 h post-infection with vaccinia (C and D) at similar stages of the cell cycle. All images from the same experiment were collected with identical camera settings, to allow comparison of fluorescence intensity. Inserts show the corresponding images with a 5-fold increase in brightness and 3-fold decrease in midtones, to facilitate visualization of the weak γ-tubulin centrosomal labelling in infected cells. The effects of a 2 h vaccinia infection on centrosomal levels of pericentrin (E and F), C-Nap 1 (G and H), Nek 2 (I and J) and centrin (K and L) are also shown. Arrowheads indicate the position of weakly labelled centrosomes in infected cells. Scale bar = 10 µm.
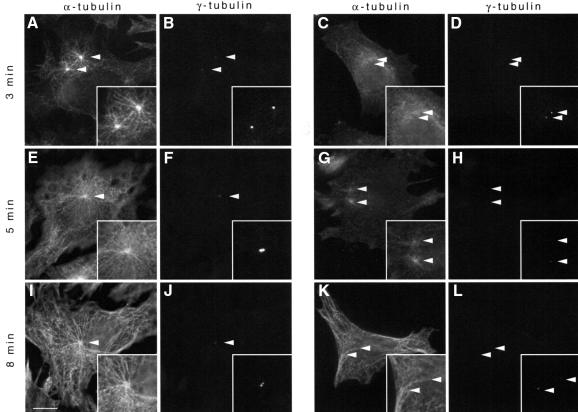
The dramatic reduction of γ-tubulin from the centrosome implies that vaccinia infection perturbs centrosome function. To test this hypothesis, we examined whether the centrosome in vaccinia-infected cells could re-nucleate microtubules, following their depolymerization by nocodazole. We found that by 2 h post-infection, when we already see a reduction in γ-tubulin, microtubule nucleation from the centrosome was very inefficient, as compared with uninfected controls, indicating that vaccinia has disrupted ‘normal’ centrosome function (Figure 13). At later times post-infection, microtubule re-nucleation efficiency from the centrosome was even lower (data not shown). However, following nocodazole washout, microtubules eventually are repolymerized throughout the cytoplasm of infected cells but do not display any organization from the MTOC, as do controls (compare Figure 13I and K).
Fig. 13. Vaccinia infection reduces centrosome microtubule nucleation efficiency. In uninfected cells, microtubules (A, E and I) nucleate from centrosomes (B, F and J) after nocodazole washout for the times indicated. In contrast, 2 h after infection with vaccinia, microtubules (C, G and K) are nucleated inefficiently from centrosomes (D, H and L). All images were collected with identical camera settings, to allow comparison of fluorescence intensity between centrosomes. Inserts (B, D, F, H, J and L) are adjusted as in Figure 12 to facilitate visualization of the weak γ-tubulin centrosomal labelling. Arrowheads indicate the position of the centrosome. Scale bar = 10 µm.
Discussion
The size of virus particles is such that they are unlikely to move within and between cells by diffusion alone, suggesting that their movements will require interactions with the host cytoskeleton. Previous data have shown that vaccinia virus both disrupts and hijacks the actin cytoskeleton to facilitate movement of the intracellular enveloped form of vaccinia virus (Cudmore et al., 1995; Röttger et al., 1999) and of the infected cell itself (Sanderson and Smith, 1998; Sanderson et al., 1998b). The data described here now show that vaccinia also uses and subsequently disrupts the microtubule cytoskeleton during its infection cycle.
It is clear from our experiments and the previous observations of Ulaeto et al. (1995) that microtubules are required to maintain the integrity of the Golgi apparatus which is in turn required for IMV wrapping to form IEV (Schmelz et al., 1994). In contrast, IMV are assembled in the absence of microtubules, albeit at reduced levels. While microtubules are not required for IMV assembly, they are required together with the dynein–dynactin complex for virion accumulation in the vicinity of the microtubule aster. One can envisage that minus end-directed microtubule-dependent movements of IMV particles from their site of assembly in the viral factory towards the MTOC, by the dynein–dynactin complex, would enhance the possibility of wrapping with the Golgi apparatus and subsequent IEV formation. Recently it has been shown that the IMV protein A27L and microtubules are required for efficient IMV dispersion from the viral factories (Sanderson et al., 2000). In the absence of A27L, mature IMV particles accumulate at the periphery of the virus factory but do not subsequently wrap to form IEV, presumably because they are unable to move on microtubules (Sanderson et al., 2000).
The microtubule- and dynein–dynactin-dependent accumulation of vaccinia in the vicinity of the MTOC is analogous to the microtubule-dependent movements required for herpes simplex virus 1 (HSV-1) and adenovirus to reach their site of replication in the nucleus (Sodeik et al., 1997; Suomalainen et al., 1999; Leopold et al., 2000). In the case of HSV-1, the UL34 protein, which is associated with the incoming nucleocapsids, interacts with the intermediate chain of cytoplasmic dynein (IC-1a) (Ye et al., 2000). It has also been reported that incoming nucleocapsids of pseudorabies virus, an alphaherpes virus closely related to HSV-1, are associated with and dependent on microtubules for their movement to the nucleus (Kaelin et al., 2000). This interaction may be mediated by the UL25 protein, a minor but essential component of the capsid, which co-localizes with microtubules and accumulates at the MTOC (Kaelin et al., 2000). The accumulation of UL25 at the MTOC is consistent with a possible interaction with the dynein–dynactin motor complex which is known to be localized at the MTOC (Echeverri et al., 1996). It would not be surprising, based on observations with HSV-1 and pseudorabies, if microtubules and dynein–dynactin were also involved in establishing the infection cycle of cytomegalovirus (CMV), Epstein–Barr virus and varicella-zoster virus, all of which are herpes viruses. The other clear example of microtubule-dependent virus movements during the establishment of infection is that of incoming human foamy virus (HFV) which is dependent on microtubules and presumably a minus end-directed microtubule motor to get to its nuclear replication site (Saib et al., 1997). In the absence of protein expression, HFV Gag proteins, which are associated with the viral genome, accumulate at the centrosome in a microtubule-dependent fashion prior to nuclear import (Saib et al., 1997). The centrosomal accumulation of Gag proteins of HFV, however, appears to be unique for this class of retroviruses as no similar localization has been reported for human immunodeficiency virus (HIV) or other retroviruses. On the other hand, the Gag protein of murine leukaemia virus and HIV has been shown to interact with KIF4, a microtubule plus end-directed kinesin motor, both in vitro and in vivo (Kim et al., 1998; Tang et al., 1999), suggesting that additional roles may exist for microtubules and motors during the outward movement of virus particles. Indeed, vaccinia virus particles are able to reach the cell periphery in the absence of actin-based motility (see images in Wolffe et al., 1997; Sanderson et al., 1998a, 2000; Röttger et al., 1999), suggesting that viral particles can also move out on microtubules (Sanderson et al., 2000). Microtubule-dependent motor-driven movements of virus particles represent an efficient mechanism to achieve a peri-nuclear localization, required to facilitate entry into the nucleus during establishment of infection. They also provide an excellent way for newly assembled virus particles to reach the cell periphery, facilitating the continued spread of infection.
Our data show that although vaccinia virus uses the microtubule cytoskeleton to achieve a peri-nuclear localization, microtubule and Golgi organization becomes disrupted later during the infection process. Interestingly, HSV-1 and CMV have also been reported to disrupt the microtubule cytoskeleton and Golgi organization in their infection cycles (Avitabile et al., 1995; Fish et al., 1996). While disruption of the microtubule network might at first sight not appear to be beneficial to the virus, it may not actually hinder viral spread but could enhance it. First, extensive virus assembly and spread to the cell periphery have already occurred by the time the microtubule cytoskeleton and Golgi organization are disrupted. Secondly, disruption of microtubule organization may overcome potential microtubule motor anchoring effects at the MTOC, thus allowing viral spread to the periphery to occur more easily. Lastly, the formation of long projections of up to 200 µm supported by extensive microtubule bundles provides a means to achieve long range spread of virus particles (Sanderson et al., 1998b).
It is clear that disruption and reorganization of the microtubule cytoskeleton by vaccinia virus is mediated by the combined effects of viral proteins with MAP-like properties and loss of microtubule-organizing function from the MTOC. The same may also be true for HSV-1, although disruption of centrosome function remains to be established, as late in infection microtubules are organized in bundles around the nucleus and do not show MTOC-orchestrated organization (Avitabile et al., 1995). The identification of viral proteins with MAP-like properties is not unique to vaccinia virus. The VP22 tegument protein from HSV-1 co-localizes with microtubules in infected cells and induces microtubule bundles when expressed in uninfected cells (Elliott and O’Hare, 1998). Other examples of viral MAPs based on their in vivo localization or in vitro association with microtubules are the N protein from murine coronavirus (Kalicharran and Dales, 1996), the movement protein from tobamovirus (Heinlein et al., 1995), the aphid transmission factor from cauliflower mosaic virus (Blanc et al., 1996), the UL25 protein from pseudorabies virus (Kaelin et al., 2000), the VP4 spike protein from rotavirus (Nejmeddine et al., 2000) and the M protein of vesicular stomatitis virus (VSV) (Melki et al., 1994). The identification of A10L and L4R, two viral core proteins, as MAP-like proteins was, however, unexpected given their previously characterized role in viral morphogenesis (Vanslyke and Hruby, 1994). The interaction of A10L and L4R with microtubules in vivo, together with the in vitro microtubule-binding data, suggest a potential mechanism for the association of viral cores with microtubules. One could envisage that viral cores which are released into the cytoplasm at the beginning of infection (Ichihashi, 1996; Vanderpasschen et al., 1998; Pedersen et al., 2000) bind directly to microtubules in a manner analogous to adenovirus or HSV-1 nucleocapsids. Further work is required to determine whether incoming cores do in fact move towards the MTOC by the dynein–dynactin complex and/or use the complex for anchoring on microtubules.
The loss of centrosome function must enhance disruption of the microtubule cytoskeleton during infection. Indeed, the loss of microtubule organization from the MTOC precedes detectable association of A10L and L4R with microtubules, which occurs from ∼8 h post-infection. Vaccinia-induced loss of centrosomal proteins is inhibited by cycloheximide, indicating that viral protein expression is required for disruption of the centrosome microtubule nucleation activity. To our knowledge, vaccinia virus infection represents the first example of virus-induced disruption of centrosome function, although we would predict that HSV-1 may have a similar effect. The mechanism by which vaccinia virus disrupts the centrosome requires further study; nevertheless, it is clear that understanding the molecular basis of this disruption will provide important insights into the regulation and stability of centrosome function which currently is the subject of intense research (Ohta et al., 1993; Lane and Nigg, 1997; Karsenti, 1999).
Materials and methods
Infections, drug treatments and transfections
HeLa cells (ATCC CCL2) were infected with the wild-type vaccinia virus strain Western Reserve (WR) or with the vaccinia deletion mutants ΔF13L (vRB12) (Blasco and Moss, 1991) or ΔA36R (Parkinson and Smith, 1994) at a multiplicity of infection of 1 p.f.u. (plaque-forming unit) per cell, as described previously (Röttger et al., 1999). Nocodazole dissolved in dimethyl sulfoxide (DMSO) and brefeldin A dissolved in ethanol were added to the culture medium to final concentration of 10 µM and 5 µg/ml, respectively unless otherwise stated. In non-treated controls, an equal volume of DMSO or ethanol was added. Cells transfected with a myc-tagged p50/dynamitin expression construct (Echeverri et al., 1996) were infected 24 h later with WR and subsequently fixed 6 h post-infection. All experiments described have been repeated 3–10 times.
Antibodies and immunofluorescence microscopy
The following antibodies were kindly provided: anti-α-tubulin by Dr E.Karsenti, anti-centrin (20H5) by Professor J.L.Salisbury (Sanders and Salisbury, 1994; Paoletti et al., 1996), anti-Nek2 and anti-C-Nap1 by Professor E.Nigg (Fry et al., 1998a,b), anti-myc and anti-gp27 by Dr T.Nilsson (Füllekrug et al., 1999) and antibodies against the corresponding vaccinia proteins: A3L, A10L and L4R by Professor D.Hruby (Vanslyke and Hruby, 1994), I1L by Professor P.Traktman (Klemperer et al., 1997), A27L (C3) by Dr M.Esteban (Rodriguez et al., 1985), A33R, A34R and A36R (Röttger et al., 1999). In addition, the following antibodies were obtained from commercial sources: anti-α-tubulin (N356) (Amersham International, UK), anti-acetylated α-tubulin (6-11B-1) (Sigma, USA), anti-γ-tubulin (GTU-88; Sigma), anti-pericentrin and anti-TGN46 (BAbCO, USA), and rabbit IgG (Sigma). Actin was visualized with fluorescently labelled phalloidin derivatives (Molecular Probes, USA).
Cells were fixed in –20°C methanol or in 5% paraformaldehyde in BRB80 (80 mM PIPES pH 6.8, 1 mM MgCl2, 1 mM EGTA) followed by 0.1% Triton X-100 permeabilization. Fixed cells were processed for immunofluorescence, viewed and images recorded as described previously (Röttger et al., 1999).
Virus stock preparation, in cells with or without microtubules
HeLa cells were pre-incubated with 25 µM nocodazole in the medium for 1 h to depolymerize microtubules, prior to infection with vaccinia ΔF13L at 1 p.f.u./cell. Nocodazole was kept in the medium throughout the infection, while an equal volume of DMSO was added to the controls. At 24 h post-infection, the cells were scraped from the flasks into the medium and sedimented by centrifugation (300 g, 7 min, 4°C). The cell membranes were disrupted and the nuclei were removed by centrifugation. The resulting post-nuclear supernatant was centrifuged through a 36% sucrose cushion (76 000 g, 30 min, 4°C). The virus pellet was resuspended in 10 mM Tris pH 9; the virus was collected by centrifugation (76 000 g, 30 min, 4°C), resuspended in 10 mM Tris pH 9 and stored at –80°C. The concentration of the virus (elemental bodies) was determined by OD260 measurement (Joklik, 1962).
In vitro microtubule binding assays
Purified EEV particles were prepared as described previously (Röttger et al., 1999) and subsequently were used to prepare virus cores following the method of Cudmore et al. (1996). Rhodamine-labelled microtubules were prepared according to Hyman et al. (1991). Vaccinia virus cores were incubated with rhodamine-labelled microtubules in BRB80 buffer containing 10 µM taxol for 5 min at room temperature. 4′,6-diamidino-2-phenylindole (DAPI) was added subsequently to a final concentration of 0.1 µg/ml to label the virus cores. Finally, the mixture was diluted 1:1–1:10 with antifade solution (0.1 mg/ml catalase, 0.1 mg/ml glucose oxidase, 10 mM glucose) and viewed without fixation. Proteinase K or trypsin treatment of core particles prior to incubation with microtubules was performed as described previously (Roos et al., 1996). Anti-A3L, A10L, L4R or control IgG antibodies were incubated with purified cores for 1 h at room temperature prior to incubation with microtubules.
Cell extracts and microtubule co-sedimentation assay
Extracts from HeLa cells infected for 24 h or uninfected controls maintained in the presence of 0.1 mg/ml rifampicin were prepared as described previously (Röttger et al., 1999). The extract was clarified by centrifugation at 150 000 g for 20 min at 4°C and cytochalasin D added to a final concentration of 1 µg/ml to depolymerize actin filaments. Endogenous tubulin in the extract was polymerized in a two-step procedure. First, the extract supernatant was supplemented with protease inhibitors, 2 mM MgGTP and 5 µM taxol and incubated for 5 min at room temperature; subsequently, an additional 15 µM taxol was added to the mix and the reaction incubated at 33°C for 30 min. For controls, no taxol was added at any stage and microtubule polymerization was inhibited either by the addition of nocodazole to a final concentration of 40 µM or by maintaining the extract at 4°C throughout the experiment. Following microtubule assembly, each 400 µg extract reaction was diluted 5-fold in BRB80 buffer (containing protease inhibitors and 20 µM taxol) and centrifuged through a 10% sucrose cushion containing protease inhibitors and 20 µM taxol at 165 000 g for 20 min at 25°C. The microtubule pellet was solubilized in SDS–PAGE sample buffer and analysed by SDS–PAGE.
Mass spectrometry and protein identification
In-gel proteolytic cleavage was performed automatically in the ‘Progest’ as described (Houthaeve et al., 1997) [Genomic Solutions Cambridge (http://www.genomicsolutions.com)] and the peptides obtained were analysed on a Bruker REFLEX MALDI mass spectrometer (Bruker Analytik, Germany) (Jensen et al., 1996b). Proteins were identified by peptide mass fingerprinting (Jensen et al., 1997) using the pro gram PeptideSearch (http://www.narrador.embl-heidelberg.de/Services/PeptideSearch/PeptideSearchIntro.html.
Microtubule aster regrowth assay
At 1 h post-infection, nocodazole was added to the culture medium to a final concentration of 25 µM to depolymerize microtubules. At 2 h post-infection, the cells were washed 3–4 times in warm medium to remove nocodazole. Washed cells were incubated in medium without nocodazole for the indicated time at 37°C to re-initiate microtubule polymerization; they were then washed briefly in warm phosphate-buffered saline (PBS) and immediately fixed. In parallel, samples were also removed at the same time point, briefly rinsed in ice-cold PBS, fixed and processed for immunofluorescence to confirm complete microtubule depolymerization before initiation of microtubule assembly. Uninfected control HeLa cells were treated and processed in an identical fashion. The same numbers of images were integrated using identical camera settings to allow direct comparison between infected and uninfected samples from the same experiment.
Acknowledgments
Acknowledgements
We would like to thank Drs M.Esteban (Centro Nacional de Biotecnologia, Madrid, Spain), D.Hruby (Oregon State University, Corvallis, OR), E.Karsenti and T.Nilsson (EMBL, Heidelberg, Germany), E.Nigg (Max Planck Institute for Biochemistry, Martinsried, Germany), J.L.Salisbury (Mayo Clinic Foundation, Rochester, MI) and P.Traktman (Medical College of Wisconsin, Milwaukee, WI) for providing antibodies; Dr R.Blasco (Centro de Investigacion en Sanidad Animal, Madrid, Spain) and Professor G.Smith (University of Oxford, Oxford, UK) for recombinant vaccinia mutants; Drs C.Echeverri (EMBL) and R.Vallee (University of Massachusetts Medical School, Worcester, MA) for the p50 dynamitin construct; Drs A.Khodjakov and C.Rieder (Wadsworth Center, New York State Department of Health, Albany, NY) for the GFP-γ-tubulin PtK1 cell line; Dr G.Griffiths and S.Schleich (EMBL) for help with the electron microscopy experiments; Drs T.Wittmann and F.Severin (EMBL) for help and advice with in vitro microtubule assays; Drs G.Griffiths and J.Krijnse-Locker (EMBL) for many discussions; and Drs A.Desai, F.Frischknecht, E.Karsenti, B.Lange, I.Vernos and T.Wittmann (EMBL) for critical reading of the manuscript and helpful suggestions. A.P. and V.M. were supported in part by European Commission Marie Curie Individual Fellowships.
References
- Avitabile E., Di Gaeta,S., Torrisi,M.R., Ward,P.L., Roizman,B. and Campadelli-Fiume,G. (1995) Redistribution of microtubules and Golgi apparatus in herpes simplex virus-infected cells and their role in viral exocytosis. J. Virol., 69, 7472–7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betakova T., Wolffe,E.J. and Moss,B. (2000) The vaccinia virus A14.5L gene encodes a hydrophobic 53-amino acid virion membrane protein that enhances virulence in mice and is conserved among vertebrate poxviruses. J. Virol., 74, 4085–4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc S., Schmidt,I., Vantard,M., Scholthof,H.B., Kuhl,G., Esperandieu,P., Cerutti,M. and Louis,C. (1996) The aphid transmission factor of cauliflower mosaic virus forms a stable complex with microtubules in both insect and plant cells. Proc. Natl Acad. Sci. USA, 93, 15158–15163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco R. and Moss,B. (1991) Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000-dalton outer envelope protein. J. Virol., 65, 5910–5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco R. and Moss,B. (1992) Role of cell-associated enveloped vaccinia virus in cell-to-cell spread. J. Virol., 66, 4170–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt J.K., Echeverri,C.J., Nilsson,T. and Vallee,R.B. (1997) Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J. Cell Biol., 139, 469–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cudmore S., Cossart,P., Griffiths,G. and Way,M. (1995) Actin-based motility of vaccinia virus. Nature, 378, 636–638. [DOI] [PubMed] [Google Scholar]
- Cudmore S., Blasco,R., Vincentelli,R., Esteban,M., Sodeik,B., Griffiths,G. and Krijnse-Locker,J. (1996) A vaccinia virus core protein, p39, is membrane associated. J. Virol., 70, 6909–6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cudmore S., Reckmann,I. and Way,M. (1997) Viral manipulations of the actin cytoskeleton. Trends Microbiol., 5, 142–148. [DOI] [PubMed] [Google Scholar]
- Dramsi S. and Cossart,P. (1998) Intracellular pathogens and the actin cytoskeleton. Annu. Rev. Cell Dev. Biol., 14, 137–166. [DOI] [PubMed] [Google Scholar]
- Duncan S.A. and Smith,G.L. (1992) Identification and characterization of an extracellular envelope glycoprotein affecting vaccinia virus egress. J. Virol., 66, 1610–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri C.J., Paschal,B.M., Vaughan,K.T. and Vallee,R.B. (1996) Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J. Cell Biol., 132, 617–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott G. and O’Hare,P. (1998) Herpes simplex virus type 1 tegument protein VP22 induces stabilization and hyperacetylation of microtubules. J. Virol., 72, 6448–6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelstad M. and Smith,G.L. (1993) The vaccinia virus 42kD envelope protein is required for the envelopement and egress of extracellular virus and for virus virulence. Virology, 194, 627–637. [DOI] [PubMed] [Google Scholar]
- Engelstad M., Howard,S.T. and Smith,G.L. (1992) A constitutively expressed vaccinia gene encodes a 42-kDa glycoprotein related to complement control factors that forms part of the extracellular virus envelope. Virology, 188, 801–810. [DOI] [PubMed] [Google Scholar]
- Finlay B.B. and Cossart,P. (1997) Exploitation of mammalian host cell functions by bacterial pathogens. Science, 276, 718–725. [DOI] [PubMed] [Google Scholar]
- Fish K.N., Britt,W. and Helson,J.A. (1996) A novel mechanism for persistence of human cytomegalovirus in macrophages. J. Virol., 70, 1855–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry A.M., Mayor,T., Meraldi,P., Stierhof,Y.D., Tanaka,K. and Nigg,E.A. (1998a) C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J. Cell Biol., 141, 1563–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry A.M., Meraldi,P. and Nigg,E.A. (1998b) A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J., 17, 470–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Füllekrug J., Suganuma,T., Tang,B.L., Hong,W., Storrie,B. and Nilsson,T. (1999) Localization and recycling of gp27 (hp24γ3): complex formation with other p24 family members. Mol. Biol. Cell, 10, 1939–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinlein M., Epel,B.L., Padgett,H.S. and Beachy,R.N. (1995) Interaction of tobamovirus movement proteins with the plant cytoskeleton. Science, 270, 1983–1985. [DOI] [PubMed] [Google Scholar]
- Hirt P., Hiller,G. and Wittek,R. (1986) Localization and fine structure of a vaccinia virus gene encoding an envelope antigen. J. Virol., 58, 757–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houthaeve T., Gausepohl,H., Ashman,K., Nillson,T. and Mann,M. (1997) Automated protein preparation techniques using a digest robot. J. Protein Chem., 16, 343–348. [DOI] [PubMed] [Google Scholar]
- Hyman A.A., Drexel,D., Kellog,D., Salser,S., Sawin,K., Stephen,P., Wordeman,L. and Mitchison,T.J. (1991) Preparation of modified tubulins. Methods Enzymol., 196, 478–485. [DOI] [PubMed] [Google Scholar]
- Ichihashi Y. (1996) Extracellular enveloped vaccinia virus escapes neutralization. Virology, 217, 478–485. [DOI] [PubMed] [Google Scholar]
- Isaacs S.N., Wolffe,E.J., Payne,L.G. and Moss,B. (1992) Characterization of a vaccinia virus-encoded 42-kilodalton class I membrane glycoprotein component of the extracellular virus envelope. J. Virol., 66, 7217–7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen O.N., Houthaeve,T., Shevchenko,A., Cudmore,S., Ashford,T., Mann,M., Griffiths,G. and Krijnse,L.J. (1996a) Identification of the major membrane and core proteins of vaccinia virus by two-dimensional electrophoresis. J. Virol., 70, 7485–7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen O.N., Podtelejnikov,A. and Mann,M. (1996b) Delayed extraction improves specificity in database searches by MALDI peptide maps. Rapid Commun. Mass Spectrom., 10, 1371–1378. [DOI] [PubMed] [Google Scholar]
- Jensen O.N., Podtelejnikov,A.V. and Mann,M. (1997) Identification of the components of simple protein mixtures by high-accuracy peptide mass mapping and database searching. Anal. Chem., 69, 4741–4750. [DOI] [PubMed] [Google Scholar]
- Johnson G.P., Goebel,S.J. and Paoletti,E. (1993) An update on the vaccinia virus genome. Virology, 196, 381–401. [DOI] [PubMed] [Google Scholar]
- Joklik W.K. (1962) The purification of four strains of poxvirus. Virology, 18, 9–18. [DOI] [PubMed] [Google Scholar]
- Kaelin K., Dezelee,S., Masse,M.J., Bras,F. and Flamand,A. (2000) The UL25 protein of pseudorabies virus associates with capsids and localizes to the nucleus and microtubules. J. Virol., 74, 474–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalicharran K. and Dales,D. (1996) The murine coronavirus as a model of trafficking and assembly of viral proteins in neural tissue. Trends Microbiol., 4, 264–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenti E. (1999) Centrioles reveal their secrets. Nature Cell Biol., 1, E62–E64. [DOI] [PubMed] [Google Scholar]
- Khodjakov A. and Rieder,C.L. (1999) The sudden recruitment of γ-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle, do not require microtubules. J. Cell Biol., 146, 585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W. et al. (1998) Binding of murine leukemia virus Gag polyproteins to KIF4, a microtubule-based motor protein. J. Virol., 72, 6898–6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemperer N., Ward,J., Evans,E. and Traktman,P. (1997) The vaccinia virus I1 protein is essential for the assembly of mature virions. J. Virol., 71, 9285–9294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane H.A. and Nigg,E.A. (1997) Antibody microinjection reveals an essential role for the human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J. Cell Sci., 110, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leopold P.L., Kreitzer,G., Miyazawa,N., Rempel,S., Pfister,K.K., Rodriguez-Boulan,E. and Crystal,R.G. (2000) Dynein- and microtubule-mediated translocation of adenovirus serotype 5 occurs after endosomal lysis. Hum. Gene Ther., 11, 151–156. [DOI] [PubMed] [Google Scholar]
- Massung R.F. et al. (1993) Potential virulence determinants in terminal regions of variola smallpox virus genome. Nature, 366, 748–751. [DOI] [PubMed] [Google Scholar]
- Melki R., Gaudin,Y. and Blondel,D. (1994) Interaction between tubulin and the viral matrix protein of vesicular stomatitis virus: possible implications in the viral cytopathic effect. Virology, 202, 339–347. [DOI] [PubMed] [Google Scholar]
- Morgan C. (1976) Vaccinia virus reexamined: development and release. Virology, 73, 43–58. [DOI] [PubMed] [Google Scholar]
- Moss B., Rosenblum,E.N. and Katz,E. (1969) Rifampicin: a specific inhibitor of vaccinia virus assembly. Nature, 224, 1280–1284. [DOI] [PubMed] [Google Scholar]
- Nejmeddine M., Trugnan,G., Sapin,C., Kohli,E., Svensson,L., Lopez,S. and Cohen,J. (2000) Rotavirus spike protein VP4 is present at the plasma membrane and is associated with microtubules in infected cells. J. Virol., 74, 3313–3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta K., Shiina,N., Okumura,E., Hisanaga,S., Kishimoto,T., Endo,S., Gotoh,Y., Nishida,E. and Sakai,H. (1993) Microtubule nucleating activity of centrosomes in cell-free extracts from Xenopus eggs: involvement of phosphorylation and accumulation of pericentriolar material. J. Cell Sci., 104, 125–137. [DOI] [PubMed] [Google Scholar]
- Paoletti A., Moudjou,M., Paintrand,M., Salisbury,J.L. and Bornens,M. (1996) Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J. Cell Sci., 109, 3089–3102. [DOI] [PubMed] [Google Scholar]
- Parkinson J.E., and Smith,G.L. (1994) Vaccinia virus gene A36R encodes a Mr. 43–50 K protein on the surface of extracellular enveloped virus. Virology, 204, 376–390. [DOI] [PubMed] [Google Scholar]
- Payne L.G. (1980) Significance of extracellular enveloped virus in the in vitro and in vivo dissemination of vaccinia. J. Gen. Virol., 50, 89–100. [DOI] [PubMed] [Google Scholar]
- Payne L.G. and Norrby,E. (1976) Presence of hemagglutinin in the envelope of extracellular vaccinia virus particles. J. Gen. Virol., 32, 63–72. [DOI] [PubMed] [Google Scholar]
- Pedersen K., Snijder,E.J., Schleich,S., Roos,N., Griffiths,G. and Krijnse Locker,J. (2000) Characterization of vaccinia virus intracellular cores: implications for viral uncoating and core structure. J. Virol., 74, 3525–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J.F., Janeczko,R. and Esteban,M. (1985) Isolation and characterization of neutralizing monoclonal antibodies to vaccinia virus. J. Virol., 56, 482–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos N., Cyrklaff,M., Cudmore,S., Blasco,R., Krijnse-Locker,J. and Griffiths,G. (1996) The use of a novel immunogold cryoelectron microscopic method to investigate the structure of the intracellular and extracellular forms of vaccinia virus. EMBO J., 15, 2343–2355. [PMC free article] [PubMed] [Google Scholar]
- Roper R.L., Payne,L.G. and Moss,B. (1996) Extracellular vaccinia virus envelope glycoprotein encoded by the A33R gene. J. Virol., 70, 3753–3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper R.L., Wolffe,E.J., Weisberg,A. and Moss,B. (1998) The envelope protein encoded by the A33R gene is required for formation of actin-containing microvilli and efficient cell-to-cell spread of vaccinia virus. J. Virol., 72, 4192–4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röttger S., Frischknecht,F., Reckmann,I., Smith,G.L. and Way,M. (1999) Interactions between vaccinia virus IEV membrane proteins and their roles in IEV assembly and actin tail formation. J. Virol., 73, 2863–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saib A., Puvion-Dutilleul,F., Schmid,M., Peries,J. and De The,H. (1997) Nuclear targeting of incoming human foamy virus gag proteins involves a centriolar step. J. Virol., 71, 1155–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M.A. and Salisbury,J.L. (1994) Centrin plays an essential role in microtubule severing during flagellar excision in Clamydomonas reinhardtii. J. Cell Biol., 124, 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson C.M. and Smith,G.L. (1998) Vaccinia virus induces calcium-independent cell–matrix adhesion during the motile phase of infection. J. Virol., 72, 9924–9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson C.M., Frischknecht,F., Way,M., Hollinshead,M. and Smith,G.L. (1998a) Roles of vaccinia virus EEV-specific proteins in intracellular actin tail formation and low pH-induced cell–cell fusion. J. Gen. Virol., 79, 1415–1425. [DOI] [PubMed] [Google Scholar]
- Sanderson C.M., Way,M. and Smith,G.L. (1998b) Virus-induced cell motility. J. Virol., 72, 1235–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson C.M., Hollinshead,M. and Smith,G.L. (2000) The vaccinia virus A27L protein is needed for the microtubule-dependent transport of intracellular mature virus particles. J. Gen. Virol., 81, 47–58. [DOI] [PubMed] [Google Scholar]
- Schmelz M., Sodeik,B., Ericsson,M., Wolffe,E.J., Shida,H., Hiller,G. and Griffiths,G. (1994) Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J. Virol., 68, 130–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shida H. (1986) Nucleotide sequence of the vaccinia virus hemagglutinin gene. Virology, 150, 451–462. [DOI] [PubMed] [Google Scholar]
- Sodeik B., Doms,R.W., Ericsson,M., Hiller,G., Machamer,C.E., van’t Hof,W., van Meer,G., Moss,B. and Griffiths,G. (1993) Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J. Cell Biol., 121, 521–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodeik B., Ebersold,M.W. and Helenius,A. (1997) Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol., 136, 1007–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T. and Kirschner,M. (1994) In vitro reconstitution of centrosome assembly and function: a central role of γ tubulin. Cell, 76, 623–637. [DOI] [PubMed] [Google Scholar]
- Suomalainen M., Nakano,M.Y., Keller,S., Boucke,K., Stidwill,R.P. and Greber,U.F. (1999) Microtubule-dependent plus- and minus end-directed motilities are competing processes for nuclear targeting of adenovirus. J. Cell Biol., 144, 657–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K.B. and McAuslan,B.R. (1970) Effect of rifampicin on poxvirus protein synthesis. J. Virol., 6, 326–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Winkler,U., Freed,E.O., Torrey,T.A., Kim,W., Li,H., Goff,S.P. and Morse,H.C.,III (1999) Cellular motor protein KIF-4 associates with retroviral gag. J. Virol., 73, 10508–10513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togel M., Wiche,G. and Propst,F. (1998) Novel features of the light chain of microtubule-associated protein MAP1B: microtubule stabilization, self interaction, actin filament binding and regulation by heavy chain. J. Cell Biol., 143, 695–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulaeto D., Grosenbach,D. and Hruby,D.E. (1995) Brefeldin A inhibits vaccinia virus envelopment but does not prevent normal processing and localisation of the putative envelopment receptor P37. J. Gen. Virol., 76, 103–111. [DOI] [PubMed] [Google Scholar]
- Vanderpasschen A., Hollinshead,M. and Smith,G.L. (1998) Intracellular and extracellular vaccinia virions enter cells by different mechanisms. J. Gen. Virol., 79, 877–887. [DOI] [PubMed] [Google Scholar]
- Vanslyke J.K. and Hruby,D.E. (1994) Immunolocalisation of vaccinia virus structural proteins during virion formation. Virology, 198, 624–635. [DOI] [PubMed] [Google Scholar]
- Weisshaar B., Doll,T. and Matus,A. (1992) Reorganization of the microtubular cytoskeleton by embryonic microtubule-associated protein 2 (MAP2c). Development, 116, 1151–1161. [DOI] [PubMed] [Google Scholar]
- Wolffe E.J., Isaacs,S.N. and Moss,B. (1993) Deletion of the vaccinia virus B5R gene encoding a 42kD membrane glycoprotein inhibits extracellular virus envelope formation and dissemination. J. Virol., 67, 4732–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe E.J., Katz,E., Weisberg,A. and Moss,B. (1997) The A34R glycoprotein gene is required for induction of specialized actin-containing microvilli and efficient cell-to-cell transmission of vaccinia virus. J. Virol., 71, 3904–3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe E.J., Weisberg,A.S. and Moss,B. (1998) Role for the vaccinia virus A36R outer envelope protein in the formation of virus-tipped actin-containing microvilli and cell-to-cell virus spread. Virology, 244, 20–26. [DOI] [PubMed] [Google Scholar]
- Ye G.-J., Vaughan,K.T., Vallee,R.B. and Roizman,B. (2000) The herpes simplex virus 1 UL34 protein interacts with a cytoplasmic dynein intermediate chain and targets nuclear membrane. J. Virol., 74, 1355–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]